Abstract
Background
Carbapenemase-producing Enterobacteriaceae (CPE) are an emerging threat for healthcare providers worldwide.
Objectives
To determine CPE carriage rates and risk factors in an unselected hospital cohort at the time of admission.
Methods
We approached 4567 patients within 72 h of admission to provide a rectal swab and answer a questionnaire on risk factors for carriage. Rectal swabs were cultured for carbapenem-resistant organisms on chromogenic and non-chromogenic agar, and tested for carbapenemase production by PCR (Check-Direct CPE). The study was approved by the NHS Research Ethics Committee.
Results
Only 6 CPE were cultured from 5 (0.1%) of 4006 patients who provided a rectal swab; only 1 was cultured using non-chromogenic media. An additional 76 culture-negative rectal swabs were initially PCR positive, but none grew a carbapenem-resistant organism despite enrichment culture and only two were positive when retested several months later by Check-Direct and a second PCR assay (Cepheid GeneXpert® Carba-R). A modified Ct cut-off of <35 would have resolved these apparent false-positives. 40% of patients had a risk factor that should prompt screening and pre-emptive isolation as defined by UK CPE guidelines but only 8.1% and 20.2% of these patients had been screened and pre-emptively isolated by clinical teams, respectively. Overseas hospitalization was the only significant risk factor for CPE carriage (P < 0.001, OR 64.3, 95% CI 7.3–488.5).
Conclusions
This study highlights a very low carriage rate of CPE. Hospitalization abroad is the most important risk factor to guide admission screening in this low-prevalence setting.
Introduction
Carbapenemase-producing organisms (CPOs), including both Enterobacteriaceae such as Klebsiella pneumoniae and non-fermenters such as Acinetobacter baumannii, are emerging worldwide.1,2 Although carbapenemase-producing non-fermenters present important challenges in some hospital specialties, especially ICUs and burns units, carbapenemase-producing Enterobacteriaceae (CPE) are a threat across the healthcare and potentially community setting, combining multidrug resistance, virulence and the potential for rapid spread.1,2
CPE is now endemic in some parts of the world, notably South-East Asia and southern Europe, but has been comparatively rare in the UK.3–6 For example, the diagnostic laboratory at our institution in London identified only 25 carbapenem-resistant Enterobacteriaceae from clinical samples between 2011 and 2013, during which time there was no formal screening policy.6 The prevalence of CPE elsewhere in the UK is unknown; it is of concern that KPC-producing Klebsiella pneumoniae have become endemic in the Manchester region, and there is evidence that the prevalence of CPE is increasing in the UK.4,6–8 Outbreaks of carbapenem-resistant A. baumannii are also reported from UK critical care units.9,10
In response to this emerging threat, Public Health England (PHE) published a CPE Toolkit for all NHS hospitals in 2013.11,12 It proposed pre-emptive isolation and screening of patients who have undergone overseas hospitalization in ‘high-risk’ countries abroad or ‘high-risk’ UK hospitals, and patients who have had CPE previously.8 However, these risk factors are not evidence based due to limited UK epidemiological data on prevalence and risk factors for CPE. The prevalence of CPE carriage at the time of hospital admission varies widely13–15 and this has implications for the cost effectiveness of admission screening.16 Another challenge is the laboratory diagnosis of CPE,16,17 particularly in a low-prevalence setting. Conventional culture-based approaches, including chromogenic agar, have good sensitivity and specificity, but results take 24–48 h, which prolongs unnecessary isolation for the vast majority of patients who will be negative.17,18 Meanwhile, PCR-based methods, which can be applied directly to a rectal swab, have faster turnaround times and may offer improved sensitivity;19,20 however, few studies have compared PCR with culture for detecting CPE at the time of hospital admission.
This universal adult admission screening study was performed to obtain an accurate prevalence and risk factor profile for CPO carriage, and to evaluate the performance of various laboratory approaches to detection of CPO including carriage of carbapenemase genes. The study was intended to support an evidence-based approach to introducing cost-effective CPO screening.
Patients and methods
Ethics
The study was approved by the Camberwell St Giles NHS Research Ethics Committee (14/LO/2085).
Patients
A team of research nurses and healthcare assistants approached adult patients within the first 72 h of their admission between February and May 2015. We approached patients on all adult inpatient wards during day shifts between Monday and Friday, though including some Saturdays. Exclusions included paediatrics, the emergency department, daycare wards and maternity wards. Subsequent episodes of admission for the same patient were excluded. Consent was obtained and a rectal swab was collected using an Eswab™ (Copan, Brescia, Italy). Eswabs have superior recovery compared with traditional swabs, and provide a liquid suspension that can be used for multiple downstream tests.21,22 During the screening consultation, each patient was asked the questions detailed in Table S1 (available as Supplementary data at JAC Online).
Swab treatment and culture
Rectal swabs were transported to the research laboratory on the day of collection and stored at 4°C until processed. 10 μL aliquots from the Eswab fluid were plated onto MacConkey with a 10 μg ertapenem disc and incubated at 35–37°C for 24 h, and both halves of chromID® CARBA SMART (bioMérieux, Basingstoke, UK), and incubated at 35–37°C for 24 h. Broth enrichment was performed for specimens that were negative by culture but positive by PCR: 100 μL aliquots from the Eswab fluid were inoculated into tryptone soya agar (TSA) broth plus 16 mg/L vancomycin (to inhibit Gram-positive organisms) and 0.12 mg/L ertapenem, and TSA broth plus 16 mg/L vancomycin and 0.25 mg/L cefotaxime. Broths were incubated at 35–37°C for 24 h. Each broth was plated out onto MacConkey and CARBA SMART and incubated at 35–37°C for 24 h. Presumptive colonies were identified using MALDI-TOF (Bruker, Coventry, UK) and antimicrobial susceptibility tested using a Vitek 2 (bioMérieux). CPE identified locally were confirmed by the national reference laboratory.
CPE detection
Check-Direct CPE (Check-Points, Wageningen, the Netherlands) was performed on the BD MAX™ platform (Becton Dickinson, Oxford, UK) on the same day as setting up the agar plates using 25 μL of the Eswab fluid. Additionally, several months after the original specimen had been collected, GeneXpert® Carba-R (Cepheid, Sunnyvale, CA, USA) was performed on specimens that either grew a CPO or were positive by the Check-Direct CPE PCR, in parallel with a repeat Check-Direct CPE test from the original Eswab fluid (which had been stored at −20°C).
Data analysis
A patient-level database was constructed to include relevant details, including laboratory test results, questionnaire data provided at the time of specimen collection, clinical details such as medical specialty, reason for admission, and patient demographics such as age, gender and underlying medical conditions. Risk factors for CPE carriage were determined through χ2 tests for categorical variables and t-tests for continuous variables.
The sample size was calculated for a range of expected proportions of CPO carriage using the standard random sampling formula and assuming 0.01 precision, 95% confidence (Z = 1.96) and 20% refusal to participate. The rate of refusal to participate was estimated from previous studies requiring rectal samples performed at Guy's and St Thomas' NHS Foundation Trust (GSTT). The sample size calculation indicated a sample of 4184 patients would provide an accurate measure of CPO prevalence in patients admitted to GSTT. Therefore, we set a target sample size of 4500 patients to recruit to allow for incomplete sample or data collection for some patients.
Results
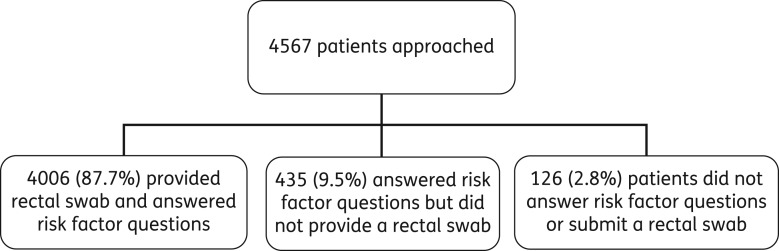
4567 patients were approached to participate in the study: 4006 (87.7%) provided a rectal swab and answered the risk factor questionnaire; 561 (12.3%) did not provide a rectal swab (Figure 1). Of the 403 patients who did not submit a rectal swab and answer the questionnaire, 76 (18.8%) declined because of the need for a rectal swab, 93 (23.1%) for no given reason and 234 declined (58.1%) for some other reason [of which 130 (55.3%) were due to feeling too unwell to participate, and 86 (36.6%) were not interested in the study]. There were no significant differences in ethnic group between patients who completed a risk factor questionnaire but did not submit a rectal swab, and those who completed a risk factor questionnaire and submitted a rectal swab (P = 0.110, data not shown).
Figure 1.
Study flow chart.
Six CPE were cultured from 5 (0.1%) of 4006 patients who submitted a rectal swab (Table 1); four were Escherichia coli and the only carbapenemases detected were OXA-48 and NDM. 5/6 CPE from 5/5 patients were detected by GeneXpert PCR, whereas 4/6 CPE from 4/5 patients were detected by Check-Direct. One hundred and twenty samples (3.0%) were initially unresolved by Check-Direct PCR but all resolved after one (n = 117), two (n = 2) or three (n = 1) repeats. 5/6 CPE grew on the CARBA SMART chromogenic media, whereas only 1/6 were identified by the standard laboratory protocol using MacConkey agar. None of the CPE culture-positive samples had visible faecal matter. No carbapenemase-producing non-Enterobacteriaceae were identified.
Table 1.
CPE: bacteria, carbapenemases and Ct values
| Check-Direct (Ct) | GeneXpert (Ct) | Direct culture/PCR |
|---|---|---|
| OXA-48 (31.8) | OXA-48 (30.2) | E. coli OXA-48 |
| OXA-48 (29.0) | OXA-48 (27.8) | E. coli OXA-48 |
| NDM (23.1) | NDM (25.6) | E. coli NDM |
| Negative | OXA-48 (34.1) | C. freundii OXA-48 |
| NDM (28.2) | NDM (28.2) | E. coli NDM |
| K. pneumoniae OXA-48 |
Samples from an additional 76 (1.9%) patients were PCR positive by Check-Direct but culture negative at the time of sample collection. Eswab aliquots stored at −20°C were retested between 1 and 2 months after sample collection by Check-Direct and tested for the first time by GeneXpert, but only 2/76 were positive, both by Check-Direct and GeneXpert. The mean Check-Direct Ct values were significantly lower for culture-positive samples compared with culture-negative samples (29.3 versus 41.0; P = 0.012). The Ct values for culture-positive samples using the GeneXpert tests was similar to Check-Direct (29.3 versus 28.3). If we applied a modified Ct cut-off of <35 to identify a PCR-positive sample using Check-Direct,19,20 then 74 of the 76 would become PCR-negative, and carbapenemase genes would be detected in samples from all four patients that were detected using the manufacturer's recommended cut-off.
The prevalence of risk factors for the 4441 patients who answered a risk factor questionnaire is summarized in Table 2. 41.9% of patients had an overnight stay in a UK hospital in the 12 months prior to their admission; 31.1% were re-admissions from our hospitals and 37.0% were from within the Greater London area. Only 1.0% of patients had an overnight stay in an overseas hospital in the 12 months prior to their admission, and 1.3% were overseas residents. 54.1% of admissions had taken antibiotics in the last 6 months, and 24.9% had taken more than one course. 64.5% of admissions had at least one risk factor.
Table 2.
Prevalence of risk factors (total number of patients 4441)
| Risk factor | Patients [n (%)] |
|---|---|
| Non-UK residents | 57 (1.3) |
| Overseas travel in the past 12 months | 1455 (32.8) |
| Overnight hospital stay in the past 12 months | |
| GSTT | 1381 (31.1) |
| Within M25 | 1644 (37.0) |
| North-West | 8 (0.2) |
| Any UK hospital (including London) | 1862 (41.9) |
| Overseas hospital (CRE risk countries as defined by PHE8) | 21 (0.5) |
| Overseas hospital (any country) | 45 (1.0) |
| Antibiotics in the past 6 months—any | 2404 (54.1) |
| Antibiotics in the past 6 months—one course | 1295 (29.2) |
| Antibiotics in the past 6 months—more than one course | 1107 (24.9) |
| At least one risk factor | 3573 (80.5) |
| At least one risk factor (excluding antibiotics) | 2865 (64.5) |
Despite the small number of culture-positive cases (n = 5), hospitalization abroad was significantly associated with CPE carriage (Table 3). Hospitalization abroad was not significantly associated with patients who were PCR positive/culture negative (Table S2).
Table 3.
Risk factors for CPE
| Risk factors | CPE− with risk factor | CPE− without risk factor | CPE+ with risk factor | CPE+ without risk factor | %CPE− with risk factor | %CPE+ with risk factor | P valuea | OR | |
|---|---|---|---|---|---|---|---|---|---|
| 95% CI | |||||||||
| Overnight hospital stay in the past 12 months | |||||||||
| overseas hospital (any country) | 40 | 3966 | 2 | 3 | 1.0 | 40.0 | <0.001 | 64.3 | 7.3–488.5 |
| overseas hospital (CPE risk countriesb) | 13 | 3993 | 1 | 4 | 0.4 | 20.0 | 0.002 | 57.5 | 2.3–594.6 |
| any UK hospital (including London) | 1704 | 2302 | 4 | 1 | 47.1 | 80.0 | 0.306 | — | — |
| within M25 | 1496 | 2510 | 1 | 4 | 42.4 | 20.0 | 0.574 | — | — |
| GSTT | 1253 | 2753 | 1 | 4 | 36.3 | 20.0 | 0.769 | — | — |
| North-West | 6 | 4000 | 0 | 5 | 0.2 | 0.0 | 1.000 | — | — |
| Overseas travel in the past 12 months | 1311 | 2695 | 2 | 3 | 32.5 | 40.0 | 1.000 | — | — |
| Non–UK residents | 48 | 3958 | 0 | 5 | 1.2 | 0.0 | 1.000 | — | — |
| Antibiotics in the past 6 months | |||||||||
| any | 2205 | 1801 | 5 | 0 | 56.2 | 100.0 | 0.128 | — | — |
| one course | 1192 | 2814 | 2 | 3 | 29.9 | 40.0 | 0.998 | — | — |
| more than one course | 1012 | 2994 | 3 | 2 | 26.2 | 60.0 | 0.228 | — | — |
| Risk factors excluding antibiotics | 2630 | 1376 | 5 | 0 | 67.7 | 100.0 | 0.287 | — | — |
| Any risk factor | 3295 | 711 | 5 | 0 | 83.1 | 100.0 | 0.683 | — | — |
aP values ≤0.05 are shown in bold text.
bAs defined by the PHE Toolkit.8
Only 13.6% of 2865 patients with risk factors defined in the PHE Toolkit were screened and 6.2% were isolated in single rooms by the clinical services during the first 48 h of admission (Table 4). Thirteen of 45 patients (28.9%) who had been hospitalized overseas in the 12 months before their admission were screened within 48 h of their admission through the clinical services, but only 15.6% were in single rooms.
Table 4.
Standard of care screens, and isolation (n = 4441)
| Risk factors | Number of approached patients with risk factor | Patients in isolation [n (%)] | Standard of care screening [n (%)]a |
|---|---|---|---|
| Number of patient with at least one risk factor (out of all approached patients) as defined by the PHE Toolkit8 | 2865 | 178 (6.2) | 389 (13.6) |
| Non-UK residents | 57 | 3 (5.3) | 4 (7.0) |
| Overseas travel in the past 12 months | 1455 | 71 (4.9) | 151 (10.4) |
| Overnight hospital stay in the past 12 months | |||
| GSTT | 1381 | 112 (8.1) | 330 (23.9) |
| within M25 | 1644 | 129 (7.8) | 377 (22.9) |
| North-West | 8 | 1 (12.5) | 2 (25.0) |
| any UK hospital (including London) | 1862 | 142 (7.6) | 443 (23.8) |
| overseas hospital (CRE risk countries) | 21 | 2 (9.5) | 11 (52.4) |
| overseas hospital (any country) | 45 | 7 (15.6) | 13 (28.9) |
aWithin the first 48 h of admission.
Patients who participated in the study were significantly older (median 58 versus 52 years), and more likely to be elective rather than non-elective admissions (45.1% versus 25.5%), but there were no significant differences in gender or ethnic group (Table S3). The difference in age and admission method probably reflects the way that patients were recruited, with older patients admitted via elective pathways likely to stay longer on inpatient wards.
Discussion
This study identified an extremely low prevalence of CPE carriage in an unselected cohort of patients at the time of hospital admission [5/4006 admissions (0.1%) who submitted a rectal swab] and no non-Enterobacteriaceae CPOs. There was therefore no hidden burden of CPE in this low-prevalence setting where only sporadic cases are identified by routine clinical sampling, despite a high level of risk factors frequently associated with CPE carriage. This is comparable to screening studies that targeted high-risk patients in low-prevalence settings at another London hospital (0.3%)15 and in Seoul, South Korea (0.3%).23 In contrast, screening studies in CPE endemic settings have identified a much higher level. For example, a study in New York found 5% of admissions to a range of wards carried CPE,24 and an even higher proportion of carriers was identified in patients attending a military hospital in Pakistan (18%)25 and post-acute care hospitals in Chicago, USA (30%).26
An important focus of the study was to compare PCR with culture-based detection of CPOs. PCR can offer more rapid turnaround time, if logistics and laboratory processes are optimized, but is more expensive on a per-test basis and does not provide data on organism identity and phenotypic susceptibility. Furthermore, it requires modification to detect newly emerging carbapenemase genes or potential genetic mutations. The sensitivity of PCR direct from rectal swabs for the detection of CPE has been 100% in most published studies.19,20,27 However, in our study, Check-Direct missed a CPE-colonized patient (Citrobacter freundii OXA-48) and both Check-Direct and GeneXpert correctly identified an NDM, but failed to identify an OXA-48 CPE in the same specimen, which is concerning.
We found that a further 76 patients were positive by a Check-Direct PCR at the time of sample collection, but no CPO could be cultured despite sensitive enrichment culture. Curiously, when we re-tested the same specimens 1–2 months later, all but two of these had reverted to negative and these two samples were also PCR-positive by the GeneXpert system. Whilst it is possible that sample degradation, laboratory error or faulty reagents explain the initial positive result and subsequent negative results on Check-Direct, other studies suggest that a modified lower Ct value may be more suitable for this system. Lau et al.19 found that only 7 of 43 positive samples that tested positive by Check-Direct could be cultured, and all culture-positive samples had a Ct value of <35. Similarly, Huang et al.20 found that 38/394 patients tested positive by Check-Direct but only 17 CPE were cultured. In this study, 5/21 ‘false positives’ had grown a previous CPO and 6/9 OXA-48 PCR-positive/culture-negative specimens were from a centre that had experienced a recent outbreak of OXA-48 CPE. These samples had a Ct value of <35, whereas the other ‘false positives’ had a Ct value of >35. An in vitro study of a panel of organisms found a high rate of false positivity from the Check-Direct assay when using the manufacturer's recommended Ct value, which was eliminated when a modified Ct value of ≤31 was applied.27 The limit of detection of Check-Direct has been found to be broadly comparable to culture through in vitro serial dilution studies.19,28 Importantly, we did not identify any of the risk factors associated with culture-positive specimens for these PCR-positive/culture-negative samples (Table S2). Whilst we cannot rule out the possibility that the PCR assay was more sensitive than culture for detecting CPE, the combined laboratory and epidemiological evidence supports recommending a modified Ct value of <35 to reduce the rate of ‘false positives’.19,20 Further laboratory and epidemiological evidence is required to make a firm conclusion of interpreting PCR-positive results with a high Ct value using this molecular assay.
Hospitals need to develop polices to detect carriers in order to guide infection control measures to prevent transmission. Although outbreaks and increasing endemic levels are reported in some hospitals in the UK and in many parts of the world, most are currently likely to have low admission prevalence.6–8,23,24 The number of cases of CPE identified in our study (five) was very low, but we were able to establish that overseas hospitalization was a significant risk factor for CPE, which has been found in other studies.1,8 However, if this study had been performed in a region of the UK where CPE was endemic, such as in parts of the north-west,4,8 then the rate of carriage in patients without overseas healthcare contact would be expected to be higher. Thus, all organizations need policies that are proportionate, effective and achievable and the ability to adapt to epidemiological changes. In our setting, the very low carriage rate questions the value of screening a large group of patients for CPE, particularly using PCR, which is more expensive than culture on a per-test basis.16 We therefore suggest, based on evidence from this large universal screening study, that targeting patients with recent overseas hospitalization is the most important group for screening in organizations currently only identifying isolated sporadic cases. However, it is also vital to remain vigilant for any changes in local or national epidemiology. Furthermore, detailed cost-effectiveness analyses are required using data from a range of prevalence estimates.
Our study highlighted a number of important practical challenges. Although the hospital had introduced risk-factor-based screening in line with the PHE Toolkit recommendations, we found this would have required pre-emptive isolation of 64% of patients, and only 13% of these were screened and 6% placed in pre-emptive isolation. We also identified a number of key issues from a laboratory perspective. Although some guidelines recommend that swabs should be faecally stained,8,12 we found that all of our CPE culture-positive specimens were not faecally stained. Finally, the standard method used by the laboratory at the time of the study identified only one of the CPE, which is of considerable concern.
Our study has a number of important strengths. While most published CPE screening studies focus on selected ‘high-risk’ patient groups, we screened and collected risk factor data from all patients admitted to hospital from a diverse multiracial community that has a high level of risk factors with the potential for a hidden burden of CPE carriage. We also evaluated culture-based and PCR methods to detect CPO in parallel. However, limitations include the very low rate of carriage, which limited the value of our planned risk factor analysis. Also, the risk factor data that we collected relied on patients remembering details of previous healthcare contact and antibiotic use. It is also possible that the carriage rate was underestimated because we took a single sample, rather than three 48 h apart as recommended in the PHE Toolkit.8 Although the sample size was unselected and there were no differences in gender or ethnic group, patients who participated in the study were older and more likely to be admitted via an elective pathway. The carriage rate may have been different in younger patients admitted through non-elective pathways, but it seems likely that the carriage would be even lower in this group, many of whom would lack traditional risk factors for healthcare-associated multidrug-resistant organisms. Finally, 12% of patients approached did not submit a rectal swab, but only 20% of these declined because of the need for a rectal swab. Furthermore, there was no significant difference in ethnic group between those patients who submitted rectal swab and those who did not. This suggests that widespread rectal admission screening is practically feasible, and is unlikely to provide skewed data due to patients from certain ethnic groups being more likely to decline to provide a rectal swab.
Conclusions
CPE is an emerging threat to healthcare systems worldwide. Our analysis of laboratory methods suggests that chromogenic media are more useful than non-chromogenic media for detecting CPE. Whilst PCR may provide a faster turnaround time, the criteria for defining a positive specimen needs to be reviewed and PCR may not be cost effective in a low-prevalence setting. Implementing current UK guidelines would require screening and isolation of around 40% of admissions, which may not be feasible. We identified an extremely low rate of CPE carriage at the time of hospital admission, with overseas hospitalization the only significant risk factor. Therefore, the value of screening a large group of patients aside from those hospitalized overseas is questionable locally, but may be justified in regions of higher prevalence.
Funding
This study was funded by a grant from the Guy's and St Thomas' Charity (Grant Number: EFT140606). The study was supported by the NIHR Collaboration for Leadership in Applied Health Research and Care South London at King's College Hospital NHS Foundation Trust. PCR kits were provided at discounted rates by CheckPoints.
Transparency declarations
J. A. O. is a consultant to Gama Healthcare. All other authors have none to declare.
Supplementary Material
Acknowledgements
This study was presented in part at ECCMID 2015 (abstract #4278) and the Infection Prevention Society 2015 (abstract #3866) meeting in Liverpool, UK, September 2015.
References
- 1. Nordmann P, Naas T, Poirel L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 2011; 17: 1791–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Karah N, Sundsfjord A, Towner K et al. Insights into the global molecular epidemiology of carbapenem non-susceptible clones of Acinetobacter baumannii. Drug Resist Updat 2012; 15: 237–47. [DOI] [PubMed] [Google Scholar]
- 3. Schwaber MJ, Carmeli Y. An ongoing national intervention to contain the spread of carbapenem-resistant Enterobacteriaceae. Clin Infect Dis 2014; 58: 697–703. [DOI] [PubMed] [Google Scholar]
- 4. Jain A, Hopkins KL, Turton J et al. NDM carbapenemases in the United Kingdom: an analysis of the first 250 cases. J Antimicrob Chemother 2014; 69: 1777–84. [DOI] [PubMed] [Google Scholar]
- 5. Walsh TR, Weeks J, Livermore DM et al. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis 2011; 11: 355–62. [DOI] [PubMed] [Google Scholar]
- 6. Hughes J, Goldenberg SD, Tosas Auguet O et al. Recent emergence of carbapenem-resistant organisms in a low prevalence UK setting in London. J Infect Prevent 2016; 17: 130–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Drew RJ, Turton JF, Hill RL et al. Emergence of carbapenem-resistant Enterobacteriaceae in a UK paediatric hospital. J Hosp Infect 2013; 84: 300–4. [DOI] [PubMed] [Google Scholar]
- 8. PHE. Acute trust toolkit for the early detection, management and control of carbapenemase-producing Enterobacteriaceae. 2013. https://www.gov.uk/government/uploads/system/uploads/attachment_data/file/329227/Acute_trust_toolkit_for_the_early_detection.pdf.
- 9. Lewis T, Loman NJ, Bingle L et al. High-throughput whole-genome sequencing to dissect the epidemiology of Acinetobacter baumannii isolates from a hospital outbreak. J Hosp Infect 2010; 75: 37–41. [DOI] [PubMed] [Google Scholar]
- 10. Turton JF, Kaufmann ME, Gill MJ et al. Comparison of Acinetobacter baumannii isolates from the United Kingdom and the United States that were associated with repatriated casualties of the Iraq conflict. J Clin Microbiol 2006; 44: 2630–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tacconelli E, Cataldo MA, Dancer SJ et al. ESCMID guidelines for the management of the infection control measures to reduce transmission of multidrug-resistant Gram-negative bacteria in hospitalized patients. Clin Microbiol Infect 2014; 20Suppl 1: 1–55. [DOI] [PubMed] [Google Scholar]
- 12. Otter JA, Mutters NT, Tacconelli E et al. Controversies in guidelines for the control of multidrug-resistant Gram-negative bacteria in EU countries. Clin Microbiol Infect 2015; 21: 1057–66. [DOI] [PubMed] [Google Scholar]
- 13. Birgand G, Armand-Lefevre L, Lepainteur M et al. Introduction of highly resistant bacteria into a hospital via patients repatriated or recently hospitalized in a foreign country. Clin Microbiol Infect 2014; 20: O887–90. [DOI] [PubMed] [Google Scholar]
- 14. Girlich D, Bouihat N, Poirel L et al. High rate of faecal carriage of extended-spectrum β-lactamase and OXA-48 carbapenemase-producing Enterobacteriaceae at a university hospital in Morocco. Clin Microbiol Infect 2014; 20: 350–4. [DOI] [PubMed] [Google Scholar]
- 15. Mack D, Scott J, Pang V et al. One year's experience of carbapenemase screening at a London Teaching Hospital. HIS Conference 2014.
- 16. Venanzio V, Gharbi M, Moore LS et al. Screening suspected cases for carbapenemase-producing Enterobacteriaceae, inclusion criteria and demand. J Hosp Infect 2015; 71: 493–5. [DOI] [PubMed] [Google Scholar]
- 17. Viau R, Frank KM, Jacobs MR et al. Intestinal carriage of carbapenemase-producing organisms: current status of surveillance methods. Clin Microbiol Rev 2016; 29: 1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Willems E, Cartuyvels R, Magerman K et al. Evaluation of 3 different agar media for rapid detection of extended-spectrum β-lactamase-producing Enterobacteriaceae from surveillance samples. Diag Microbiol Infect Dis 2013; 76: 16–9. [DOI] [PubMed] [Google Scholar]
- 19. Lau AF, Fahle GA, Kemp MA et al. Clinical performance of Check-Direct CPE, a multiplex PCR for direct detection of blaKPC, blaNDM/VIM, and blaOXA48 from Perirectal Swabs. J Clin Microbiol 2015; 53: 3729–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang TD, Bogaerts P, Ghilani E et al. Multicentre evaluation of the Check-Direct CPE(R) assay for direct screening of carbapenemase-producing Enterobacteriaceae from rectal swabs. J Clin MIcrobiol 2015; 70: 1669–73. [DOI] [PubMed] [Google Scholar]
- 21. Van Horn KG, Audette CD, Tucker KA et al. Comparison of 3 swab transport systems for direct release and recovery of aerobic and anaerobic bacteria. Diag Microbiol Infect Dis 2008; 62: 471–3. [DOI] [PubMed] [Google Scholar]
- 22. Smismans A, Verhaegen J, Schuermans A et al. Evaluation of the Copan ESwab transport system for the detection of methicillin-resistant Staphylococcus aureus: a laboratory and clinical study. Diag Microbiol Infect Dis 2009; 65: 108–11. [DOI] [PubMed] [Google Scholar]
- 23. Kim J, Lee JY, Kim SI et al. Rates of fecal transmission of extended-spectrum β-lactamase-producing and carbapenem-resistant Enterobacteriaceae among patients in intensive care units in Korea. Ann Lab Med 2014; 34: 20–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Swaminathan M, Sharma S, Poliansky Blash S et al. Prevalence and risk factors for acquisition of carbapenem-resistant Enterobacteriaceae in the setting of endemicity. Infect Control Hosp Epidemiol 2013; 34: 809–17. [DOI] [PubMed] [Google Scholar]
- 25. Day KM, Ali S, Mirza IA et al. Prevalence and molecular characterization of Enterobacteriaceae producing NDM-1 carbapenemase at a military hospital in Pakistan and evaluation of two chromogenic media. Diag Microbiol Infect Dis 2013; 75: 187–91. [DOI] [PubMed] [Google Scholar]
- 26. Lin MY, Lyles-Banks RD, Lolans K et al. The importance of long-term acute care hospitals in the regional epidemiology of Klebsiella pneumoniae carbapenemase-producing Enterobacteriaceae. Clin Infect Dis 2013; 57: 1246–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Findlay J, Hopkins KL, Meunier D et al. Evaluation of three commercial assays for rapid detection of genes encoding clinically relevant carbapenemases in cultured bacteria. J Antimicrob Chemother 2015; 70: 1338–42. [DOI] [PubMed] [Google Scholar]
- 28. Nijhuis R, Samuelsen O, Savelkoul P et al. Evaluation of a new real-time PCR assay (Check-Direct CPE) for rapid detection of KPC, OXA-48, VIM, and NDM carbapenemases using spiked rectal swabs. Diag Microbiol Infect Dis 2013; 77: 316–20. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.