Abstract
Objectives: There is an urgent need for accurate and fast diagnostic tests capable of identifying carbapenemase producers. Here, we assessed the performance of a new multiplex lateral flow assay (OKN K-SeT) for the rapid detection of OXA-48-like, KPC and NDM carbapenemase-producing Enterobacteriaceae from culture colonies.
Methods: Two hundred collection isolates with characterized β-lactamase content and 183 non-duplicate consecutive isolates referred to two National Reference Centres over a 2 month period in 2016 were used to evaluate the OKN K-SeT assay.
Results: The assay correctly detected all 42 OXA-48-like-, 27 KPC- and 30 NDM-producing isolates from the collection panel, including 7 isolates that co-produced NDM and OXA-181 carbapenemases. No cross-reactivity was observed with non-targeted carbapenemases (n = 41) or with non-carbapenemase producers (n = 60). Prospectively, all OXA-48-like (n = 69), KPC (n = 9) and NDM (n = 19) carbapenemase-producing Enterobacteriaceae isolates were correctly detected, while 11 carbapenemase producers not targeted by the assay went undetected [VIM (n = 8) and OXA-23/OXA-58-like (n = 3)]. Overall, the sensitivity and specificity of the assay were 100%.
Conclusions: The OKN assay is efficient, rapid and easy to implement in the workflow of a clinical microbiology laboratory for the confirmation of OXA-48, NDM and KPC carbapenemases. This test represents a powerful diagnostic tool as it enables the rapid detection of the most clinically important carbapenemases without the need for more costly and less frequently available molecular assays.
Introduction
The emergence and global dissemination of carbapenemase-producing Enterobacteriaceae (CPE) constitutes a major public health concern. Accurate and timely detection of CPE is essential for patient management and for rapid implementation of appropriate infection control measures.1,2 In addition, the most promising therapies to treat infection caused by CPE involve combinations of a broad-spectrum β-lactam and a β-lactamase inhibitor (e.g. ceftazidime/avibactam) that are only active against certain classes of carbapenemases [e.g. avibactam inhibits Ambler class A and D carbapenemase but not metallo-β-lactamases (MBLs)]. Accordingly, it becomes crucial not only to detect a carbapenemase activity but also to characterize the enzyme. However, the detection of CPE constitutes one of the most challenging issues for diagnostic microbiological laboratories as it is extremely difficult to detect them based only on the resistance phenotype.3
Since 2010, various tests have been developed for the detection of carbapenemase activity in cultured bacteria. These include inhibitor-based tests,3 tests based on carbapenem hydrolysis including the colorimetric assays (e.g. the Carba NP tests or derivatives),4,5 MS,6 electrochemical assays (e.g. the BYG test)7 and the carbapenem inactivation method.8 All these tests are able to detect the presence of carbapenemase activity and sometimes to discriminate between Ambler class A, B and D carbapenemases (e.g. Carba NP test II, combined disc test inhibition tests).3–5 However, one of their major limitations relates to the absence of reliable phenotypical tests for detection of OXA-48 producers. This is mostly due to (i) the lack of suitable inhibitor compounds that might be used in confirmatory tests and (ii) weak carbapenemase activity, which might affect their detection by biochemical methods9–12 and results in relatively low MICs of carbapenems.
Recently, lateral flow assays based on monoclonal antibodies generated by immunization in mice have been developed for easy and rapid detection of OXA-48-like and KPC carbapenemases.13,14 This technology was shown by our group14 and confirmed by several other investigators in different countries15–18 to be a powerful means (100% sensitivity and 100% specificity) to identify OXA-48 and KPC producers within 15 min directly from bacterial colonies. The potential of the OXA-48-K-Se T® assay (Coris BioConcept) for the direct detection of OXA-48-producing strains in biological samples such as blood or urine (detection limit of 106 cfu/mL) was also highlighted in some of the studies.15,18
Here we present the evaluation of a new multiplex assay (the OKN K-SeT), which incorporates specific antibodies against OXA-48, NDM and KPC in a single test and aims to detect these three carbapenemases directly from solid cultures.
Materials and methods
Strain collection
Two hundred enterobacterial isolates with characterized β-lactamase content, referred to the Associated French National Reference Centre for Antibiotic Resistance between 2008 and 2014, were selected to validate the OKN K-SeT assay. This strain collection included 60 non-carbapenemase producers with various resistance mechanisms to carbapenems (including ESBL and/or acquired AmpC cephalosporinases ± impermeability and extended-spectrum oxacillinases OXA-163 and OXA-405) and 140 carbapenemase producers, including 36 Ambler class A carbapenemase producers [KPC (n = 27), IMI (n = 3), GES (n = 2), SME (n = 2), NMC-A (n = 1) and FRI-1 (n = 1)], 61 metallo-β-lactamase producers (Ambler class B) [NDM (n = 30), VIM (n = 18), IMP (n = 12) and GIM (n = 1)], 42 OXA-48-like producers (Ambler class D) [OXA-48 (n = 22), OXA-162 (n = 1), OXA-181 (n = 10), OXA-204 (n = 5), OXA-232 (n = 2) and OXA-244 (n = 2)] and 1 OXA-372 carbapenemase not related to OXA-48. Seven of the isolates from the collection panel harboured both blaNDM and blaOXA-181 genes. All isolates in the collection panel had been previously verified for the presence of carbapenemase using the updated version of the Carba NP test.19 PCR/sequencing results for β-lactamases genes were used as reference gold standard as previously described.20
Prospective analysis
From 15 July to 15 September 2016, all non-duplicate clinical isolates with decreased susceptibility to at least one carbapenem referred to the Belgian National Reference Centre were included (183 isolates). All isolates were verified for the presence of carbapenemase using the updated version of the BYG Carba test (BYG Carba v2.0).21 Carbapenemase resistance genes were sought in all tested strains by two in-house ISO15189-certified multiplex PCRs targeting blaOXA-48-like, blaNDM, blaKPC, blaVIM and blaIMP,22 and the amplicons were sequenced using external Sanger sequencing services (Macrogen, Seoul, Korea) for allele identification.
Susceptibility testing
MICs of carbapenems (imipenem, meropenem and ertapenem) were determined using the Etest (bioMérieux, La Balme les Grottes, France) and the results were interpreted according to the EUCAST guidelines (version 6.0, January 2016; http://www.eucast.org).
OKN K-SeT assay
The OKN K-SeT assay is a multiplex lateral flow assay developed by Coris BioConcept (Gembloux, Belgium) for the detection of OXA-48-like, KPC and NDM carbapenemases in a single test. For this study, the strains to be tested were grown on trypticase soy agar supplemented with 5% sheep blood (bioMérieux, Marcy-l’Étoile, France) for 16–24 h at 37°C. The tests were performed according to the manufacturer’s recommendations.14
Statistical analysis
The sensitivity and specificity of the assay were calculated on all tested strains (collection strains and prospective consecutively obtained isolates), whereas the positive and negative predictive values were calculated only on the consecutive isolates that had been referred prospectively to the Belgian National Reference Centre.
Results
Performance of the OKN K-SeT assay on collection strains
The OKN K-SeT assay was able to detect correctly all OXA-48-like-producing isolates with carbapenemase activity (n = 42), as well as those producing KPC [n = 27; including KPC-2 (n = 22) and KPC-3 (n = 5)] and NDM [n = 30; including NDM-1 (n = 24), NDM-4 (n = 2), NDM-5 (n = 1), NDM-6 (n = 1), NDM-7 (n = 1) and NDM-9 (n = 1)] (Table S1, available as Supplementary data at JAC Online). The OXA-48-like carbapenemases included the following variants: OXA-48 (n = 22), OXA-162 (n = 1), OXA-181 (n = 10), OXA-204 (n = 5), OXA-232 (n = 2) and OXA-244 (n = 2). Although most of the OXA-48-like isolates presented only slightly elevated MICs of carbapenem, they were perfectly detected by the test (Table S1). On the other hand, all CPE strains that produced an enzyme not belonging to any of these three major carbapenemase families yielded negative results with the test. These isolates included 9 Ambler class A carbapenemase producers [IMI (n = 3), GES (n = 2), SME (n = 2), NMC-A (n = 1) and FRI-1 (n = 1)], 31 metallo-β-lactamase producers of Ambler class B [VIM (n = 18), IMP (n = 12) and GIM (n = 1)] and 1 OXA-372, a novel class D carbapenem-hydrolysing β-lactamase unrelated to OXA-48.23 Likewise, all 60 non-carbapenemase-producing isolates gave negative results with the test.
No cross-reaction was observed with strains that produced a narrow-spectrum oxacillinase, such as OXA-1 (n = 15), OXA-2 (n = 1), OXA-9 (n = 14) and OXA-10 (n = 3), or with strains that produced an extended-spectrum oxacillinase of OXA-48 type devoid of any significant carbapenemase activity, such as OXA-163 (n = 2) and OXA-405 (n = 1). These data are in line with results from previous evaluations by the OXA-48 K-SeT assay and they confirm the excellent specificity of this new triplex assay, which recognizes only carbapenem-hydrolysing OXA-48-like variants.
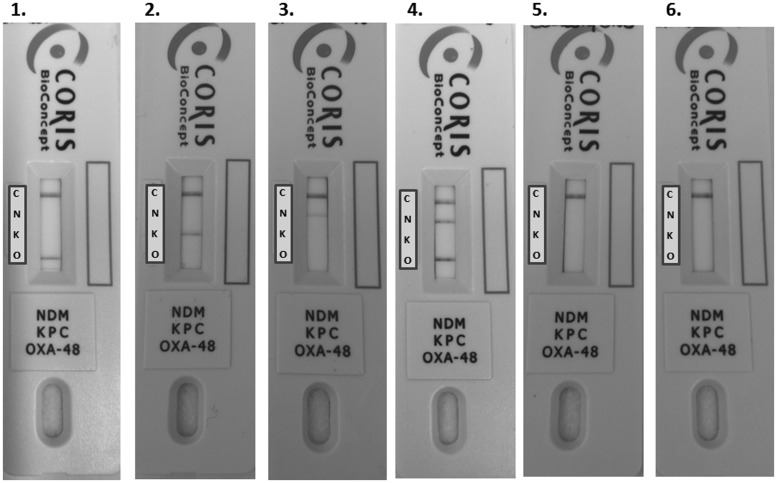
Remarkably, the OKN K-SeT assay perfectly detected the targeted carbapenemases even in rare CPE isolates that lacked or presented a very weak carbapenemase activity, as revealed with the biochemical tests. Indeed, the Carba NP test yielded equivocal results for three isolates [OXA-181 (n = 1), OXA-244 (n = 1) and GES-5 (n = 1)] and it failed to detect three other strains [OXA-181 (n = 1), OXA-244 (n = 1) and IMI-2 (n = 1)]. Besides these 6 strains, the Carba NP test was able to detect the other 134 carbapenemase producers regardless of the carbapenemase type. None of the 60 control strains gave false-positive results. Examples of positive and negative results for the detection of OXA-48, KPC and NDM by the OKN assay are shown in Figure 1.
Figure 1.
Multiplex lateral flow immunochromatographic assay for the detection of NDM (N), KPC (K) and OXA-48-like (O) carbapenemases. For a negative result a single line appears at the position of the control line (C). Examples of positive (1, 2, 3 and 4) and negative (5 and 6) results are shown. 1, Klebsiella pneumoniae OXA-48 positive; 2, K. pneumoniae KPC-3; 3, Escherichia coli NDM-1; 4, E. coli NDM-1 + OXA-181; 5, K. pneumoniae OXA-163; 6, K. pneumoniae ESBL (CTX-M-15) and impermeability.
Prospective evaluation of the OKN K-SeT assay
During the study period, 183 consecutive enterobacterial clinical isolates with decreased susceptibility to at least one carbapenem were referred by 57 clinical laboratories to the Belgian National Reference Centre for CPE. Using the BYG Carba test, a carbapenemase activity was detected in 57% (105/183) of the isolates (Table 1). During this prospective study, the BYG Carba failed to detect one OXA-48-positive Enterobacter cloacae and one VIM-1-producing Citrobacter freundii, both of which were detected by PCR. For the 76 non-carbapenemase-producing isolates (negative both by BYG Carba and by PCR testing), the decreased susceptibility to carbapenems resulted from ESBLs and/or cephalosporinase production associated with reduced outer membrane permeability (Table 1).
Table 1.
Results of the OKN K-SeT assay and of the BYG Carba test for enterobacterial isolates prospectively referred to the Belgian National Reference Centre
| β-Lactamase content | Species | No. of isolates | RESIST-3 OKN K-SeT assaya | BYG Carba testa |
|---|---|---|---|---|
| Carbapenemase producers | ||||
| OXA-48 | Klebsiella pneumoniae | 42 | OXA-48 | P |
| E. cloacae | 12 | OXA-48 | P | |
| E. cloacae | 1 | OXA-48 | N | |
| Escherichia coli | 7 | OXA-48 | P | |
| Klebsiella oxytoca | 3 | OXA-48 | P | |
| C. freundii | 2 | OXA-48 | P | |
| Kluyvera georgiana | 1 | OXA-48 | P | |
| OXA-48 + NDM-1 | E. coli | 1 | OXA-48+NDM | P |
| NDM-1 | K. pneumoniae | 11 | NDM | P |
| E. cloacae | 3 | NDM | P | |
| E. coli | 2 | NDM | P | |
| Enterobacter aerogenes | 1 | NDM | P | |
| NDM-5 | E. coli | 1 | NDM | P |
| KPC-3 | K. pneumoniae | 7 | KPC | P |
| E. coli | 1 | KPC | P | |
| KPC-3 + VIM-1 | C. freundii | 1 | KPC | P |
| VIM-1 | E. cloacae | 4 | N | P |
| E. coli | 1 | N | P | |
| C. freundii | 1 | N | N | |
| Serratia marcescens | 1 | N | P | |
| VIM-4 | K. oxytoca | 1 | N | P |
| OXA-23 | P. mirabilis | 2 | N | P |
| OXA-58 | P. mirabilis | 1 | N | P |
| Non-carbapenemase producers | ||||
| ESBL + impermeability | K. pneumoniae | 19 | N | N |
| E. cloacae | 11 | N | N | |
| E. coli | 8 | N | N | |
| C. freundii | 1 | N | N | |
| K. oxytoca | 1 | N | N | |
| cephalosporinase + impermeability | E. cloacae | 13 | N | N |
| E. aerogenes | 3 | N | N | |
| C. freundii | 1 | N | N | |
| Hafnia alvei | 1 | N | N | |
| S. marcescens | 1 | N | N | |
| DHA-1 + impermeability | K. pneumoniae | 1 | N | N |
| cephalosporinase + ESBL + impermeability | E. cloacae | 4 | N | N |
| E. aerogenes | 1 | N | N | |
| K. pneumoniae | 1 | N | N | |
| E. coli | 1 | N | N | |
| C. freundii | 1 | N | N | |
| other | E. coli | 3 | N | N |
| K. pneumoniae | 1 | N | N | |
| K. oxytoca | 3 | N | N | |
| E. cloacae | 1 | N | N |
For the RESIST-3 OKN K-SeT assay, the detected carbapenemases are indicated by their family name (OXA-48, NDM and KPC).
P, positive result; N, negative result.
The OKN K-SeT assay was able to detect all the isolates that produced one of the three targeted carbapenemase families, including 69 OXA-48-like [OXA-48 (n = 68) and OXA-181 (n = 1)], 9 KPC (all KPC-3) and 19 NDM [NDM-1 (n = 18) and NDM-5 (n = 1)]. All isolates that produced a carbapenemase different from those targeted by the assay gave a negative result {namely, 11 CPE isolates [VIM-1 (n = 8), VIM-4 (n = 1), OXA-23 (n = 1) and OXA-58 (n = 1)]}, as did the 76 non-CPE (Table 1).
Interestingly, two OXA-23-producing Proteus mirabilis isolates and one OXA-58-producing P. mirabilis isolate yielded a positive BYG Carba test but went undetected by our routine set of PCRs targeting the five major carbapenemase families in Enterobacteriaceae. These enzymes were detected through another multiplex PCR assay targeting other class D oxacillinases (data not shown) and by WGS.24
Global performance of the OKN K-SeT assay
Based on the overall results (collection strains and prospective study), the sensitivity and specificity of the OKN K-SeT assay reached 100% (95% CI 96.7%–100%) and 100% (95% CI 98.6%–100%), respectively, for the detection of OXA-48-like, 100% (95% CI 90.4%–100%) and 100% (95% CI 98.9%–100%) for KPC, and 100% (95% CI 92.7%–100%) and 100% (95% CI 98.9%–100%) for NDM. Interestingly, it correctly detected eight isolates that contained a combination of NDM-1 and OXA-181 (seven in the collection panel and one in the prospective study). For the prospective study, the OKN K-SeT assay yielded a positive and negative predictive value of 100% for the detection of the three targeted carbapenemases.
Discussion
The OKN K-SeT assay is an immunochromatographic assay designed to detect OXA-48 and its variants that possess carbapenem-hydrolysing activities, and the NDM and KPC carbapenemase families. Globally, the performance of the OKN K-SeT assay was 100% for sensitivity and 100% for specificity without any false positives, false negatives or ambiguous results. In the prospective study, the assay allowed direct rapid detection of 97 of the 109 carbapenemases (89%) that were found in 107 CPE isolates. Thus, the use of additional confirmatory tests would have been required only for the detection of 12 CPE isolates (i.e. 9 VIM producers, including 1 VIM in combination with a KPC enzyme, and 2 OXA-23-producing P. mirabilis isolates and 1 OXA-58-producing P. mirabilis isolate).
The OKN K-SeT assay can be easily performed by untrained personnel in a routine laboratory workflow and gives easy-to-read results after only ≤15 min of incubation, providing a very quick answer to whether the strain produces an OXA-48-, KPC- or NDM-like carbapenemase, or possibly a combination of more than one of these enzymes. In countries like Belgium or France, where these three enzyme families represent 90%–95% of all carbapenemases in Enterobacteriaceae (L. Dortet and Y. Glupczynski, unpublished results) the OKN assay could be suitable for general testing for carbapenemase production in the following settings: (i) on all enterobacterial isolates with decreased susceptibility to carbapenems (inhibition diameters <28 and <25 mm for meropenem and ertapenem, respectively), according to EUCAST guidelines for detection of resistance mechanisms of clinical and/or epidemiological importance and presenting an inhibition zone diameter of temocillin <12 mm; (ii) on colonies growing on selective CPE screening media; or (iii) directly on clinical samples such as urine or blood, as previously proposed by Wareham et al.15 and Pasteran et al.18
Since the OKN assay does not detect certain carbapenemases (mostly the VIM family), the use of other diagnostic methods will still be required in cases of positive screening results based on the detection of carbapenem hydrolytic activity and a negative result by the immunochromatographic test. An update of the current assay with the addition of VIM-type monoclonal antibodies is currently under development and should in the near future fill the gap, thus making it possible to detect in a single rapid test the four major families of carbapenemases that are usually encountered in Enterobacteriaceae in Europe.
Meanwhile, the availability of the OKN K-SeT assay provides a powerful tool to detect carbapenemase producers cheaply (recommended price of €10 per test) in a local hospital setting and it represents a cost-effective alternative to more costly and less widely available characterization by molecular amplification methods.
The speed and ease of use of this multiplex assay represents a significant technical advance and makes it particularly attractive as it would decrease the need for molecular methods to confirm the results. Additional potential benefits would possibly also include better patient management, e.g. more rapid switching to ceftazidime/avibactam treatment in cases of infection caused by a KPC- or OXA-48-like CPE, and a reduction in the escalation of antibiotic resistance through better infection control.
Supplementary Material
Acknowledgements
We thank our Belgian microbiologist colleagues for referring their clinical isolates to the National Reference Centre. We would also like to thank Coris BioConcept for providing the OKN K-SeT assays.
Funding
The Belgian National Reference Centre is partially supported by the Belgian Ministry of Social Affairs through a fund within the health insurance system.
This study was supported in part by a research grant from the Region Wallonne under the CWALity convention n°1318265, project FEAR (Fighting Enterobacteriaceae Antibiotic Resistance), a grant from the French Ministère de l’Education Nationale et de la Recherche (EA7361), Université Paris Sud and in part by a grant from Joint Programme Initiative on Antimicrobial Resistance (ANR-14-JAMR-0002). A. J., R. B., L. D. and T. N. are members of the Laboratory of Excellence LERMIT supported by a grant from ANR (ANR-10-LABX-33).
Transparency declarations
L. D. is co-inventor of the Carba NP test, the patent of which has been licensed to bioMérieux (La Balmes les Grottes, France). All other authors: none to declare.
Supplementary data
Table S1 is available as Supplementary data at JAC Online (http://jac.oxfordjournals.org/).
References
- 1. Nordmann P, Poirel L.. The difficult-to-control spread of carbapenemase producers in Enterobacteriaceae worldwide. Clin Microbiol Infect 2014; 20: 821–30. [DOI] [PubMed] [Google Scholar]
- 2. Patel G, Bonomo RA.. “Stormy waters ahead”: global emergence of carbapenemases. Front Microbiol 2013; 4: 48.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aguirre-Quiñonero A, Martínez-Martínez L.. Non-molecular detection of carbapenemases in Enterobacteriaceae clinical isolates. J Infect Chemother 2017; 23: 1–11. [DOI] [PubMed] [Google Scholar]
- 4. Dortet L, Poirel L, Nordmann P.. Rapid identification of carbapenemase types in Enterobacteriaceae and Pseudomonas spp. by using a biochemical test. Antimicrob Agents Chemother 2012; 56: 6437–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dortet L, Agathine A, Naas T. et al. Evaluation of the RAPIDEC® CARBA NP, the Rapid CARB Screen® and the Carba NP test for biochemical detection of carbapenemase-producing Enterobacteriaceae. J Antimicrob Chemother 2015; 70: 3014–22. [DOI] [PubMed] [Google Scholar]
- 6. Lasserre C, De Saint Martin L, Cuzon G. et al. Efficient detection of carbapenemase activity in Enterobacteriaceae by matrix-assisted laser desorption ionization-time of flight mass spectrometry in less than 30 minutes. J Clin Microbiol 2015; 53: 2163–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bogaerts P, Yunus S, Massart M. et al. Evaluation of the BYG Carba test, a new electrochemical assay for rapid laboratory detection of carbapenemase-producing Enterobacteriaceae. J Clin Microbiol 2016; 54: 349–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tijet N, Patel SN, Melano RG.. Detection of carbapenemase activity in Enterobacteriaceae: comparison of the carbapenem inactivation method versus the Carba NP test. J Antimicrob Chemother 2016; 71: 274–6. [DOI] [PubMed] [Google Scholar]
- 9. Poirel L, Potron A, Nordmann P.. OXA-48-like carbapenemases: the phantom menace. J Antimicrob Chemother 2012; 67: 1597–606. [DOI] [PubMed] [Google Scholar]
- 10. Woodford N, Eastaway A, Ford M. et al. Comparison of BD Phoenix, Vitek 2, and MicroScan automated systems for detection and inference of mechanisms responsible for carbapenem resistance in Enterobacteriaceae. J Clin Microbiol 2010; 48: 2999–3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pasteran F, Veliz O, Ceriana P. et al. Evaluation of the Blue-Carba test for rapid detection of carbapenemases in gram-negative bacilli. J Clin Microbiol 2015; 53: 1996–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kabir MH, Meunier D, Hopkins KL. et al. A two-centre evaluation of RAPIDEC® CARBA NP for carbapenemase detection in Enterobacteriaceae, Pseudomonas aeruginosa and Acinetobacter spp. J Antimicrob Chemother 2016; 71: 1213–6. [DOI] [PubMed] [Google Scholar]
- 13. Ote I, Bogaerts P, Denorme L. et al. Development of a novel immunochromatographic confirmatory test for the detection of OXA-48 carbapenemase in Enterobacteriaceae In: Abstracts of the Twenty-fifth European Congress of Clinical Microbiology and Infectious Diseases, Copenhagen, Denmark. Abstract O047. European Society of Clinical Microbiology and Infectious Diseases, Basel, Switzerland. [Google Scholar]
- 14. Glupczynski Y, Evrard S, Ote I. et al. Evaluation of two new commercial immunochromatographic assays for the rapid detection of OXA-48 and KPC carbapenemases from cultured bacteria. J Antimicrob Chemother 2016; 71: 1217–22. [DOI] [PubMed] [Google Scholar]
- 15. Wareham DH, Shah R, Betts JW. et al. Evaluation of an immunochromatographic lateral flow assay (OXA-48 K-SeT) for rapid detection of OXA-48 like carbapenemases in Enterobacteriaceae. J Clin Microbiol 2016; 54: 471–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dortet L, Jousset A, Sainte-Rose V. et al. Prospective evaluation of the OXA-48 K-SeT assay, an immunochromatographic test for the rapid detection of OXA-48-type carbapenemases. J Antimicrob Chemother 2016; 71: 1834–40. [DOI] [PubMed] [Google Scholar]
- 17. Meunier D, Vickers A, Pike R. et al. Evaluation of the K-SeT R.E.S.I.S.T. immunochromatographic assay for the rapid detection of KPC and OXA-48-like carbapenemases. J Antimicrob Chemother 2016; 71: 2357–9. [DOI] [PubMed] [Google Scholar]
- 18. Pasteran F, Denorme L, Ote I. et al. Rapid identification of OXA-48 and OXA-163 subfamilies in carbapenem-resistant Gram-negative bacilli with a novel immunochromatographic lateral flow assay. J Clin Microbiol 2016; 54: 2832–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dortet L, Brechard L, Poirel L. et al. Impact of the isolation medium for detection of carbapenemase-producing Enterobacteriaceae using an updated version of the Carba NP test. J Med Microbiol 2014; 63: 772–6. [DOI] [PubMed] [Google Scholar]
- 20. Dortet L, Cuzon G, Plésiat P. et al. Prospective evaluation of an algorithm for the phenotypic screening of carbapenemase-producing Enterobacteriaceae. J Antimicrob Chemother 2016; 71: 135–40. [DOI] [PubMed] [Google Scholar]
- 21. Noël A, Huang TD, Berhin C. et al. Comparative evaluation of four phenotypic tests for the detection of carbapenemase-producing Gram-negative bacteria. J Clin Microbiol 2017; 55: 510–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bogaerts P, Rezende de Castro R, de Mendonça R. et al. Validation of carbapenemase and extended-spectrum β-lactamase multiplex endpoint PCR assays according to ISO 15189. J Antimicrob Chemother 2013; 68: 1576–82. [DOI] [PubMed] [Google Scholar]
- 23. Antonelli A, D’Andrea MM, Vaggelli G. et al. OXA-372, a novel carbapenem-hydrolysing class D β-lactamase from a Citrobacter freundii isolated from a hospital wastewater plant. J Antimicrob Chemother 2016; 70: 2749–56. [DOI] [PubMed] [Google Scholar]
- 24. Girlich D, Bonnin RA, Bogaerts P. et al. Chromosomal amplification of the blaOXA-58 carbapenemase gene in a Proteus mirabilis clinical isolate. Antimicrob Agents Chemother 2017; 61: e016917–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.