Abstract
Objectives
To establish the role of specific, non-synonymous SNPs in the RNA polymerase β subunit (rpoB) gene in reducing the susceptibility of Clostridium difficile to fidaxomicin and to explore the potential in vivo significance of rpoB mutant strains.
Methods
Allelic exchange was used to introduce three different SNPs into the rpoB gene of an erythromycin-resistant derivative (CRG20291) of C. difficile R20291. The genome sequences of the created mutants were determined and each mutant analysed with respect to growth and sporulation rates, toxin A/B production and cytotoxicity against Vero cells, and in competition assays. Their comparative virulence and colonization ability was also assessed in a hamster infection model.
Results
The MIC of fidaxomicin displayed by three mutants CRG20291-TA, CRG20291-TG and CRG20291-GT was substantially increased (>32, 8 and 2 mg/L, respectively) relative to that of the parent strain (0.25 mg/L). Genome sequencing established that the intended mutagenic substitutions in rpoB were the only changes present. Relative to CRG20291, all mutants had attenuated growth, were outcompeted by the parental strain, had lower sporulation and toxin A/B production capacities, and displayed diminished cytotoxicity. In a hamster model, virulence of all three mutants was significantly reduced compared with the progenitor strain, whereas the degree of caecum colonization was unaltered.
Conclusions
Our study demonstrates that particular SNPs in rpoB lead to reduced fidaxomicin susceptibility. These mutations were associated with a fitness cost in vitro and reduced virulence in vivo.
Introduction
Clostridium difficile is a spore-forming Gram-positive anaerobic bacillus that causes gastrointestinal infections in humans. C. difficile infection (CDI) is associated primarily with the use of antibiotics, which disrupts the resident gut microbiota and permits colonization.1 Over the past decade, so-called ‘hypervirulent’ strains of C. difficile have become increasingly represented among clinical isolates, spreading throughout North America and Europe, and have been associated with outbreaks of increased disease severity, higher relapse rates and an expanded repertoire of antibiotic resistance.2,3 Such strains may have contributed to a rising incidence of CDI—particularly in elderly patients—and an increase in recurrent CDI.4 The US CDC recently identified CDI as an ‘urgent threat’.3
Primary treatment of CDI often involves either metronidazole or vancomycin, which both cause further damage to the colonic microbiota;5 recurrent infection following treatment is seen in 20%–30% of patients.6C. difficile is intrinsically resistant to many broad-spectrum antibiotics, which facilitates its spread within the hospital environment,3 and C. difficile clinical isolates with reduced susceptibility to metronidazole have been identified.7 Furthermore, an increased number of strains show reduced susceptibility to rifamycin antibiotics, which have been used to treat relapsing CDI.8 Efforts to understand the nature of these susceptibility changes are warranted to control the growing threat of antibiotic resistance.
Fidaxomicin is a macrocyclic antibiotic that exhibits potent activity against C. difficile by inhibiting bacterial RNA synthesis.9 It interferes with the formation of the DNA–RNA polymerase complex before the initiation of transcription.10 Contrary to the bacteriostatic action of the macrolides and rifamycins, it is bactericidal and has been shown to be narrow-spectrum. Only moderate activity has been reported against Staphylococcus aureus and Enterococcus, and no activity reported against Bacteroides and other Gram-negative bacteria found in the normal human gut microflora.11 In clinical trials, fidaxomicin has been shown to be non-inferior to vancomycin as an initial clinical cure for CDI, with a dosing regimen of 200 mg twice daily for 10 days. It had minimal side effects, was well tolerated and in addition, as it is hardly adsorbed, led to very low concentrations in the plasma of patients, but high concentrations in their faeces.12,13 Importantly, fidaxomicin significantly reduces recurrences compared with vancomycin.14
C. difficile strains with point mutations with reduced susceptibility to fidaxomicin were previously isolated using either a single-step or multiple-passage approach (and one strain was isolated from a patient in a clinical trial).15 The single-step approach generated mutant strains with point mutations in the gene encoding the β subunit of RNA polymerase, rpoB, although any potential variations in other genes that may have influenced susceptibility were not investigated at the time. Different SNPs led to replacement by three different amino acids, which in turn led to variable fidaxomicin MIC elevations (Table 1).15 As fidaxomicin had a decreased ability to inhibit transcription by these RNA polymerase mutants, RNA polymerase target modification was suggested as the mechanism by which C. difficile could develop resistance to fidaxomicin,15 although this requires further verification. Polymorphisms in other genes may also be associated with reduced susceptibility to fidaxomicin, such as marR/CD22120.16 The MarR (multiple antibiotic resistance regulator) family of transcription factors is ubiquitous in bacteria and archaea. Many are implicated in the regulation of an operon encoding an efflux pump. Mutations within the operon can lead to multiple antibiotic resistances.17 The clinical impact of fidaxomicin MIC elevations is unknown.
Table 1.
Random mutants in rpoB conferring reduced susceptibility to fidaxomicin10
| Nucleotide change | Amino acid change | MIC (mg/L) of fidaxomicin |
|---|---|---|
| T3428A (TA) | Val1143Asp | >64 |
| T3428G (TG)a | Val1143Gly | 16 |
| G3427T (GT) | Val1143Phe | 2 |
Clinical isolate collected from a patient presenting with CDI recurrence 6 days after completion of fidaxomicin therapy.
This study aimed to use an allelic exchange approach to recreate, in situ within rpoB, the point mutations potentially responsible for elevated fidaxomicin MICs, but within a clean, isogenic background. The subsequent phenotypic characteristics, competitive fitness and virulence of the isogenic mutants in the in vivo hamster model of CDI were investigated to understand the potential clinical significance of polymorphisms responsible for reduced C. difficile susceptibility to fidaxomicin.
Materials and methods
Ethics
All animal procedures were carried out in accordance with the United Kingdom Home Office Inspectorate under the Animals (Scientific Procedures) Act 1986. All experiments were approved by the University of Nottingham Ethics committee and carried out under licence number 40/3950.18,19
Antibiotics, bacterial strains and growth conditions
C. difficile strain CRG2029119 is a deliberately engineered erythromycin-resistant derivative of the PCR-ribotype 027 R20291 strain isolated from a UK outbreak.2 It was grown anaerobically at 37°C in a Don Whitley workstation in brain heart infusion medium supplemented with 5 mg/mL yeast extract and 0.1% (w/v) l-cysteine (BHIS) or in tryptone yeast (TY) broth. C. difficile medium was supplemented with 250 mg/L d-cycloserine, 8 mg/L cefoxitin and/or 15 mg/L thiamphenicol, when appropriate. Escherichia coli TOP10 (Invitrogen) and E. coli CA434 were cultured aerobically at 37°C in LB broth, supplemented with 25 mg/L chloramphenicol,20 when appropriate.
Generation of C. difficile mutant strains by allelic exchange
The desired single nucleotide changes were introduced into the C. difficile genome using the previously described vector pMTL-YN4,21 but with a number of key differences. Plasmid pMTL-YN4 carries a functional copy of a Clostridium sporogenes-derived pyrE gene, which can be used as a counter-selection marker for double crossover mutants, provided the C. difficile recipient is a pyrE mutant. However, we made use of an R20291pyrE-derivative strain (CRG20291) that was both macrolide, lincosamide, streptogramin (MLS) resistant and prototrophic through the genomic integration of an erm(B) gene at the pyrE locus,19 concomitant with restoration of the pyrE gene to WT. The C. difficile strain CRG20291 was chosen because the use of R20291 in the Golden Syrian hamster is complicated by the susceptibility of R20291 to clindamycin (MIC, 16 mg/L).19 Clindamycin treatment disrupts the hamsters’ normal gut flora prior to infection with C. difficile. However, clindamycin can persist at inhibitory levels (4–6 μg/g) 11 days following administration,22 and can affect the time from infection to colonization and death of strains with different clindamycin MICs.23 CRG20291 has been shown to provide more reliable results in the in vivo hamster infection model.19
Directed mutagenesis
Directed mutagenesis was performed in the progenitor strain, C. difficile CRG20291, to generate three rpoB SNPs (Table 1).15 An allelic exchange cassette was synthesized (Biomatik) complementary to the rpoB region comprising 500 bp on either side of rpoBT3428A and containing the promoter region upstream of rpoB (Figure 1). The cassette also incorporated flanking restriction sites for SacI and Mlul. Following cleavage with the corresponding restriction enzymes, DNA fragments were cloned into the knock-out vector, pMTL-YN4,21 to give pMTL-YN4: rpoBT3428A. The construct was verified for the inclusion of the mutated region by restriction digest and sequencing. Site-directed mutagenesis (Q5® site-directed mutagenesis kit NEB) was used to create the other two exchange vectors (pMTL-YN4: rpoBT3428G and pMTL-YN4: rpoBG3427T). These were also verified by Sanger sequencing.
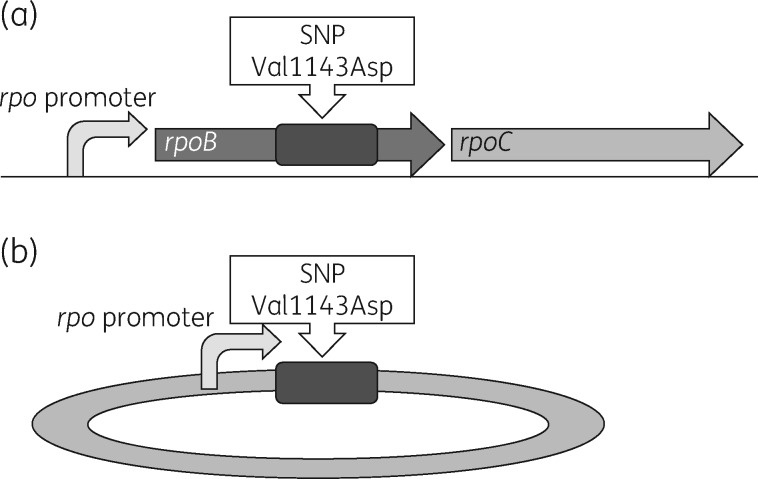
Figure 1.
Graphic representation of the directed mutagenesis approach used in the study. (a) A section of the C. difficile genome. (b) The allelic exchange plasmid. (a) Arrangement of rpoB and rpoC in the C. difficile chromosome and the promoter that drives gene expression of rpoB and rpoC. The dark grey rectangle represents the 1 kb exchange cassette containing the nucleotide that was replaced. (b) The rpoB promoter and the 1 kb homology arms containing the SNP. Using homologous recombination, valine is turned into aspartic acid when rpoB is translated.
The three verified plasmids were transformed into E. coli CA434 and conjugated into C. difficile CRG20291.19 After conjugation, transconjugants were selected on BHIS thiamphenicol and purified colonies carrying the modified rpoB gene conferring resistance were restreaked on BHIS agar supplemented with 0.5 mg/L fidaxomicin (equivalent to 2 × MIC). Purified colonies were checked for plasmid loss, and purified DNA used for PCR amplification of a region containing the changed nucleotide.19 Mutations were verified by Sanger sequencing.
WGS
Three clones of each mutant were verified by whole-genome sequencing, genomic DNA concentrations were measured using the Qubit dsDNA BR assay kit (Q32850, Thermo Fisher Scientific) and DNA integrity was assessed using conventional gel electrophoresis. Standard fragment sequencing libraries were prepared according to the Illumina TruSeq PCR free library preparation kit (#FC-121-3001, Illumina). Genomic DNA (2 μg) was fragmented to 400–600 bp using the Covaris S2 sonicator. Fragmented DNA was purified using Agencourt AMPure XP beads (#A63881, Beckman Coulter) at 1:1.6 DNA:bead ratio and end-repaired at 30°C for 30 min. End-repaired DNA was size-selected using AMPure XP beads at the desired fragment length. Illumina sequencing Y adapters were ligated at 30°C for 10 min and incubated at 70°C for 5 min. Unligated adapters were removed with a further two rounds of purification at 1:1 DNA:bead ratio. Purified adapter-ligated DNA was quantified by qPCR (#KK4824, Kapa Biosystems, Inc.), and libraries were pooled at equimolar concentrations. The library pool was loaded at a final concentration of 10 pM on the MiSeq sequencer for cluster generation and sequencing using 2 × 250 bp paired-end reads on a full MiSeq run.
The genome sequences were compared with CRG20291 using the quality-based variant detection workflow from CLC Genomics Workbench.
In vitro testing of C. difficile parental and mutant strains
MICs of fidaxomicin
The MICs of fidaxomicin for the parental strain and the three mutant strains were determined by agar dilution according to CLSI standards.24 All assays were performed in triplicate.
Phenotypic characterization
The three generated strains were characterized and compared with the parental strain for: growth; growth in competition; sporulation rate and capacity; toxin A/B production; and cytotoxicity. All in vitro experiments were performed in triplicate.
Growth curves Mutant and parental strains were grown overnight, the OD600 was standardized and a 1:10 dilution of each strain inoculated into fresh BHIS broth. Growth was analysed using a plate reader, with measurements taken every hour for 24 h.
Competition assays Mutant and parental strains were grown overnight, the OD600 standardized to the lowest reading and a 1:100 dilution of each strain inoculated together into BHIS broth. Samples were taken at 0, 2, 4, 6, 8, 10 and 24 h, diluted and plated onto BHIS-CC agar [BHIS supplemented with cefoxitin (8 mg/L) and cycloserine (250 mg/L)] for total viable count and onto BHIS-CC-FDX agar (BHIS-CC + 2 × MIC of fidaxomicin; 0.5 mg/L) for mutant count. cfu/mL were calculated from the colony counts after a 24 h incubation.
Measurement of sporulation rates We analysed spore production over a 120 h period. Sporulation rates were determined by measuring the development of spores that formed cfu after heat shock over time.2
C. difficile strains were grown from frozen stocks on BHIS-CC agar. Sporulation was achieved through incubation of cultures in BHIS broth in anaerobic conditions at 37°C for 5 days. Overnight cultures of C. difficile isolates were used to inoculate a starter culture in BHIS broth diluted 1:100, which was grown to an OD600 of between 0.2 and 0.5 to ensure low levels of spores on inoculation of the main sporulation culture. The main culture was then inoculated 1:100 with the starter culture (timepoint 0).
To measure colony formation, 500 μL samples of the sporulation medium were removed (at 0, 24, 48, 72, 96 and 120 h) from the anaerobic chamber and heated at 65°C for 30 min to kill vegetative cells, but not spores. Samples were returned to the anaerobic chamber, serially diluted in phosphate buffered saline and plated onto BHIS agar supplemented with the bile salt taurocholate (0.1%, Sigma) to induce germination and enhance recovery of C. difficile spores. Plates were incubated for 24 h before heat-resistant cfu were enumerated. For all measurements of heat-resistant cfu, a spo0A mutant strain, in which the master regulator of sporulation has been inactivated, was included as a sporulation-negative control.25
Measurement of toxin A/B production via ELISA Strains were grown in TY broth, overnight cultures were subcultured 1:100 into fresh media and supernatants of mutants and parental strains were collected at 24, 48 and 72 h. The production of toxin A/B was quantified by a standard curve using a toxin A/B ELISA assay (Techlab TOX A/B ELISA™). The standard curve was produced using C. difficile toxin A and B standards (The Native Antigen Company), serially diluted 1:2 from 125 to 1.95 ng/mL.
Measurement of cytotoxicity C. difficile strains were cultured in TY broth. Overnight cultures were subcultured 1:100 into fresh media and sampled at 0, 24, 48 and 72 h. At each timepoint, OD600 was measured, samples centrifuged and the supernatant filter sterilized and stored at −20°C. The supernatants were serially diluted, added to confluent monolayers of Vero (African green monkey kidney) cells and incubated for 24 h before scoring under a microscope. The toxin endpoint titre was the dilution at which the Vero cells resembled a negative control (no cell rounding).
In vivo testing of mutant strains
Virulence testing using a hamster infection model
In vitro findings were correlated with actual virulence and colonization assays in vivo. Infectivity was assessed in infection assays using female Golden Syrian hamsters, as described previously.19 Groups of eight hamsters were pretreated with clindamycin for 7 days prior to being challenged with 105 C. difficile spores. The primary output was survival or prolonged survival. Hamsters were monitored for disease symptoms and general condition, and euthanized when a predetermined threshold of symptom severity and/or worsening of condition was met, according to a previously published scoring system.26
Enumeration of caecal colonization
As a secondary assessment, the adherence properties of the mutant and parental strains were compared by enumerating the bacteria obtained from the caeca of infected hamsters. This was deemed to represent colonization. In brief, homogenized samples were centrifuged and the resulting supernatant heat-treated at 65°C prior to serial dilution and plating on cycloserine–cefoxitin fructose agar. The number of colonies was counted after 2–3 days of incubation.
Statistical methods
All statistical analysis was carried out using GraphPad Prism. One-way or two-way ANOVA, followed by Dunnett’s multiple comparison test or Sidak’s multiple comparison test, were used. All P values reported are multiplicity adjusted.
Results
Generation of C. difficile mutant strains by allelic exchange
The desired single nucleotide changes were introduced into the C. difficile genome to create the CRG20291 derivative strains. In all three instances, fidaxomicin-resistant colonies were obtained and the presence of the anticipated SNP confirmed in each strain. In CRG20291-TA, the thymine at rpoB position 3428 was changed to adenine, substituting valine with aspartic acid (Val1143Asp). In CRG20291-TG, the thymine at rpoB position 3428 was replaced with guanine, substituting valine with glycine (Val1143Gly). In CRG20291-GT, the guanine at position 3427 was replaced with thymine, substituting valine with phenylalanine (Val1143Phe). Sequencing confirmed that mutants did not contain additional spontaneous genomic changes compared with the parental strain.
In vitro testing of mutant strains
MICs
All constructed mutants had reduced susceptibility to fidaxomicin compared with the parental strain. CRG20291-TA had the lowest susceptibility (MIC >32 mg/L), followed by CRG20291-TG (8 mg/L) and lastly CRG20291-GT (2 mg/L), versus the parental strain (0.25 mg/L).
Strain phenotypic characterization
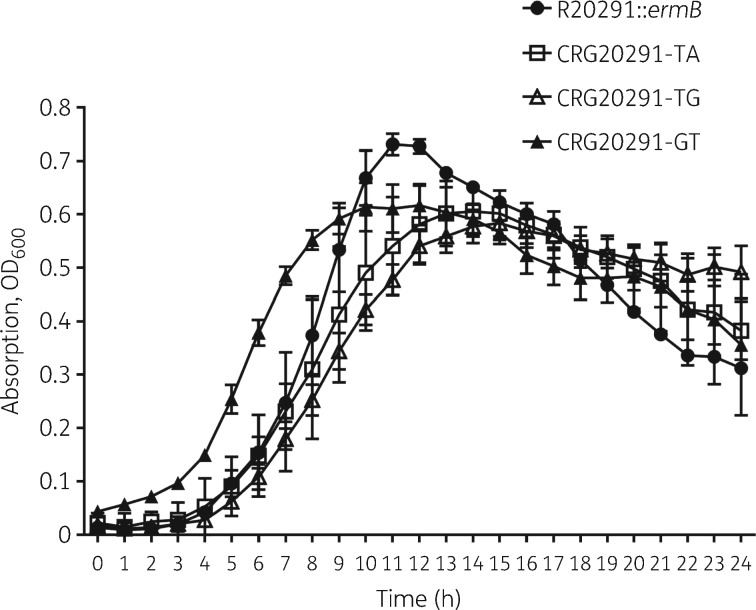
GrowthAll mutants grew differently compared with the parental strain (Figure 2), with all three reaching lower peak OD600 values than their progenitor. CRG20291-GT exhibited a shorter lag phase versus the parental and the other two mutant strains. The time to peak OD600 was shorter (∼11 h) for the parental strain compared with ∼14 h for both CRG20291-TA and CRG20291-TG. The generation time was similar between CRG20291-GT and the parental strain, but marginally impaired in the CRG20291-TA and CRG20291-TG mutants.
Figure 2.
Growth curves over 24 h for mutants CRG20291-TA (open squares), CRG20291-TG (open triangles) and CRG20291-GT (filled triangles) compared with the parental strain of C. difficile (filled circles). Values are plotted as mean ± SD.
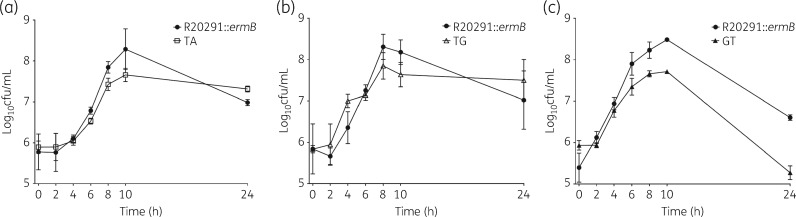
Growth in competition All three mutants were outcompeted by the parental strain in vitro (Figure 3). This was statistically significant for CRG20291-TA at the 10 h timepoint (P = 0.03) and for CRG20291-GT from the 6 h timepoint onwards (P values from < 0.0001 to 0.0031). Mutant CRG20291-TA showed a similar generation time and time-to-peak cfu count (∼10 h) to the parental strain, but the parental strain showed an 8% higher total viable count (P = 0.03). Mutant CRG20291-TG exhibited similar growth, generation time and time-to-peak count (both 8 h) to the parental strain. When mutant CRG20291-GT was compared with the parental strain, the time-to-peak count was 10 h for both, but the generation time for the mutant lagged behind that of the parental strain (41 min, parental strain; 51 min, GT). Notably, the mutant declined more rapidly versus the parental strain and, again, peak viable count was 10% higher with the parental strain (P < 0.0001). The parental strain appeared to have a competitive growth advantage over all three mutants, most notably compared with CRG20291-GT.
Figure 3.
In vitro growth in competition assay for the three mutant strains (a) CRG20291-TA (open squares), (b) CRG20291-TG (open triangles) and (c) CRG20291-GT (filled triangles) compared with the parental strain of C. difficile (filled circles). Each mutant was grown in competition with the parental strain and fitness was evaluated over 24 h. Values are plotted as mean ± SD.
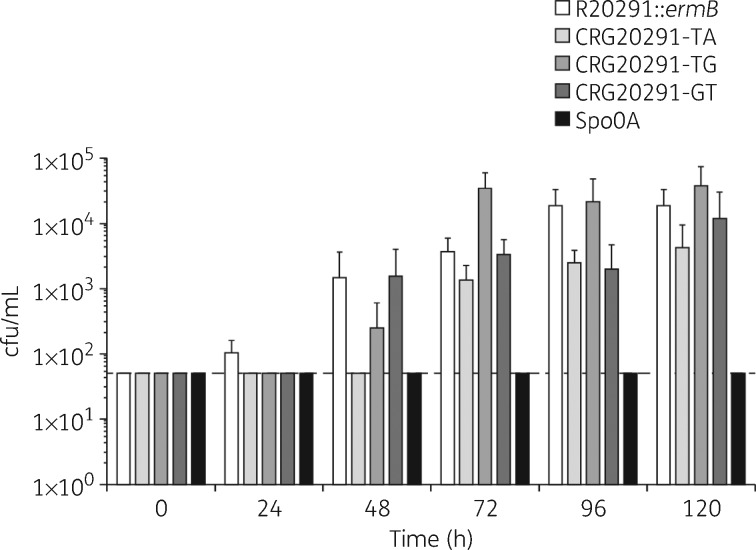
Sporulation rate and capacity The initiation of sporulation was delayed in all three mutants compared with the parental strain, where no spores could, in contrast to the WT, be detected after 24 h (Figure 4). Strain CRG20291-TA showed the longest delay, with heat-resistant cfu only becoming detectable after 72 h. In fact, the number of spores present at all timepoints tested remained significantly below that of the parental strain, even at 120 h. CRG20291-GT showed a reduced total sporulation capacity versus the parental strain at 24, 96 and 120 h, and equalled it at 48 and 72 h. Strain CRG20291-TG also formed no heat-resistant cfu at 24 h and fewer than the parental strain at 48 h, but produced more spores than the parental strain (albeit not statistically significant) from 72 h onwards. The majority of differences, apart from that between CRG20291-TG and the parental strain at 72 h (P = 0.0014), were not statistically significant.
Figure 4.
Rates of sporulation in the parental strain, mutant strains and sporulation-negative control. Values are plotted as mean ± SD. The broken line represents the detection limit of the assay (50 cfu/mL).
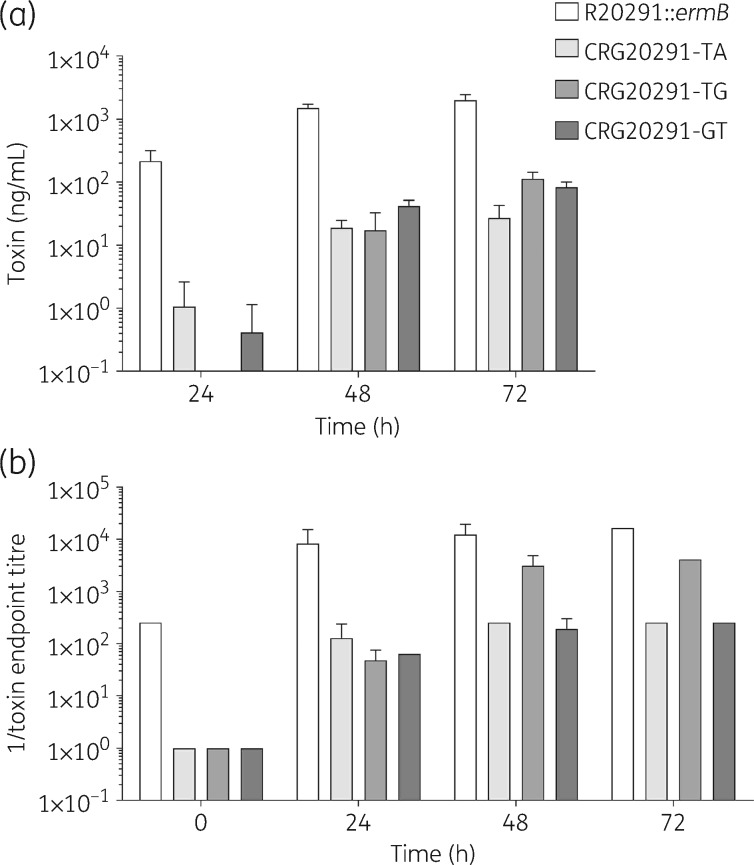
Toxin A/B production In all three mutants, toxin A/B production was attenuated compared with the parental strain at all timepoints measured (Figure 5a). For strain CRG20291-TG, the lag in toxin A/B production was particularly evident, with no toxin detected at 24 h. At 72 h, the difference in level of toxin A/B production between mutant and parental strains was 74-fold for CRG20291-TA, 18-fold for CRG20291-TG and 24-fold for CRG20291-GT (P ≤ 0.0001 for all measurements).
Figure 5.
Toxin A/B production and cytotoxicity. (a) Toxin production by ELISA assay in TY broth. (b) Cytotoxity in Vero cells. Values are plotted as mean ± SD.
CytotoxicityAll three mutants showed reduced cytotoxicity in Vero cells at all timepoints compared with the parental strain (Figure 5b). At 48 and 72 h post-incubation, the culture supernatant of strain CRG20291-TG demonstrated higher cytotoxicity than the other two mutant strains, although it was still 4-fold less cytotoxic than the parental strain’s culture supernatant.
In vivo virulence and caecum colonization in the hamster infection model
Virulence and colonization
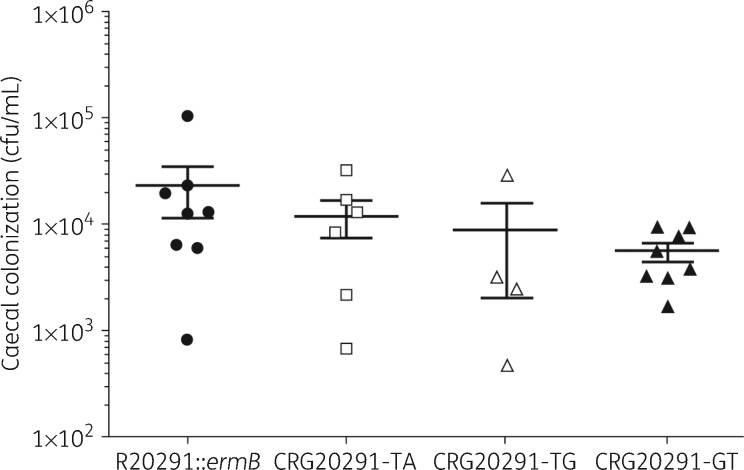
All three mutant strains showed a statistically significant reduction in virulence compared with the parental strain in the hamster infection model (P values from 0.0001 to 0.0087; Figure 6). CRG20291-TA showed the lowest level of virulence (with a mean of 53.5 h until clinical endpoint), followed by CRG20291-GT (mean 51.25 h) and then CRG20291-TG (mean 40 h). The parental strain was the most virulent, with a mean of 31.2 h until clinical endpoint. There was no significant difference in colonization levels of hamster caecum between the parental and the mutant strains (Figure 7). The mutant strains did colonize slightly less with averages of 1.20E + 04 (CRG20291-TA), 8.96E + 03 (CRG20291-TG) and 5.55E + 03 (CRG20291-GT) cfu/mL compared with 2.34E + 04 cfu/mL for the parental strain.
Figure 6.
Time from spore infection to clinical endpoint (sign of infection) using the hamster model of infection for mutants CRG20291-TA (open squares), CRG20291-TG (open triangles) and CRG20291-GT (filled triangles) compared with the parental strain of C. difficile (filled circles). Values are plotted as mean ± SD.
Figure 7.
Caecum colonization in the hamster model of infection. After the endpoint, caecal colonization was evaluated, by plating on cycloserine–cefoxitin fructose agar. The mutants CRG20291-TA (open squares), CRG20291-TG (open triangles) and CRG20291-GT (filled triangles) were compared with the parental strain of C. difficile (filled circles). Values are plotted as mean ± SD.
Discussion
In the present study, we confirmed a link between the genotypic changes in the RNA polymerase β subunit rpoB and reduced susceptibility of C. difficile to fidaxomicin using directed mutagenesis in C. difficile. The created mutant strain CRG20291-TA had an MIC of >32 mg/L, which is by far the highest resistance exhibited by the three strains.
A random fidaxomicin mutant with the equivalent TA amino-acid substitution that was previously isolated in the laboratory was reported to have an MIC of >64 mg/L.15 The single clinical isolate with reduced fidaxomicin susceptibility that has been reported has an MIC of 16 mg/L. The deliberately created mutant strain, CRG20291-TG, generated in the current study, has the same amino-acid changes as the clinical isolate and an MIC of 8 mg/L. Finally, strain CRG20291-GT, created here, has an MIC of 2 mg/L, which is the same as a previously isolated random GT mutant. These data show that the fidaxomicin susceptibilities of the mutant strains generated in the present study are broadly in line with the previously isolated random mutants of Seddon et al.15 An in-depth phenotypic characterization comparing parental strain and mutants revealed changes in virulence-associated traits, such as competitive growth, sporulation and toxin production. First, they showed altered growth over a 24 h period in vitro, and less competitive growth fitness versus the parental strain. Sporulation characteristics, an important feature of C. difficile that plays a significant role in the transmission and recurrence of CDI, particularly in a hospital setting, were also marginally affected in the mutant strains.2 Toxin production and cytotoxicity were also reduced in the mutants compared with the parental strain.
Furthermore, we found reduced virulence of the rpoB mutants compared with the parental strain in the hamster infection model. The virulence of strains CRG20291-TA and GT were most greatly reduced (53.5 and 51.25 h, respectively, to the clinical endpoint compared with 31.2 h for the parental strain). Interestingly, CRG20291-GT was also most impaired in the competition assay and, albeit not statistically significant, seemed to exhibit slightly reduced colonization. The in vivo assessment of virulence might suggest that C. difficile strains with reduced susceptibility to fidaxomicin may be less able to cause disease and would, as a consequence, be less likely to persist in a clinical setting. It would be interesting to investigate further whether these mutants produce less toxin in vivo, which could play a role in their ability to cause disease. Additionally, it would be illuminating to investigate the sporulation ability of these mutants, given the crucial role of spores in persistence and transmission. So far, only one resistant clinical isolate has been characterized,27 which may indicate that such strains are not particularly prevalent or widespread in the clinical setting. Nevertheless, further investigations and evaluation are required.
Despite the high faecal concentrations of fidaxomicin achieved during therapy for CDI (> 1 mg/g),9,28 the clinical relevance of spontaneous rpoB mutants is unknown. In the case of rifamycin resistance, studies have shown that no fitness cost is associated with the mutation. Equally, it has been shown that these mutations do persist in vivo, in the clinical setting.29 In the case of fidaxomicin during all of these trials, there was only one report of a strain with reduced susceptibility, which was isolated from a relapse patient during a clinical trial.27
Interestingly, rpoB gene mutations that are clinically relevant have been identified in other bacteria. For example, in Mycobacterium tuberculosis, such mutations are thought to be associated with varying fitness costs and some mutations have been associated with altered cell wall permeability. These M. tuberculosis mutations are a clinical concern and can even accumulate in the latent stage of the disease.30,31 In Neisseria meningitidis, different mutations within rpoB have been associated with a range of fitness costs, and it has been suggested that the impact on virulence is such that they are only very rarely seen in the clinical setting.32 Conversely, a study of rpoB mutations conferring reduced susceptibility of C. difficile to rifamycin29 found that the majority of mutations generated in that study lacked in vitro fitness costs. Among these was the Arg505Lys substitution, which was as virulent as the WT, providing a possible explanation for its dominance among rifamycin-resistant clinical isolates.
Our findings suggest that the specific rpoB mutations isolated and analysed in this study that were responsible for fidaxomicin resistance also impose a fitness cost. It might then be conceivable that fidaxomicin is less likely to cause a rapid spread of resistant isolates compared with other antibiotics that inhibit RNA polymerase, such as the rifamycins. This suggestion is supported by the paucity (just one) of fidaxomicin-resistant strains isolated from clinical settings to date.27
Our study had a number of limitations, the primary one being the small number of replicates in the samples. The use of OD (turbidity) as a measurement of viable bacterial cell count is also a limitation, as OD has been shown not to be truly congruent with viable count,33 although our competition experiments were based on viable counts. However, we do not perceive the limitations to be considerable, as we assessed relative differences between the strains, which would be disposed to the same limitations. Further work is required to explore the frequency of these rpoB mutations in clinical situations.
In conclusion, in the present study, rpoB mutations in C. difficile strains were recreated using directed mutagenesis and shown to confer reduced susceptibility to fidaxomicin. The mutants were additionally shown to exhibit reduced competitive fitness in vitro and to be less virulent in vivo compared with the parental strain. The changes in phenotype that result from these alterations in the primary sequence of rpoB may have consequences for the mutant cell lines carrying these amino-acid substitutions. The reduced fitness of such mutants might explain, at least in part, why so few cases of fidaxomicin resistance have been reported in the literature so far.
Acknowledgements
We would like to thank Tina Morley and Rhian Harper Owen from iS LifeScience for their editorial support, funded by Astellas Pharma, Inc., in the preparation of the article. We would also like to thank Sharla McTavish for assistance with strain characterization.
Funding
This work was supported by Astellas Pharma, Inc. This funding covered the initiation of the study, conduct of the research and editorial support for the manuscript.
Transparency declarations
S. A. K. and N. P. M. have received grants from Summit plc. C. M. L. was a full-time salaried employee of Astellas Pharma Europe Ltd at the time of this study. He also has an international patent application, WO2015EP00965, in relation to a treatment regimen for fidaxomicin. All other authors: none to declare.
This study was initiated by Astellas Pharma, Inc. Under the direction of the authors, Tina Morley from iS LifeScience drafted the initial version of the manuscript and Rhian Harper Owen provided editorial support throughout its development.
Author contributions
All authors met ICMJE criteria for authorship, were involved in revising the article critically for important intellectual content and approved the final version of the manuscript for submission. All others who contributed to this document, but who did not meet authorship criteria, are acknowledged.
References
- 1. Hutton ML, Mackin KE, Chakravorty A. et al. Small animal models for the study of Clostridium difficile disease pathogenesis. FEMS Microbiol Lett 2014; 352: 140–9. [DOI] [PubMed] [Google Scholar]
- 2. Burns DA, Heeg D, Cartman ST. et al. Reconsidering the sporulation characteristics of hypervirulent Clostridium difficile BI/NAP1/027. PLoS One 2011; 6: e24894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. US Department of Health and Human Services, CDC. Antibiotic Resistance Threats in the United States, 2013 http://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf.
- 4. Cocanour CS. Best strategies in recurrent or persistent Clostridium difficile infection. Surg Infect (Larchmt) 2011; 12: 235–9. [DOI] [PubMed] [Google Scholar]
- 5. Rafii F, Sutherland JB, Cerniglia CE.. Effects of treatment with antimicrobial agents on the human colonic microflora. Ther Clin Risk Manag 2008; 4: 1343–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cornely OA, Miller MA, Louie TJ. et al. Treatment of first recurrence of Clostridium difficile infection: fidaxomicin versus vancomycin. Clin Infect Dis 2012; 55 Suppl 2: S154–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lynch T, Chong P, Zhang J. et al. Characterization of a stable, metronidazole-resistant Clostridium difficile clinical isolate. PLoS One 2013; 8: e53757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Spigaglia P. Recent advances in the understanding of antibiotic resistance in Clostridium difficile infection. Ther Adv Infect Dis 2016; 3: 23–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhanel GG, Walkty AJ, Karlowsky JA.. Fidaxomicin: a novel agent for the treatment of Clostridium difficile infection. Can J Infect Dis Med Microbiol 2015; 26: 305–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Artsimovitch I, Seddon J, Sears P.. Fidaxomicin is an inhibitor of the initiation of bacterial RNA synthesis. Clin Infect Dis 2012; 55 Suppl 2: S127–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Louie TJ, Emery J, Krulicki W. et al. OPT-80 eliminates Clostridium difficile and is sparing of Bacteroides species during treatment of C. difficile infection. Antimicrob Agents Chemother 2009; 53: 261–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goldstein JC, Citron DM, Sears P. et al. Comparative susceptibilities to fidaxomicin (OPT-80) of isolates collected at baseline, recurrence, and failure from patients in two phase III trials of fidaxomicin against Clostridium difficile infection. Antimicrob Agents Chemother 2011; 55: 5194–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Citron DM, Babakhani F, Goldstein EJ. et al. Typing and susceptibility of bacterial isolates from the fidaxomicin (OPT-80) phase II study for Clostridium difficile infection. Anaerobe 2009; 15: 234–6. [DOI] [PubMed] [Google Scholar]
- 14. Drekonja DM, Butler M, MacDonald R. et al. Comparative effectiveness of Clostridium difficile treatments: a systematic review. Ann Intern Med 2011; 155: 839–47. [DOI] [PubMed] [Google Scholar]
- 15. Seddon J, Babakhani F, Gomez A. et al. RNA polymerase target modification in Clostridium difficile with reduced susceptibility to fidaxomicin. In: Abstracts of the Fifty-first Interscience Conference on Antimicrobial Agents and Chemotherapy, Chicago, IL, USA, 2011. Abstract C1-1274. American Society for Microbiology, Washington, DC, USA.
- 16. Leeds JA, Sachdeva M, Mullin S. et al. In vitro selection, via serial passage, of Clostridium difficile mutants with reduced susceptibility to fidaxomicin or vancomycin. J Antimicrob Chemother 2014; 69: 41–4. [DOI] [PubMed] [Google Scholar]
- 17. Grove A. MarR family transcription factors. Curr Biol 2013; 23: R142–3. [DOI] [PubMed] [Google Scholar]
- 18. Kuehne SA, Collery MM, Kelly ML. et al. Importance of toxin A, toxin B, and CDT in virulence of an epidemic Clostridium difficile strain. J Infect Dis 2014; 209: 83–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kelly ML, Ng YK, Cartman ST. et al. Improving the reproducibility of the NAP1/B1/027 epidemic strain R20291 in the hamster model of infection. Anaerobe 2016; 39: 51–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Williams DR, Young DI, Young M.. Conjugative plasmid transfer from Escherichia coli to Clostridium acetobutylicum. J Gen Microbiol 1990; 136: 819–26. [DOI] [PubMed] [Google Scholar]
- 21. Ng YK, Ehsaan M, Philip S. et al. Expanding the repertoire of gene tools for precise manipulation of the Clostridium difficile genome: allelic exchange using pyrE alleles. PLoS One 2013; 8: e56051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Larson HE, Borriello SP.. Quantitative study of antibiotic-induced susceptibility to Clostridium difficile enterocecitis in hamsters. Antimicrob Agents Chemother 1990; 34: 1348–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sambol SP, Tang JK, Merrigan MM. et al. Infection of hamsters with epidemiologically important strains of Clostridium difficile. J Infect Dis 2001; 183: 1760–6. [DOI] [PubMed] [Google Scholar]
- 24. Clinical and Laboratory Standards Institute. Methods for Antimicrobial Susceptibility Testing of Anaerobic Bacteria—Eighth Edition: Approved Standard M11-A8. CLSI, Wayne, PA, USA, 2012. [Google Scholar]
- 25. Heap JT, Kuehne SA, Ehsaan M. et al. The ClosTron: mutagenesis in Clostridium refined and streamlined. J Microbiol Methods 2010; 80: 49–55. [DOI] [PubMed] [Google Scholar]
- 26. Kuehne SA, Cartman ST, Heap JT. et al. The role of toxin A and toxin B in Clostridium difficile infection. Nature 2010; 467: 711–4. [DOI] [PubMed] [Google Scholar]
- 27. Goldstein EJ, Babakhani F, Citron DM.. Antimicrobial activities of fidaxomicin. Clin Infect Dis 2012; 55 Suppl 2: S143–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sears P, Crook DW, Louie TJ. et al. Fidaxomicin attains high fecal concentrations with minimal plasma concentrations following oral administration in patients with Clostridium difficile infection. Clin Infect Dis 2012; 55 Suppl 2: S116–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dang UT, Zamora I, Hevener KE. et al. Rifamycin resistance in Clostridium difficile is generally associated with a low fitness burden. Antimicrob Agents Chemother 2016; 60: 5604–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lahiri N, Shah RR, Layre E. et al. Rifampin resistance mutations are associated with broad chemical remodeling of Mycobacterium tuberculosis. J Biol Chem 2016; 291: 14248–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ford CB, Lin PL, Chase MR. et al. Use of whole genome sequencing to estimate the mutation rate of Mycobacterium tuberculosis during latent infection. Nat Genet 2011; 43: 482–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Colicchio R, Pagliuca C, Pastore G. et al. Fitness cost of rifampin resistance in Neisseria meningitidis: in vitro study of mechanisms associated with RpoB H553Y mutation. Antimicrob Agents Chemother 2015; 59: 7637–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Augustin JC, Rosso L, Carlier V.. Estimation of temperature dependent growth rate and lag time of Listeria monocytogenes by optical density measurements. J Microbiol Methods 1999; 38: 137–46. [DOI] [PubMed] [Google Scholar]