Abstract
Background
Ciprofloxacin, a fluoroquinolone, targets two essential bacterial enzymes, DNA gyrase and topoisomerase IV. Plasmid-borne qnr genes, encoding proteins that protect DNA gyrase and topoisomerase IV from inhibition by fluoroquinolones, contribute to resistance development. However, the presence of a plasmid-borne qnr gene alone is insufficient to confer clinical resistance.
Objectives
We asked whether the level of expression of qnr was a limiting factor in its ability to confer clinical resistance and whether expression could be increased without reducing fitness or viability.
Methods
qnrB and qnrS were recombineered onto the chromosome of Escherichia coli under the control of constitutive promoters of various strengths. Expression was measured by qPCR, MIC and relative fitness as a function of expression level were determined.
Results
For both qnr genes there was a positive relationship between the level of qnr mRNA and the MIC of ciprofloxacin. The highest MICs achieved with qnrB or qnrS as the sole resistance determinant were 0.375 and 1 mg/L, respectively, and were reached at expression levels that did not affect growth rate or viability. The qnrS-mediated MIC is above the EUCAST clinical breakpoint for resistance to ciprofloxacin. In the absence of Lon protease activity, overexpression of qnr genes was associated with high fitness cost, possibly explaining observations of toxicity in other genetic backgrounds.
Conclusions
The ability to generate a high MIC without incurring a fitness cost shows that, in an appropriate genetic context, qnrS has the potential to generate clinical resistance to ciprofloxacin in one step.
Introduction
Ciprofloxacin is a fluoroquinolone with a range of indications, including urinary tract infections frequently caused by Escherichia coli.1–3 Fluoroquinolones target DNA gyrase and topoisomerase IV, which cleave and re-ligate DNA and are required for altering supercoiling levels during replication, and for chromosome decatenation.4 When ciprofloxacin binds to either enzyme it traps it in a stable complex with cleaved DNA, leading eventually to cell death.5
Despite being a synthetic and highly potent drug with dual-targeting activity, resistance to ciprofloxacin has reached high frequencies among clinical isolates worldwide.1 In the decade after its introduction all resistance reported in Enterobacteriaceae was associated with chromosomal mutations.6 Recently, plasmid-mediated quinolone resistance (PMQR) has become a frequent component of clinical resistance.2,7 The first reported PMQR gene reducing susceptibility to ciprofloxacin in Gram-negatives was qnrA in Klebsiella pneumoniae.8 Subsequently, several classes of qnr genes (qnrB, qnrC, qnrD, qnrS and qnrVC) were identified that reduce susceptibility to fluoroquinolones.7,9 Qnr proteins bind to DNA gyrase and topoisomerase IV and protect the enzymes from inhibition by quinolones.10,11
EUCAST sets the clinical breakpoint defining resistance for ciprofloxacin in Enterobacteriaceae at >0.5 mg/L,12 whereas CLSI defines the breakpoint at >4 mg/L.13 No single genetic alteration increases the MIC for susceptible WT E. coli above either breakpoint.2,14 Ciprofloxacin-resistant isolates of E. coli carry multiple genetic alterations, usually mutations in genes encoding the drug targets, mutations that up-regulate drug efflux and/or a PMQR gene.2,15 In general, the presence of a qnr gene in WT E. coli leads to an MIC of ciprofloxacin of 0.125–0.25 mg/L.7–9,14,15 A recent study showed that the MIC associated with plasmid-borne qnr genes increased from 0.25 to 32 mg/L when bacteria were grown in urine at pH 5.16 Under the same conditions, the MIC for the susceptible WT increased from 0.015 to 1 mg/L. This shows that MIC can be strongly influenced by environmental conditions though the significance of this very interesting observation for clinical outcomes remains to be determined.
qnr homologues exist on the chromosomes of many bacterial species but their spread into Gram-negative pathogens occurred by carriage on multidrug resistance plasmids.9 However, the chromosomal location of many qnr genes, and their association with mobile genetic elements in Gram-negatives, suggests there is in principle no barrier to their transfer onto chromosomes in Gram-negatives.
We addressed the potential for chromosomally located qnr in E. coli to confer a clinically relevant resistance level to ciprofloxacin and asked whether the level of resistance would be constrained by associated fitness costs. We report that qnrS can be expressed at levels that increase the MIC above the clinical breakpoint, without incurring significant loss in fitness.
Materials and methods
Media
LB was used as liquid medium for bacterial growth and for solid medium the LB was supplemented with 1.5% agar (LA). Chloramphenicol was used at 30 mg/L where indicated and counter-selection of sacB was done on LA lacking NaCl, supplemented with 5% sucrose. BBLTM Mueller–Hinton II (Becton, Dickinson & Company, France) was used for MIC determinations.
Strains
A set of isogenic strains was constructed, carrying constitutive promoters of different strengths17 upstream of a cat–sacB cassette at the galK locus in MG1655. Coding sequences for qnrB and qnrS were amplified from clinical isolates of E. coli15 and transcriptionally fused by recombineering to each of the promoters replacing the cat–sacB cassette.18 A lexA box, located between the native qnrB promoter and the coding sequence,19 was incorporated in the qnrB constructs. Strains in which lon was inactivated were constructed by P1-mediated transduction of the lon::kan allele from the Keio collection.20 PCR and DNA sequencing (Macrogen Europe Laboratory, Amsterdam, The Netherlands) were used to confirm the genetic constructions. Genotypes of bacterial strains and oligonucleotide sequences are given in Table S1 and Table S2 (available as Supplementary data at JAC Online).
qPCR
RNA extractions were made in three biological replicates. RNA was isolated at OD600 0.25–0.4 using an RNeasy Mini Kit (Qiagen) and quantified using a Nanodrop NO-1000 spectrophotometer. A DNase Turbo Free (Ambion, Life Technologies) kit was used to remove DNA. cDNA was made using a High Capacity Reverse Transcription kit (Applied Biosystems). qPCRs contained 1 μL of cDNA (diluted 1:10, 1:100, 1:1000), 12.5 μL of PerfeCTa SYBR Green FastMix (Quanta Biosciences), 1.25 μL of 10 μM forward and reverse primers (Table S2) and ddH2O up to a final volume of 20 μL. Eco Real-Time PCR System (Illumina) was used for qPCR. Control housekeeping genes were idnT, cysG and hcaT.21
MIC
A bacterial colony was dispersed in 1 mL of 0.9% NaCl and spread evenly over a Mueller–Hinton II agar plate using a cotton swab; an Oxoid™ M.I.C.Evaluator™ Strip (Thermo Fisher Scientific, Basingstoke, UK) was applied and incubated for 18 h at 37 °C.
Growth measurements
Overnight cultures (four biological replicates/strain) were diluted 1:1000 in LB. Growth was measured using Bioscreen C (Oy Growth Curves AB Ltd, Finland) with continuous shaking at 37 °C and readings at OD600 every 5 min. The natural logarithm (ln) of each OD600 value was plotted against time. Slopes of linear regression lines for 10 subsequent OD600 readings were computed. Exponential growth rate was calculated by dividing ln(2) with the value of the maximum slope. To evaluate whether qnr overexpression affected bacterial viability, cfu were quantified after plating serial dilutions from stationary-phase cultures onto LA.
Results and discussion
qnr mRNA levels and MIC increase as a function of promoter strength
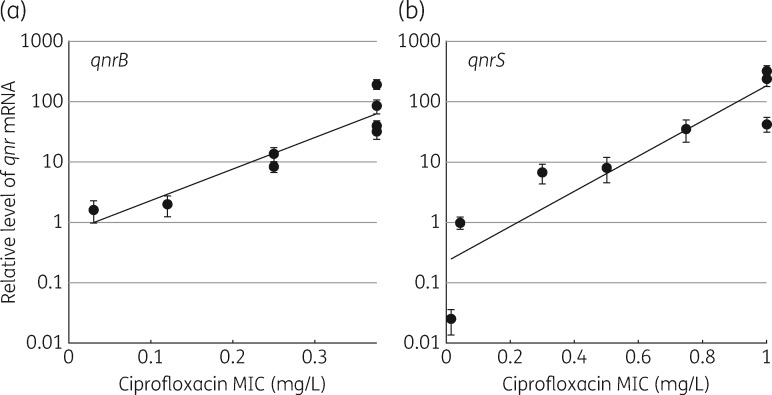
Isogenic strains were constructed with qnrB or qnrS transcriptionally fused to each of eight constitutively expressed promoters with different strengths (Table S1). The levels of qnrB and qnrS mRNA associated with the different promoters varied over a >100-fold range, with the strongest promoters producing 190-fold (qnrB) and 300-fold (qnrS) the level of the control mRNAs. Differences in mRNA levels for qnrB and qnrS as a function of the different promoters were quantified and related to the MIC of ciprofloxacin (Figure 1). The MIC of ciprofloxacin also increased as a function of the increase in mRNA, up to 0.375 mg/L for qnrB and up to 1 mg/L for qnrS. The maximal MICs remained constant with the four (qnrB) and three (qnrS) strongest promoters (Figure 1). The highest MICs achieved are significantly higher than those associated with the same qnr genes expressed on plasmids in the original clinical isolate, or equivalent genes on plasmids in other genetic backgrounds.9,15 For qnrS the MIC achieved with the three strongest promoters is above the level of the clinical resistance breakpoint defined by EUCAST.12
Figure 1.
Ciprofloxacin MIC increases as a function of the level of qnr mRNA. MIC of ciprofloxacin as a function of qnrB (a) and qnrS (b) mRNA levels, relative to the levels of three control housekeeping genes measured by qPCR. Standard deviations in mRNA level are indicated. The sloped line indicates the regression fit.
Increased MIC without an incurred fitness cost
In order to assess if the increased expression of the qnr genes incurred a biological fitness cost, exponential growth rates, and bacterial viability counts from the stationary phase, were measured (Table 1). This was done to test whether the metabolic cost of producing Qnr proteins, or an increased level of interactions between Qnr and DNA topoisomerases, would reduce growth rate or bacterial viability. For qnrB, there was no significant variation in either parameter, regardless of promoter strength, mRNA level or MIC value. For qnrS, the two strongest promoters indicated a growth rate reduction of ∼5%, but this was not statistically significant (P > 0.05). However, the maximal MIC of 1 mg/L was achieved with the third strongest promoter without reducing the growth rate (Table 1). We conclude that overexpression of these genes, resulting in the case of qnrS in an MIC of up to 1 mg/L, can be achieved without any significant fitness cost in terms of growth rate or viability.
Table 1.
Strain phenotype as a function of qnr expression level
| Strain | Relevant genotypea | mRNA (±SD)b | MICc | Growth rate (±SD)d | cfu × 109 (±SD)e |
|---|---|---|---|---|---|
| CH1464 | MG1655 | — | 0.015 | 1.00 (0.03) | — |
| CH7243 | J23113-qnrB | 1.62 (0.65) | 0.03 | 1.01 (0.01) | 4.61 (0.78) |
| CH7244 | J23117-qnrB | 1.96 (0.73) | 0.12 | 0.98 (0.04) | 4.22 (0.54) |
| CH7245 | J23115-qnrB | 8.31 (1.64) | 0.25 | 1.04 (0.04) | 4.12 (0.51) |
| CH7246 | J23105-qnrB | 13.59 (3.49) | 0.25 | 1.01 (0.05) | 3.64 (0.26) |
| CH7247 | J23110-qnrB | 39.48 (8.78) | 0.375 | 0.96 (0.07) | 3.73 (0.61) |
| CH7248 | J23118-qnrB | 32.68 (8.89) | 0.375 | 0.99 (0.03) | 3.91 (0.59) |
| CH7470 | J23101-qnrB | 84.54 (21.62) | 0.375 | 1.02 (0.05) | 4.55 (0.56) |
| CH7471 | J23100-qnrB | 193.12 (33.34) | 0.375 | 1.04 (0.03) | 3.78 (0.98) |
| CH7249 | J23113-qnrS | 0.02 (0.01) | 0.015 | 1.02 (0.06) | 3.55 (0.32) |
| CH7250 | J23117-qnrS | 1.00 (0.23) | 0.045 | 1.02 (0.02) | 4.03 (0.63) |
| CH7251 | J23115-qnrS | 6.73 (2.42) | 0.3 | 0.99 (0.05) | 3.81 (0.37) |
| CH7252 | J23105-qnrS | 8.15 (3.63) | 0.5 | 0.99 (0.07) | 3.29 (0.36) |
| CH7253 | J23118-qnrS | 35.36 (14.2) | 0.75 | 1.06 (0.05) | 3.82 (0.69) |
| CH7254 | J23110-qnrS | 42.61 (11.45) | 1 | 1.02 (0.02) | 3.46 (0.35) |
| CH7472 | J23101-qnrS | 328.30 (62.19) | 1 | 0.94 (0.05) | 3.37 (0.4) |
| CH7473 | J23100-qnrS | 234.37 (57.31) | 1 | 0.96 (0.04) | 3.33 (0.77) |
SD, standard deviation.
Strains are isogenic to MG1655. qnrB and qnrS were placed downstream of J23-series promoters in the galK locus.
mRNA levels of qnr relative to control genes hcaT, idnT and cysG, with standard deviations.
MIC of ciprofloxacin in mg/L.
Exponential growth rates relative to CH1464 (MG1655), with standard deviations.
cfu from serial dilution of overnight cultures, with standard deviations.
Growth rate in strains overexpressing Qnr is dependent on the activity of Lon protease
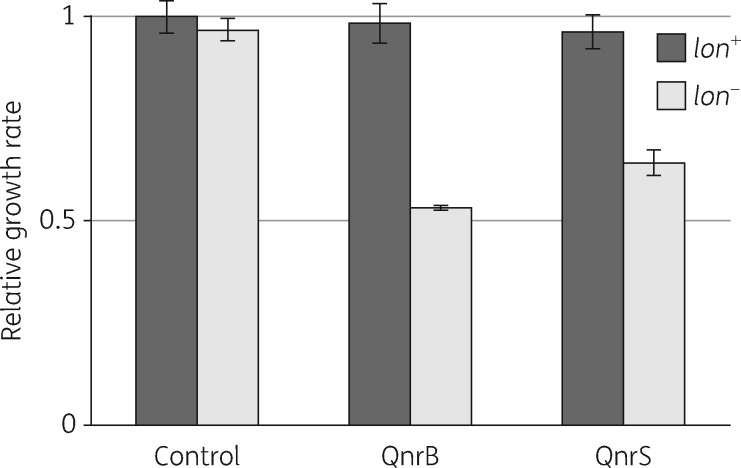
Previous in vitro biochemical studies have shown that although qnrB and qnrS protect DNA gyrase at low concentrations, they are inhibitory at higher concentrations.22,23 Although it could be argued that inhibition in vitro might not reflect the complexity of expression regulation in whole cells, there is a recent publication showing that overexpression in E. coli of different qnr genes found in clinical isolates is toxic, based on observations of strongly reduced viability.24 We asked what might explain this significant discrepancy with our study in which qnr overexpression was not observed to cause significant toxicity. Whereas we studied qnr expression in MG1655, the Machuca et al.24 study used E. coli BL21, a strain that is commonly used for protein overexpression studies and lacks two important protease activities, Lon and OmpT.25 Proteases such as Lon play an important role in E. coli in removing ‘proteins without partners’.26 We hypothesized that in MG1655, protease activities act to turn over excess Qnr when maximum protection of DNA gyrase has been reached, thus protecting the cell from toxic side effects. Accordingly, the toxicity observed with Qnr overproduction in BL21 might result from the failure to turn over excess proteins. To test this hypothesis, we constructed isogenic strains in which qnrB or qnrS were expressed from the strongest J23-series promoter, J23100, and measured growth rate as a function of lon activity (Figure 2). The growth rate of the control strain, MG1655, was unaffected by lon activity. However, in each of the strains expressing a qnr gene the growth rate decreased significantly (by 40%–50%) in the strains in which lon was inactivated (Figure 2). This shows that Lon plays an important role in preventing potential toxicity associated with Qnr overexpression and explains, at least in part, the discrepancy between our data and the BL21 study.
Figure 2.
lon activity is required to protect strains overexpressing qnr from toxic effects on growth rate. Growth rates of strains overexpressing qnr as a function of lon activity. Control, no qnr gene; QnrB, overexpressed from J23100; QnrS, overexpressed from J23100. Strain genotypes in Table S1.
Conclusions
In a recent study, decreased susceptibility to ciprofloxacin was caused by amplification of plasmid-encoded qnrA during in vitro selection.27 The observation that increased levels of qnr result in an increased MIC is in agreement with our results. However, tandem gene amplifications are inherently unstable and susceptibility returned to normal in the absence of selection, as expected. A point of interest is that qnr genes in natural isolates are known to be located on many different plasmid backbones,7 raising the possibility that there might be significant natural variation in drug susceptibility associated with differences in plasmid copy number or mechanisms controlling gene expression. Our finding that overexpression of qnr can cause significant toxicity in the absence of Lon protease activity shows that the genetic context in which a qnr gene is overexpressed will play an important role in determining the phenotype. Our study suggests that qnr genes, and qnrS in particular, when placed in a genetic context in which they are expressed at high levels, have the potential to generate genetically stable clinical resistance in one step, without incurring significant fitness costs.
Supplementary Material
Acknowledgments
Funding
This work was supported by grants to D. H. from the Swedish Research Council, Vetenskapsrådet (grant numbers: 2013-02904 and 2016-04449).
Transparency declarations
None to declare.
Supplementary data
Tables S1 and S2 are available as Supplementary data at JAC Online.
References
- 1. WHO. Antimicrobial Resistance: Global Report on Surveillance 2014. Geneva, Switzerland: WHO, 2014. [Google Scholar]
- 2. Redgrave LS, Sutton SB, Webber MA. et al. Fluoroquinolone resistance: mechanisms, impact on bacteria, and role in evolutionary success. Trends Microbiol 2014; 22: 438–45. [DOI] [PubMed] [Google Scholar]
- 3. Castro W, Navarro M, Biot C.. Medicinal potential of ciprofloxacin and its derivatives. Future Med Chem 2013; 5: 81–96. [DOI] [PubMed] [Google Scholar]
- 4. Chen CR, Malik M, Snyder M. et al. DNA gyrase and topoisomerase IV on the bacterial chromosome: quinolone-induced DNA cleavage. J Mol Biol 1996; 258: 627–37. [DOI] [PubMed] [Google Scholar]
- 5. Hooper DC. Mechanisms of action of antimicrobials: focus on fluoroquinolones. Clin Infect Dis 2001; 32 Suppl 1: S9–15. [DOI] [PubMed] [Google Scholar]
- 6. Hooper DC. Mechanisms of action and resistance of older and newer fluoroquinolones. Clin Infect Dis 2000; 31 Suppl 2: S24–8. [DOI] [PubMed] [Google Scholar]
- 7. Rodriguez-Martinez JM, Machuca J, Cano ME. et al. Plasmid-mediated quinolone resistance: two decades on. Drug Resist Updat 2016; 29: 13–29. [DOI] [PubMed] [Google Scholar]
- 8. Martinez-Martinez L, Pascual A, Jacoby GA.. Quinolone resistance from a transferable plasmid. Lancet 1998; 351: 797–9. [DOI] [PubMed] [Google Scholar]
- 9. Hooper DC, Jacoby GA.. Mechanisms of drug resistance: quinolone resistance. Ann N Y Acad Sci 2015; 1354: 12–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tran JH, Jacoby GA, Hooper DC.. Interaction of the plasmid-encoded quinolone resistance protein QnrA with Escherichia coli topoisomerase IV. Antimicrob Agents Chemother 2005; 49: 3050–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tran JH, Jacoby GA, Hooper DC.. Interaction of the plasmid-encoded quinolone resistance protein Qnr with Escherichia coli DNA gyrase. Antimicrob Agents Chemother 2005; 49: 118–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. EUCAST. Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 6.0, 2016 http://www.eucast.org.
- 13. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Sixth Informational Supplement M100-S26. CLSI, Wayne, PA, USA, 2016. [Google Scholar]
- 14. Huseby DL, Pietsch F, Brandis G. et al. Mutation supply and relative fitness shape the genotypes of ciprofloxacin-resistant Escherichia coli. Mol Biol Evol 2017; 34: 1029–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nazir H, Cao S, Hasan F. et al. Can phylogenetic type predict resistance development? J Antimicrob Chemother 2011; 66: 778–87. [DOI] [PubMed] [Google Scholar]
- 16. Martin-Gutierrez G, Rodriguez-Martinez JM, Pascual A. et al. Plasmidic qnr genes confer clinical resistance to ciprofloxacin under urinary tract physiological conditions. Antimicrob Agents Chemother 2017; 61: e02615–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. iGEM. Registry of Standard Biological Parts http://partsregistry.org.
- 18. Yu D, Ellis HM, Lee EC. et al. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci U S A 2000; 97: 5978–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Da Re S, Garnier F, Guerin E. et al. The SOS response promotes qnrB quinolone-resistance determinant expression. EMBO Rep 2009; 10: 929–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Baba T, Ara T, Hasegawa M. et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2006; 2: 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhou K, Zhou L, Lim Q. et al. Novel reference genes for quantifying transcriptional responses of Escherichia coli to protein overexpression by quantitative PCR. BMC Mol Biol 2011; 12: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jacoby GA, Walsh KE, Mills DM. et al. qnrB, another plasmid-mediated gene for quinolone resistance. Antimicrob Agents Chemother 2006; 50: 1178–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tavio MM, Jacoby GA, Hooper DC.. QnrS1 structure-activity relationships. J Antimicrob Chemother 2014; 69: 2102–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Machuca J, Diaz de Alba P, Recacha E. et al. Cytotoxic effect associated with overexpression of QNR proteins in Escherichia coli. Microb Drug Resist 2017; 23: 822–5. [DOI] [PubMed] [Google Scholar]
- 25. Jeong H, Barbe V, Lee CH. et al. Genome sequences of Escherichia coli B strains REL606 and BL21(DE3). J Mol Biol 2009; 394: 644–52. [DOI] [PubMed] [Google Scholar]
- 26. Gottesman S. Proteases and their targets in Escherichia coli. Annu Rev Genet 1996; 30: 465–506. [DOI] [PubMed] [Google Scholar]
- 27. Vinue L, Corcoran MA, Hooper DC. et al. Mutations that enhance the ciprofloxacin resistance of Escherichia coli with qnrA1. Antimicrob Agents Chemother 2016; 60: 1537–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.