Abstract
Objectives
The global spread of carbapenemase-producing Enterobacteriaceae represents a substantial challenge in clinical practice and rapid and reliable detection of these organisms is essential. The aim of this study was to develop and validate a lateral flow immunoassay (Carba5) for the detection of the five main carbapenemases (KPC-, NDM-, VIM- and IMP-type and OXA-48-like).
Methods
Carba5 was retrospectively and prospectively evaluated using 296 enterobacterial isolates from agar culture. An isolated colony was suspended in extraction buffer and then loaded on the manufactured Carba5.
Results
All 185 isolates expressing a carbapenemase related to one of the Carba5 targets were correctly and unambiguously detected in <15 min. All other isolates gave negative results except those producing OXA-163 and OXA-405, which are considered low-activity carbapenemases. No cross-reaction was observed with non-targeted carbapenemases, ESBLs, AmpCs or oxacillinases (OXA-1, -2, -9 and -10). Overall, this assay reached 100% sensitivity and 95.3% (retrospectively) to 100% (prospectively) specificity.
Conclusions
Carba5 is efficient, rapid and easy to implement in the routine workflow of a clinical microbiology laboratory for confirmation of the five main carbapenemases encountered in Enterobacteriaceae.
Introduction
The worldwide spread of carbapenemase-producing organisms is a global concern1 and an economic threat.2 Among these organisms, Enterobacteriaceae have a major role as causes of nosocomial infections (and, for Escherichia coli, also of community-acquired infections). The emergence and dissemination of carbapenemase-producing Enterobacteriaceae (CPE) is undoubtedly a matter of great public health concern considering CPE are often resistant to several if not all classes of antibiotics, and very few (or no) antibiotic options remain available for them.3,4 The rapid detection and identification of CPE is essential to help physicians quickly implement appropriate infection control measures, to adapt antibiotic treatment rapidly and to optimize care strategies and outcomes. Based on their amino acid sequence, carbapenemases are divided into different molecular classes: A, B and D of the Ambler classification. Their susceptibility to carbapenems varies from weak activity to very efficient hydrolysis. Discrimination between these carbapenemases will provide valuable information for appropriate treatment. For instance, a β-lactamase inhibitor (ceftazidime/avibactam) is now available, but does not inhibit MBL activity.5,6
Several diagnostic tests have been developed based on the detection of carbapenem-hydrolysing activity [MALDI-TOF MS7,8 or biochemical assay (e.g. Carba NP test and derivatives),9,10 the carbapenem inactivation method11 and OXA-48 disc test12,13]. In addition, several phenotypic confirmation tests have also been developed, but usually require 24 h.13,14 Although they have proven useful, it is now also crucial to identify the implicated carbapenemase. Molecular methods are undoubtedly appropriate for this purpose. End-point PCR and real-time PCR can also be used in single or multiplex formats targeting the main carbapenemases with high specificity and sensitivity.15–17 However, these methods are expensive and require a high level of expertise to obtain accurate results and are not suitable for all clinical microbiology laboratories worldwide. As an alternative, antibody-based methods such as lateral flow immunoassay (LFIA) have already been validated for carbapenemase-producer detection and carbapenemase identification.18–22 These tests yield results from cultured strains within 15 min, with 100% sensitivity and specificity, although they do not allow the simultaneous detection of the five main carbapenemases.
To meet current needs, antimicrobial drug resistance detection methods must be cheap (reduced cost of consumables and equipment) and easy to use (reduced technical complexity) for the end user. This led us to develop an LFIA, named Carba5, for the detection of the five main carbapenemase families, NDM-, IMP-, VIM- and KPC-type and OXA-48-like. Carba5 is a robust assay, easily transferable in a commercialized version, which is stable for >24 months without refrigeration, user-friendly (no need for trained staff), high in performance (sensitive and specific) and low in cost (although pricing is not yet specified, similar LFIA tests usually cost ∼10€ per test)20 compared with >30–40€ for molecular tests. Moreover, the detection results are obtained in a short time without the need for highly technical equipment for the readout. Here, we validated Carba5 on 296 agar-cultured enterobacterial isolates [180 characterized isolates for their β-lactamase content and 116 consecutive carbapenem-resistant isolates referred to the Associated French National Reference Centre (F-NRC) for expertise], including 185 isolates expressing NDM-, IMP-, VIM- or KPC-type or OXA-48-like carbapenemases, which were perfectly detected in 15 min.
Methods
Ethics statement
All experiments were performed in compliance with French and European regulations on the care of laboratory animals (European Community Directive 86/609, French Law 2001-486, 6 June 2001) and with the agreements of the Ethics Committee of the Commissariat à l’Energie Atomique (CEtEA ‘Comité d’Ethique en Expérimentation Animale’ n° 44) no. 12-026 and 15-055 delivered to S. S. by the French Veterinary Services and CEA agreement D-91-272-106 from the Veterinary Inspection Department of Essonne (France).
Strains tested
For the LFIA validation, 180 enterobacterial isolates with PCR-characterized β-lactamase content were used to evaluate Carba5. This collection represented 55 non-carbapenemase producers and 125 carbapenemase producers, including 30 Ambler class A producers (22 KPC, 3 IMI, 2 SME, 1 NMC-A, 1 FRI-1, 1 GES-5), 52 metallo-β-lactamase producers from Ambler class B (23 NDM, 17 VIM, 11 IMP, 1 GIM-1) and 38 Ambler class D producers corresponding to 37 OXA-48-like (21 OXA-48, 1 OXA-162, 3 OXA-181, 5 OXA-204, 2 OXA-232, 2 OXA-244, 1 OXA-517, 1 OXA-519, 1 OXA-535) and one OXA-372. Five isolates expressed both NDM-type and OXA-48-like carbapenemases.
For the prospective evaluation, 116 consecutive enterobacterial clinical isolates with decreased susceptibility to at least one carbapenem were used. These isolates were sent between April and May 2017 by clinical microbiology laboratories throughout France to the F-NRC for carbapenem-resistant Enterobacteriaceae.
Carbapenemase activity detection and PCR/sequencing experiments
Carbapenemase activity was evidenced in Enterobacteriaceae using the updated version of the Carba NP test and using an in-house PCR and sequencing approach directly on colonies.10
Monoclonal antibodies
Biozzi mice (10 weeks old) were immunized with the five recombinant carbapenemases, NDM-1 (G29-R270), OXA-48 (K23-P265), KPC-2 (A30-Q293), IMP-1 (L22-N246) and VIM-1 (A22-E266), corresponding to the full sequence without the signal sequence expressed with the hexa-histidine tag on their C terminus. Consecutively, monoclonal antibodies were obtained, characterized and selected for the LFIA format as previously described.19
Manufactured Carba5 and assay protocol
The required antibodies were produced on a large scale as previously described19 and provided to NG Biotech (Guipry, France) for the development of multiplex LFIA (see Figure 1). The strains to be tested were grown overnight at 37°C on URISelectTM 4 or Mueller–Hinton agar plates (Bio-Rad, Marnes la Coquette, France). One single isolated colony was collected from the plate with an inoculation loop and suspended in 150 μL of extraction buffer to perform the lysis step. Subsequently, 100 μL of this extract was loaded on the cassette and allowed to migrate for 15 min. The protocol of the commercialized kit will be similar to the protocol described here.
Figure 1.

Carba5 principle.
Data acquisition
Data could be acquired in two separate ways, i.e. (i) a naked eye reading [the carbapenemase is identified by associating the positive result (positive test line) with the name of the carbapenemase written on the plastic part of the cassette], and (ii) using a computer-assisted reader that only gives results for positive test lines with the corresponding target name. An illustration of the results obtained is shown in Figure 2.
Figure 2.
Data acquisition using a handheld reader. (a) Results obtained after 15 min of migration with carbapenemase-expressing strains. NDM, IMP, VIM, OXA and KPC, one colony resuspended in 150 µL of extraction buffer and 100 µL loaded on the cassette. For all, 25 µL from each previous extract pooled leading to a 1 to 5 dilution and 100 µL of this pool loaded on the cassette. (b) Previous IMP-expressing strain extract, picture of the cassette, positive test line analysis and corresponding value and carbapenemase name (table). (c) Previous pool of the five carbapenemase-expressing strain extracts, positive test line analysis and corresponding value (table). An asterisk indicates a test line not visible owing to camera sensitivity.
For this study, basic format cassettes were used and the carbapenemase names were not indicated, so the ‘handheld reader’ method was preferred.
Statistical analysis
The sensitivity, specificity, positive predictive value and negative predictive value were calculated with their respective 95% CI using the free software vassarStats: Website for Statistical Computation on http://vassarstats.net/. The gold standard was PCR followed by sequencing. Results were extrapolated to the global French epidemiology data obtained between 2012 and 2015.
Results
Prior to Carba5 validation, manufactured single-test LFIAs were validated with the F-NRC carbapenem-resistant Enterobacteriaceae collection (180 isolates) for the detection of NDM-type,19 IMP-type, VIM-type and OXA-48-like carbapenemases (data not shown). The manufactured single test for KPC-type carbapenemase was validated with only 60 strains from the same collection. These single tests gave 100% sensitivity and specificity according to the PCR-characterized β-lactamase content of the isolates. The manufactured Carba5 was developed with the same antibodies and tested with the F-NRC collection for validation. Routine use was successively assessed with strains showing a decreased susceptibility to at least one carbapenem.
Validation of Carba5
As described in Table S1 (available as Supplementary data at JAC Online), Carba5 was able to detect (strong signals) unambiguously all OXA-48-like-producing isolates with carbapenemase activity [n = 37; including OXA-48 (n = 21), OXA-162 (n = 1), OXA-181 (n = 3), OXA-204 (n = 5), OXA-232 (n = 2), OXA-244 (n = 2), OXA-517 (n = 1), OXA-519 (n = 1) and OXA-535 (n = 1)], as well as the KPC producers [n = 22; including KPC-2 (n = 19) and KPC-3 (n = 3)], NDM producers [n = 23; including NDM-1 (n = 16), NDM-4 (n = 2), NDM-5 (n = 1), NDM-6 (n = 1), NDM-7 (n = 1) and NDM-9 (n = 2)], IMP producers [n = 11; including IMP-1 (n = 5), IMP-8 (n = 5) and IMP-11 (n = 1)] and VIM producers [n = 17; including VIM-1 (n = 12), VIM-2 (n = 2), VIM-4 (n = 2) and VIM-19 (n = 1)]. Five strains simultaneously expressing NDM-1 and OXA-181 were also detected and thus show two positive test lines. The results obtained with a non-carbapenemase-producing strain, a carbapenemase-producing strain and a multiple carbapenemase-producing strain are shown in Figure 3. The three isolates expressing OXA-48-like broad-spectrum oxacillinases with very weak carbapenemase activity, OXA-163 (n = 2) and OXA-405 (n = 1), also gave positive results and were considered false positives.
Figure 3.
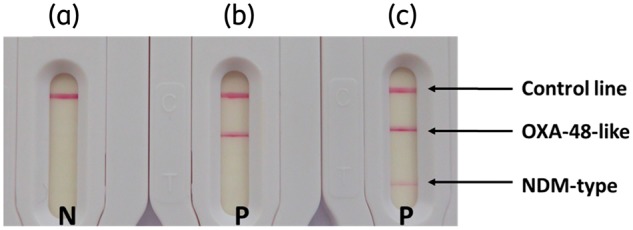
Results obtained with different strains. One colony suspended in extraction buffer and loaded on the cassette. (a) Klebsiella pneumoniae expressing CTX-M-18. (b) K. pneumoniae expressing OXA-162. (c) K. pneumoniae expressing NDM-1 and OXA-48. N, negative result; P, positive result.
All the CPE expressing a carbapenemase not related to any of the five carbapenemase families targeted gave negative results. They corresponded to eight Ambler class A [IMI (n = 3), GES-5 (n = 1), NMC-A (n = 1), SME (n = 2) and FRI-1 (n = 1)], one OXA-372 (an Ambler class D carbapenem-hydrolysing enzyme not related to OXA-48 β-lactamase)23 and one Ambler class B GIM-1. In addition, no positive signal was obtained with the 51 non-carbapenemase producers.
Prospective evaluation of Carba5
For this study, 116 consecutive isolates were blindly tested and LFIA results were compared with those of the home-made Carba NP test and PCR sequencing, which are routinely performed by the F-NRC on all isolates.10 In less than 15 min Carba5 detected 70 carbapenemase-positive isolates (Table 1) corresponding to OXA-48-like producers [n = 57; including OXA-48 (n = 52), OXA-181 (n = 3) and OXA-244 (n = 2)], NDM producers [n = 9; including NDM-1 (n = 5), NDM-5 (n = 3) and NDM-9 (n = 1)], VIM producers [n = 2; including VIM-1 (n = 1) and VIM-4 (n = 1)] and one KPC-3 producer. One strain was positive for both NDM and OXA-48 (NDM-5 and OXA-48). The 46 other isolates gave negative test results. Similar results were obtained with the Carba NP test, with 2 h incubation, for 45 isolates. The PCR sequencing results revealed 71 carbapenemase producers; 70 corresponded to those previously identified with Carba5 and the Carba NP test, and one corresponded to the IMI-producing Enterobacter cloacae, a non-targeted carbapenemase, which gave a positive result with the Carba NP test. Forty-five non-carbapenemase producers were confirmed negative by PCR.
Table 1.
Prospective evaluation of Carba5 on 116 consecutive isolates referred to the F-NRC for carbapenem-resistant Enterobacteriaceae
| Species | No. of isolates | Carba5 results |
Carbapenemase content (PCR + sequencing) | |||||
|---|---|---|---|---|---|---|---|---|
| NDM | IMP | VIM | OXA | KPC | ||||
| Carbapenemase producers | ||||||||
| NDM-type | E. coli | 1 | P | N | N | N | N | NDM-1 |
| E. coli | 3 | P | N | N | N | N | NDM-5 | |
| E. coli | 1 | P | N | N | N | N | NDM-9 | |
| K. pneumoniae | 3 | P | N | N | N | N | NDM-1 | |
| Morganella morganii | 1 | P | N | N | N | N | NDM-1 | |
| OXA-48-like | E. coli | 13 | N | N | N | P | N | OXA-48 |
| K. pneumoniae | 24 | N | N | N | P | N | OXA-48 | |
| Klebsiella oxytoca | 3 | N | N | N | P | N | OXA-48 | |
| E. cloacae | 3 | N | N | N | P | N | OXA-48 | |
| Citrobacter koseri | 2 | N | N | N | P | N | OXA-48 | |
| Citrobacter freundii | 6 | N | N | N | P | N | OXA-48 | |
| Enterobacter aerogenes | 1 | N | N | N | P | N | OXA-48 | |
| E. coli | 2 | N | N | N | P | N | OXA-181 | |
| K. pneumoniae | 1 | N | N | N | P | N | OXA-181 | |
| E. coli | 2 | N | N | N | P | N | OXA-244 | |
| KPC-type | K. pneumoniae | 1 | N | N | N | N | P | KPC-3 |
| VIM-type | K. pneumoniae | 1 | N | N | P | N | N | VIM-1 |
| E. cloacae | 1 | N | N | P | N | N | VIM-4 | |
| IMI-type | E. cloacae | 1 | N | N | N | N | N | IMI-1 |
| multiple carbapenemases | E. coli | 1 | P | N | N | P | N | NDM-1 + OXA-48 |
| Non-carbapenemase producers | ||||||||
| E. coli | 4 | N | N | N | N | N | ||
| K. pneumoniae | 15 | N | N | N | N | N | ||
| E. cloacae | 17 | N | N | N | N | N | ||
| E. aerogenes | 3 | N | N | N | N | N | ||
| Hafnia alvei | 2 | N | N | N | N | N | ||
| Proteus mirabilis | 1 | N | N | N | N | N | ||
| C. freundii | 1 | N | N | N | N | N | ||
| M. morganii | 2 | N | N | N | N | N | ||
N, negative result; P, positive result.
Global performances of Carba5
Considering the results obtained with the F-NRC collection, the sensitivity and specificity of Carba5 towards the five target-related enzymes respectively reached 100% (95% CI = 95.8%–100%) and 95.3% (95% CI = 86.0%–98.8%) (misidentifying two OXA-163 and one OXA-405 as positives). This test is able to detect strains simultaneously expressing two carbapenemases (e.g. six strains with a combination of NDM-type and OXA-48-like enzymes). Importantly, no cross-reaction was observed with the non-targeted carbapenemases (IMI, SME, NMC-A, FRI, GES, GIM and OXA-372), ESBLs (TEM, SHV and CTX-M), AmpCs (CMY-2, DHA-2 and ACC-1) or oxacillinases (OXA-1, -2, -9 and -10). During the prospective study, Carba5 reached 100% (95% CI = 93.5%–100%) sensitivity, 100% (95% CI = 90.4%–100%) specificity, 100% (95% CI = 93.5%–100%) positive predictive value and 100% (95% CI = 90.4%–100%) negative predictive value for the identification of the five main carbapenemase families. Bacterial colonies were grown on two different plates and there was no interference depending on the media considered (Mueller–Hinton/URISelectTM 4).
Discussion
The spread of carbapenemase-expressing strains is a major public health concern. From an infection control point of view, the rapid identification of such strains is essential to prevent spread in hospital settings. Furthermore, with novel inhibitor/β-lactam combinations, knowing exactly which carbapenemase is present is a prerequisite for the proper use of these novel molecules. Therefore, fast and user-friendly assays are mandatory. Carba5 fulfils all of the requirements to stratify patients.
Carba5 has been optimized to detect the KPC, NDM, VIM and IMP carbapenemase families as well as OXA-48 and its related variants. Most of the time, positive results were interpretable in less than the recommended 15 min of migration. Compared with the single-test LFIAs, no interference between the different carbapenemase detections was induced by the multiplex format. Moreover, Carba5 is able to detect and identify the simultaneous production of several of the five main carbapenemases by one strain. Overall, Carba5 detects these five carbapenemases with a sensitivity of 100% without any false negatives. It detected new variants of OXA-48, such as OXA-517, OXA-519 and OXA-535,24–26 and OXA-48-related variants without significant carbapenemase activity (OXA-163 and OXA-405). As these enzymes do not confer reduced susceptibility to carbapenems, their identification requires further testing. In our testing scheme, these peculiar OXA variants were over-represented (3 of 180), while they represent only 1 of almost 10 000 isolates in France over a 4 year period. Therefore, considering global French epidemiology, the specificity of Carba5 would be 99.98% (95% CI = 99.90%–99.99%).
Meanwhile, during the prospective evaluation, Carba5 detected 70 of the 71 (98.6%) carbapenemase producers. One IMI producer was not detected, but this should not be considered a major drawback considering the very low prevalence of this carbapenemase.
During the period 2012–15, 3320 carbapenemase producers of 9518 enterobacterial isolates were received at the F-NRC. 118 KPC, 13 IMI, 1 FRI-1, 366 NDM, 115 VIM, 10 IMP, 2491 OXA-48, 120 OXA-181, 35 OXA-204, 8 OXA-232, 10 OXA-244 and 33 multiple carbapenemase producers (including 30 NDM + OXA-48-like, 1 NDM + VIM and 2 VIM + OXA-48-like) have been identified. By extrapolating the results obtained with the LFIA, we would expect that all isolates except those producing IMI, and FRI-1 (total = 14) would have been correctly identified. In addition, the unique OXA-405-producing Serratia marcescens would give a false-positive result. Thus, the overall performances of Carba5 for the detection of the five main carbapenemase would be 100% (95% CI = 99.86%–100%) sensitivity, 99.98% (95% CI = 99.90%–99.99%) specificity, 99.97% (95% CI = 99.80%–99.99%) positive predictive value and 100% (95% CI = 99.92%–100%) negative predictive value, respectively. More generally, the overall performance of Carba5 for the detection of all CPE (considering the 14 non-targeted carbapenemases as false-negative results) would be 99.58% (95% CI = 99.27%–99.76%) sensitivity, 99.98% (95% CI = 99.90%–99.99%) specificity, 99.97% (95% CI = 99.80%–99.99%) positive predictive value and 99.77% (95% CI = 99.61%–99.87%) negative predictive value, respectively.
So far, Carba5 can detect NDM-1/4/5/6/7/9, IMP-1/8/11, VIM-1/2/4/19, KPC-2/3 and OXA-48/162/181/204/232/244/517/519/535, and could be performed in routine laboratories, yielding unequivocal results in ∼15 min. Strains with decreased susceptibility to at least one carbapenem could quickly be characterized if at least one of the main carbapenemases is expressed. Carba5 has been used at the F-NRC to confirm negative results obtained with the Carba NP test (data not shown). Some Providencia strains with negative Carba NP test results were actually NDM producers and were identified by Carba5 and later confirmed by PCR (data not shown). These false-negative Carba NP test results could be due to low expression levels of the carbapenemase or low permeability of the bacterial outer membrane. In addition, Carba5 is currently used with strains growing on CPE screening media (such as CarbaSMART)27 and shows the same performance as with the two media used for this study, making it a reliable tool for CPE confirmation.
Compared with other LFIA tests available, Carba5 targets the five main carbapenemases. It could be used whatever the carbapenemase endemic context. Its universal use could help to report sporadic outbreaks leading to epidemiological updates. We agree that the epidemiology of carbapenemases is different between countries from different continents, but the five main carbapenemases account in each country for >99.5% of the determinants encountered. The relative percentages vary between the different countries and our test is thus suitable for most. We also agree that greater validation is now necessary in the form of multicentre validation. This validation is currently in progress in countries with high prevalences of OXA-48, KPC and MBLs.
In the near future, we aim to combine Carba5 with an LFIA already developed and dedicated to the identification of CTX-M producers to provide a very powerful and broad-spectrum tool for the detection of β-lactamases and guidance of antibiotic treatments.
Conclusions
Carba5 is efficient, rapid and easy to implement in the routine workflow of a clinical microbiology laboratory for the detection of CPE. It could complete the existing panel of tests available for the detection of carbapenemases, particularly in countries with limited resources and/or high prevalence of carbapenemases. Discrimination of OXA-48-like and KPC carbapenemases could rapidly guide treatments with ceftazidime/avibactam, and more generally prevent antibiotic misuse and provide efficient tools to contain the spread of these deadly bacteria in hospital settings.
Supplementary Material
Acknowledgements
We are thankful to Quentin Baratte for his involvement in the production of the required monoclonal antibodies.
Funding
This work was supported by the Assistance Publique—Hôpitaux de Paris (AP-HP), the University Paris-Sud, the Laboratory of Excellence in Research on Medication and Innovative Therapeutics (LERMIT) supported by a grant from the French National Research Agency (ANR-10-LABX-33) and by the Joint Programming Initiative on Antimicrobial Resistance (JPIAMR) DesInMBL (ANR-14-JAMR-002).
Transparency declarations
L. D. is co-inventor of the Carba NP test, the patent of which has been licensed to bioMérieux (La Balmes les Grottes, France). All other authors: none to declare.
Supplementary data
Table S1 is available as Supplementary data at JAC Online.
References
- 1. WHO. Antimicrobial Resistance: Global Report on Surveillance 2014. http://apps.who.int/iris/bitstream/10665/112642/1/9789241564748_eng.pdf? ua=1.
- 2. Shriber DE, Baris E, Marquez PV. et al. Final Report. The World Bank, 2017; 1–172. http://documents.worldbank.org/curated/en/323311493396993758/pdf/114679-REVISED-v2-Drug-Resistant-Infections-Final-Report.pdf. [Google Scholar]
- 3. Poirel L, Pitout JD, Nordmann P.. Carbapenemases: molecular diversity and clinical consequences. Future Microbiol 2007; 2: 501–12. [DOI] [PubMed] [Google Scholar]
- 4. Doi Y, Paterson DL.. Carbapenemase-producing Enterobacteriaceae. Semin Respir Crit Care Med 2015; 36: 74–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stone GG, Bradford PA, Yates K. et al. In vitro activity of ceftazidime/avibactam against urinary isolates from patients in a Phase 3 clinical trial programme for the treatment of complicated urinary tract infections. J Antimicrob Chemother 2017; 72: 1396–9. [DOI] [PubMed] [Google Scholar]
- 6. Pfizer Launches ZaviceftaTM (Ceftazidime-Avibactam) in the U.K. and Germany, a New Antibiotic to Treat Complicated Infections Caused by Gram-Negative Bacteria 2017. http://press.pfizer.com/press-release/pfizer-launches-zavicefta-ceftazidime-avibactam-uk-and-germany-new-antibiotic-treat-co.
- 7. Lasserre C, De Saint Martin L, Cuzon G. et al. Efficient detection of carbapenemase activity in Enterobacteriaceae by matrix-assisted laser desorption ionization−time of flight mass spectrometry in less than 30 minutes. J Clin Microbiol 2015; 53: 2163–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Papagiannitsis CC, Študentová V, Izdebski R. et al. Matrix-assisted laser desorption ionization–time of flight mass spectrometry meropenem hydrolysis assay with NH4HCO3, a reliable tool for direct detection of carbapenemase activity. J Clin Microbiol 2015; 53: 1731–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dortet L, Brechard L, Poirel L. et al. Impact of the isolation medium for detection of carbapenemase-producing Enterobacteriaceae using an updated version of the Carba NP test. J Med Microbiol 2014; 63: 772–6. [DOI] [PubMed] [Google Scholar]
- 10. Dortet L, Agathine A, Naas T. et al. Evaluation of the RAPIDEC® CARBA NP, the Rapid CARB Screen® and the Carba NP test for biochemical detection of carbapenemase-producing Enterobacteriaceae. J Antimicrob Chemother 2015; 70: 3014–22. [DOI] [PubMed] [Google Scholar]
- 11. Tijet N, Patel SN, Melano RG.. Detection of carbapenemase activity in Enterobacteriaceae: comparison of the carbapenem inactivation method versus the Carba NP test. J Antimicrob Chemother 2016; 71: 274–6. [DOI] [PubMed] [Google Scholar]
- 12. Dortet L, Poirel L, Nordmann P.. Rapid identification of carbapenemase types in Enterobacteriaceae and Pseudomonas spp. by using a biochemical test. Antimicrob Agents Chemother 2012; 56: 6437–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tsakris A, Poulou A, Bogaerts P. et al. Evaluation of a new phenotypic OXA-48 disk test for differentiation of OXA-48 carbapenemase-producing Enterobacteriaceae clinical isolates. J Clin Microbiol 2015; 53: 1245–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hrabák J, Chudáčková E, Papagiannitsis CC.. Detection of carbapenemases in Enterobacteriaceae: a challenge for diagnostic microbiological laboratories. Clin Microbiol Infect 2014; 20: 839–53. [DOI] [PubMed] [Google Scholar]
- 15. van der Zee A, Roorda L, Bosman G. et al. Multi-centre evaluation of real-time multiplex PCR for detection of carbapenemase genes OXA-48, VIM, IMP, NDM and KPC. BMC Infect Dis 2014; 14: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tato M, Ruiz-Garbajosa P, Traczewski M. et al. Multisite evaluation of Cepheid Xpert Carba-R assay for detection of carbapenemase-producing organisms in rectal swabs. J Clin Microbiol 2016; 54: 1814–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ellington MJ, Findlay J, Hopkins KL. et al. Multicentre evaluation of a real-time PCR assay to detect genes encoding clinically relevant carbapenemases in cultured bacteria. Int J Antimicrob Agents 2016; 47: 151–4. [DOI] [PubMed] [Google Scholar]
- 18. Glupczynski Y, Evrard S, Ote I. et al. Evaluation of two new commercial immunochromatographic assays for the rapid detection of OXA-48 and KPC carbapenemases from cultured bacteria. J Antimicrob Chemother 2016; 71: 1217–22. [DOI] [PubMed] [Google Scholar]
- 19. Boutal H, Naas T, Devilliers K. et al. Development and validation of a lateral flow immunoassay for rapid detection of NDM-producing Enterobacteriaceae. J Clin Microbiol 2017; 55: 2018–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dortet L, Jousset A, Sainte-Rose V. et al. Prospective evaluation of the OXA-48 K-SeT assay, an immunochromatographic test for the rapid detection of OXA-48-type carbapenemases. J Antimicrob Chemother 2016; 71: 1834–40. [DOI] [PubMed] [Google Scholar]
- 21. Glupczynski Y, Jousset A, Evrard S. et al. Prospective evaluation of the OKN K-SeT assay, a new multiplex immunochromatographic test for the rapid detection of OXA-48-like, KPC and NDM carbapenemases. J Antimicrob Chemother 2017; 72: 1955–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Notake S, Matsuda M, Tamai K. et al. Detection of IMP metallo-β-lactamase in carbapenem-nonsusceptible Enterobacteriaceae and non-glucose-fermenting Gram-negative rods by immunochromatography assay. J Clin Microbiol 2013; 51: 1762–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Antonelli A, D’Andrea MM, Vaggelli G. et al. OXA-372, a novel carbapenem-hydrolysing class D β-lactamase from a Citrobacter freundii isolated from a hospital wastewater plant. J Antimicrob Chemother 2015; 70: 2749–56. [DOI] [PubMed] [Google Scholar]
- 24. Dabos L, Bogaerts P, Raczynska J. et al. OXA-517, an extended-spectrum cephalosporin- and carbapenem-hydrolysing OXA-48-like variant. In: Abstracts of the Twenty-seventh European Congress of Clinical Microbiology and Infectious Diseases, Vienna, Austria, 2017. Abstract P0234. ESCMID, Basel, Switzerland.
- 25. Dabos L, Bogaerts P, Bonnin R. et al. Genetic and biochemical characterization of OXA-519, a novel OXA-48-like β-lactamase. In: Abstracts of the Twenty-seventh European Congress of Clinical Microbiology and Infectious Diseases, Vienna, Austria, 2017. Abstract P0232. ESCMID, Basel, Switzerland.
- 26. Dabos L, Jousset A, Potron A. et al. Genetic and biochemical characterization of OXA-535, a novel OXA-48-like enzyme progenitor of OXA-436 from Shewanella bicestrii In: Abstracts of the Twenty-seventh European Congress of Clinical Microbiology and Infectious Diseases, Vienna, Austria, 2017. Abstract P0233. ESCMID, Basel, Switzerland.
- 27. Meunier D, Vickers A, Pike R. et al. Evaluation of the K-SeT R.E.S.I.S.T. immunochromatographic assay for the rapid detection of KPC and OXA-48-like carbapenemases. J Antimicrob Chemother 2016; 71: 2357–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



