Abstract
We describe mutant tissue lines of Arabidopsis that are able to grow in vitro as callus on hormone-free medium. The 14 lines presented here show different hormone autotrophic differentiation behaviors that can be classified into three categories: (a) forming roots (rooty callus), (b) forming shoots or shoot-like structures (shooty callus), or (c) growing without organ formation (callus). Three fast-growing lines showed altered steady-state mRNA levels of the Cdc2 and CycD3 cell cycle genes. Three of the six rooty callus lines contained about 20- to 30-fold higher levels of auxins than wild-type callus. These and two other lines with normal auxin content showed an increased steady-state level of IAA1 and IAA2 transcripts in the absence of exogenous auxin. Five of the six shooty callus lines had increased steady-state mRNA levels of the CKI1 gene and/or of the homeobox genes KNAT1 and STM, suggesting that the phenotype is linked to altered cytokinin signaling. Also, one cytokinin-overproducing line with only 5% of wild-type cytokinin oxidase activity was identified. These results indicate that screening for hormone-autonomous growth identifies mutants with altered hormone content or signaling.
Plant cell division, growth, and differentiation need to be precisely controlled during development to ensure coordinated growth of tissues. Most mitotic activity is restricted to the meristems and the young growing tissues surrounding them. A loss of this control could lead to cell division and growth at ectopic positions and eventually to the development of unorganized tissue growth (i.e. plant tumors).
In many instances, de-differentiation and tumor formation in plant tissue is caused by an imbalance of the hormones auxin and cytokinin. Plant tumors are often able to grow in vitro as callus on medium without auxin or cytokinin. For example, pathogenesis of the crown gall disease after infection by Agrobacterium tumefaciens, a well-studied case of plant tumor formation, is caused by the transfer and expression of auxin- and cytokinin-synthesizing genes to the plant cell (for review, see Morris, 1995). The auxin-to-cytokinin ratio influences qualitatively and quantitatively growth and differentiation of tumors in planta or callus grown in vitro. A high auxin-to-cytokinin ratio leads to root formation in calli, while a low auxin-to-cytokinin ratio favors shoot formation (Skoog and Miller, 1957).
Not only altered hormonal content, but also changes in hormone sensitivity or signal transduction can lead to the formation of tumors with a distinct differentiation behavior. The transgenic overexpression of the Agrobacterium rhizogenes rolB gene enhances auxin sensitivity and induces the formation of ectopic roots (Cardarelli et al., 1987; Maurel et al., 1991; Schmülling et al., 1993). Overproduction of the CKI1-encoded His kinase homolog mimics enhanced cytokinin signaling and induces shoot formation (Kakimoto, 1996). In Nicotiana tabacum, plants carrying the Hl-1 allele form tumors after infection with the auxin-synthesizing genes of A. tumefaciens. In wild-type plants tumors form after treatment with auxin and cytokinin (Meyer et al., 1997). This indicates that Hl-1 enhances the sensitivity of certain tissues to cytokinins or activates growth-factor-independent pathways.
Other examples of genes that deregulate proper control of cell division and growth are the oncogenes 6b and lso of A. tumefaciens T-DNA. Infection with either gene leads to the formation of undifferentiated tumors on a limited number of host plants (Hooykaas et al., 1988; Otten and Schmidt, 1998). Similarly, overexpression of the KNAT2 and CycD3 genes causes an auxin- and/or cytokinin-independent tumor formation on Arabidopsis leaves (Dockx et al., 1996; Riou-Khamlichi et al., 1999). Arabidopsis tumors that show hormone-independent growth are also formed as a consequence of somatic mutations after γ-ray irradiation (Persinger and Town, 1991).
Plant tumors also arise spontaneously in certain combinations of genotypes and in high-inbred lines. These so-called genetic tumors have been especially well studied in the genus Nicotiana, e.g. in Nicotiana glauca × Nicotiana langsdorffii hybrids (Smith et al., 1976; Ichikawa and Syono, 1991) and in radish (Buzovkina et al., 1993; Lutova et al., 1997).
We have used a genetic approach to identify elements that normally regulate cell division and differentiation and prevent tumor formation. In light of the close links between tumor formation and auxin and cytokinin metabolism and signaling, we hypothesized that this approach would lead to the identification of potential new hormone mutants. Negative control elements of cell division and differentiation that are not directly related to these hormones may also be identified using this approach. We isolated 14 Arabidopsis mutant tissue lines that showed a de-differentiation of organized tissues after germination, and subsequently grew in vitro as callus without exogenous hormones. Based on their differentiation behavior, we distinguish three mutant classes: the shooty callus class (s1-s6), the rooty callus class (r1- r6), and the callus class (c1-c2). We describe changes in hormone concentration and altered steady-state transcript levels of genes such as the IAA, CKI1, and homeobox genes, which play central roles in growth and development.
MATERIALS AND METHODS
Mutant Screening and Growth Conditions
Ethylmethane sulfonate (EMS) mutagenized seeds of Arabidopsis Heynh. ecotype Columbia (Col-1 gl1) were purchased from Lehle Seeds (Round Rock, TX). Approximately 29,000 surface-sterilized M2 individuals from four parental families were plated in vitro on hormone-free Murashige and Skoog (MS) medium (Murashige and Skoog, 1962), containing 3% (w/v) Suc and solidified with 9 g/L agar (Agar Agar reinst, Merck, Darmstadt, Germany). Plates were incubated at 24°C under light (16 h)/dark (8 h) cycles. Seedlings were screened after 4 weeks for individuals that de-differentiated and continued to grow as callus-like tissue. Calli were further propagated on hormone-free MS medium and transferred weekly to fresh medium.
Growth Measurement
For growth measurements, tumor explants (approximately 100 mg fresh weight) were placed on individual Petri dishes on hormone-free medium. The growth rate was expressed as W = (Wt − W0)/W0. W0 and Wt are the initial sample weight and the weight after 7 d, respectively. Data are the means of three experiments with five samples per data point in each experiment. Wild-type callus was grown on medium containing 1 mg/L naphthylacetic acid and 0.1 mg/L N6-(Δ2isopentenyl)adenine riboside (iP).
Hormone Analysis
The cytokinin and indole-3-acetic acid (IAA) content of callus harvested 7 d after subcultivation was measured as described previously (Prinsen et al., 1995a, 1995b). Cytokinins and IAA were extracted overnight from approximately 1 g of frozen tissue in CHCl3/methanol/water/acetic acid (Bieleski, 1964) and purified combining solid-phase extraction and immunoaffinity chromatography using a broad spectrum anti-cytokinin antibody. The stable isotopes [2H5]trans-zeatin, [2H5]zeatin riboside, [2H5]zeatin 9-glucoside, [2H5]zeatin 7-glucoside, [2H5]zeatin O-glucoside, [2H5]zeatin riboside O-glucoside, [2H5]zeatin riboside 5-′-monophosphate, [2H6]iP, [2H6]iP riboside, [2H6]iP 9-glucoside (20 ng of each, Apex Organics, Honiton, Devon, UK) and [phenyl-13C6-]indole-3-acetic acid (50 ng, Cambridge Isotope Lab, Cambridge, MA) were initially added as internal tracers for recovery and analytical purposes. The different cytokinin fractions obtained after purification were analyzed by micro liquid chromatography with column switch configuration coupled to positive ion electrospray tandem mass spectrometry using multiple reactant monitoring (Prinsen et al., 1995b, 1998). After pentafluorobenzyl derivatization of IAA, PFB-IAA was analyzed by negative ion chemical ionization gas chromatography-selected ion monitoring-MS (Epstein and Cohen, 1981). Prior to purification, IAA conjugates were converted to free IAA by alkaline hydrolysis (Bialek and Cohen, 1989).
Cytokinin Oxidase Activity
Cytokinin oxidase was extracted from 3 to 8 g of callus tissue harvested 7 d after subcultivation. The cytokinin oxidase activity was determined using the method of Chatfield and Armstrong (1986) as modified by Motyka and Kamínek (1994) based on the measurement of the rate of conversion of [2,8-3H]iP to adenine. Separation of the substrate from the product of the enzyme reaction was achieved by thin-layer chromatography on microcrystalline cellulose plates developed with the upper phase of the 4:1:2 (v/v) mixture of ethylacetate/n-propanol/water. Zones containing iP and adenine were located under UV light, cut into strips, and their radioactivity was measured using the liquid scintillation technique. Cytokinin oxidase activity determinations were repeated three times for each tissue sample. The sd averaged 8% and did not exceed 17% of the means.
RNA Analysis
Total RNA was extracted from plant tissues according to Verwoerd et al. (1989). RNA (50 μg) was separated in a denaturing 1.5% (w/v) agarose-formaldehyde gel, transferred to nylon membranes (Hybond N, Amersham, Buckinghamshire, UK), and hybridized with radioactive-labeled DNA probes generated with a random primer labeling kit (Prime-It II, Stratagene, Heidelberg). Hybridizations were carried out in hybridization solution (QuickHyb, Stratagene) according to the manufacturer's instructions. The lowest stringency wash was performed in 0.2× SSC and 0.1% (w/v) SDS at 65°C. As a control for loading, the blot was rehybridized with a 25S rDNA probe.
RESULTS
Identification and Classification of Mutant Lines
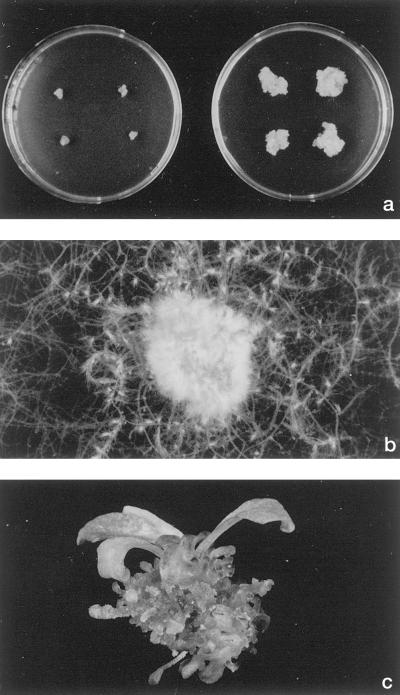
Among approximately 29,000 mutagenized M2 seedlings, we identified 14 lines that showed tissue de-differentiation after germination. In all cases tissue de-differentiation yielded callus or callus-like tissue that could, in contrast to wild-type callus, be propagated on hormone-free medium (Fig. 1a). Based on their main characteristics, we distinguished three mutant classes: (a) six lines that we called rooty callus (r1–r6) grew as a brownish, soft callus, formed roots, and were uniform in appearance (Fig. 1b); (b) six lines that we called shooty callus (s1-s6) were green, formed shoots or shoot-like structures (Fig. 1c), and differed in their capability to form shoots and in the size of the shoots; and (c) two lines that we called callus1 and callus2 (c1 and c2) formed neither shoots nor roots on hormone-free medium (Fig. 1a). Line c1 responded readily to exogenous cytokinin by forming shoots, whereas no shoots formed in c2, even after prolonged incubation with elevated cytokinin concentrations (10 mg/L iP). c2 formed roots in response to exogenous auxin. Therefore, for further investigation, c1 was examined in parallel with the shooty callus class, c2 with the rooty callus class. The phenotype of all mutant tissue lines has been stable for more than 2 years.
Figure 1.
Phenotype of Arabidopsis mutant lines cultivated in vitro on hormone-free medium. a, Growth of mutant line c1 (right) compared with growth of control calli on hormone-free MS medium (left). b, Root-forming phenotype of mutant line r2 grown on hormone-free MS medium. c, Shoot-forming phenotype of mutant line s3 grown on hormone-free MS medium.
We attempted to regenerate plants from the calli to obtain progeny and to characterize the mutant lines genetically. The rooty callus and callus lines, as well as the majority of the shooty callus lines, could not be regenerated to form plants or were infertile. From line r1 we obtained two seeds by selfing more than 200 regenerants. One of these seeds germinated in vitro and reproduced the parental phenotype. Dedifferentiation and callus formation occurred in the F2 progeny of line s1, obtained after backcrossing with the wild type, at a ratio of approximately 1:3 (122 of 466 analyzed F2 seedlings showed the mutant phenotype and 344 were phenotypically wild type), indicating that a single recessive mutation is responsible for the phenotype. The mutated locus has been mapped to chromosome 2 (data not shown). A more detailed analysis of this mutant will be described elsewhere (M. Frank, I. Lorenz-Meyer, A. Guivarch, D. Chriqui, and T. Schmülling, unpublished data).
Growth and Cell Cycle Regulator Genes
First, we compared the growth rate of the mutant lines growing on hormone-free medium with wild-type callus growing on auxin- and cytokinin-containing medium. The gain in fresh weight was for all mutant lines 2- to 13-fold faster than for wild-type callus. The highest increases in fresh weight were found in the lines s6 and r3 (Table I; data not shown).
Table I.
Growth and cell-cycle gene expression in wild-type (WT) and mutant Arabidopsis lines
| Plant Material | W | Increasea | Ratio
|
||||
|---|---|---|---|---|---|---|---|
| H4/25S | Cdc2a/25S | Cdc2a/H4 | CycD3/25S | CycD3/H4 | |||
| Seedling (WT) | ND | — | 1.1 | 0.9 | 0.8 | 0.8 | 0.7 |
| Callus | |||||||
| WT | 0.1 | — | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| s2 | 0.7 | 7 | 1.2 | 0.9 | 0.9 | 1.1 | 0.9 |
| s6 | 1.7 | 17 | 2.5 | 1.4 | 0.7 | 3.5 | 1.4 |
| c2 | 0.7 | 7 | 1.6 | 1.4 | 0.9 | 2.5 | 1.6 |
| r2 | 0.3 | 3 | 0.9 | 1.0 | 1.0 | 1.0 | 1.1 |
| r3 | 1.5 | 15 | 1.9 | 1.3 | 0.7 | 1.9 | 1.0 |
The growth coefficient (W) for 1 week of growth is expressed as (Wt − W0/W0. W0 and Wt are the initial and final average weights of calli. Data are the means of three independent experiments with five samples per point in each experiment. Intensities of hybridizations shown in Figure 2 were determined by measurement of the optical filter density. Ratios of signal strength were compared with the ratio of WT callus, which was set arbitrarily at 1. ND, Not determined.
Increase in mutant calli compared with wild-type calli.
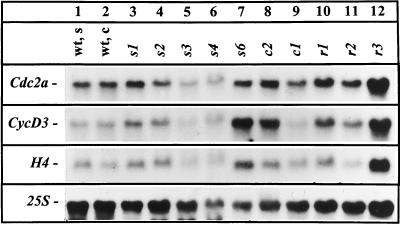
Next, we investigated steady-state mRNA levels of the cell cycle genes H4, Cdc2a, and CycD3 in the mutant lines and compared it with wild-type seedlings and callus. Deregulated expression of the homologous genes in animals is often linked to tumor formation (Hunter, 1997). Results of northern-blot analyses are shown in Figure 2, and the relative signal strength compared with the 25S control hybridization is listed for selected lines in Table I.
Figure 2.
Northern-blot analyses of H4, Cdc2, and CycD3 steady-state mRNA levels in wild-type seedlings (wt, s), wild-type calli (wt, c), and mutant calli. Total RNA (50 μg) was separated in a denaturing formaldehyde gel and, after blot transfer to a nylon filter, hybridized with 32P-labeled specific cDNA probes. Hybridization with a 25S rDNA probe served as a control for loading.
The transcript abundance of the histone H4 gene, a marker for the cell cycle S phase, was in almost all mutant calli comparable to that of wild-type seedlings and calli. The highest H4/25S ratios, 2.5 and 1.9, were found in the fastest-growing lines s6 and r3, respectively (Table I).
Expression of Cdc2a occurs in dividing cells and in cells with competence to divide (Hemerly et al., 1993). As indicated in Figure 2 and Table I, the fastest-growing mutant lines, s6, r3, and c2, showed a slightly enhanced steady-state mRNA level of Cdc2a. The Cdc2a/25S ratios were 1.3- to 1.4-fold higher than in the wild type or in the other mutant calli (Table I; data not shown).
CycD3 regulates the G1/S transition. Its overexpression causes cytokinin-independent tumor formation in plants (Riou-Khamlichi et al., 1999). Figure 2 and Table I show that CycD3 transcript levels were approximately 1.9- to 3.5-fold increased in lines s6, c2, and r3 compared with wild-type seedlings and calli.
Analysis of the Auxin and Cytokinin Content
The mutant lines mimic a hormone effect without the presence of exogenous hormones. We therefore determined the endogenous concentrations of auxin and cytokinins. Table II shows that the IAA content was approximately 10- to 25-fold higher in the lines r1, r2, and r6 than in control tissue. In the same lines, the IAA conjugate concentration was increased 5- to 33-fold. In contrast, lines r3, r4, and r5, both c lines, and all s lines contained similar auxin and auxin conjugate levels as controls (data not shown).
Table II.
Content of free and conjugated IAA in wild-type (WT) seedlings and mutant calli
| Plant Material | IAA | IAA Conjugates |
|---|---|---|
| pmol g−1 fresh wt | ||
| WT seedling | 12.7 ± 4.6 | 13,400 ± 5,300 |
| Callus | ||
| r1 | 295 ± 41 | 430,000 ± 173,000 |
| r2 | 205 ± 44 | 179,000 ± 23,000 |
| r6 | 121 ± 12 | 71,000 ± 19,000 |
The mutant calli and WT seedlings were grown in vitro on solidified, hormone-free MS medium. IAA and IAA conjugate concentrations were determined by gas chromatography-selected ion reactant-MS as described in “Materials and Methods.” The data are the means ± se of three independent biological replications.
Table III shows that line c1 contained between 5- and 80-fold higher concentrations of most of the 17 different cytokinin metabolites of the iP-, trans-zeatin, and dihydrozeatin-type cytokinins compared with wild type. The highest increases were found for zeatin riboside (20-fold) and the O-conjugates zeatin O-glucoside (81-fold) and dihydrozeatin O-glucoside (45-fold). It is noteworthy that the total content of N-conjugates was higher than that of the O-conjugates (Table III). This indicates that N-conjugation is relevant in Arabidopsis, while it is weak or does not occur in cytokinin-overproducing tobacco (Motyka et al., 1996; Faiss et al., 1997; Rupp et al., 1999). The cytokinin content of all other mutant lines was similar to wild type or did not show consistent differences (data not shown).
Table III.
Cytokinin content of mutant line c1 compared to wild-type (WT) seedlings
| Cytokinin Metabolite | WT Seedling | c1 Callus | Increaseb |
|---|---|---|---|
| pmol g−1 fresh wt | |||
| iP | 7.6a | 1.2 ± 0.1 | 0.2 |
| iP Riboside | 3.7 ± 3.3 | 22.7 ± 5.4 | 6 |
| iP Riboside 5′-monophosphate | 1.5 ± 0.1 | 12.5 ± 0.4 | 8 |
| iP 9-Glucoside | 4.7 ± 2.2 | 25.7 ± 4.3 | 6 |
| trans-Zeatin | 0.8a | 0.65 ± 0.0 | 1 |
| Zeatin riboside | 2.2 ± 1.7 | 42.9 ± 5.4 | 20 |
| Zeatin riboside 5′-monophosphate | 2.4 ± 1.3 | 11.4 ± 2.0 | 5 |
| Zeatin O-glucoside | 6.5 ± 1.0 | 525 ± 105 | 81 |
| Zeatin riboside O-glucoside | 2.3 ± 0.3 | 14.8 ± 0.0 | 6 |
| Zeatin 7-glucoside | 148 ± 26 | 751 ± 25 | 5 |
| Zeatin 9-glucoside | 21 ± 20 | 257 ± 50 | 12 |
| Dihydrozeatin | 1.4a | 0.2 ± 0.1 | 0.2 |
| Dihydrozeatin riboside | 1.1 ± 0.7 | 2.8 ± 0.3 | 3 |
| Dihydrozeatin riboside 5′-monophosphate | 1.0 ± 0.3 | 1.4 ± 0.0 | 1 |
| Dihydrozeatin O-glucoside | 7.1 ± 2.5 | 322 ± 46 | 45 |
| Dihydrozeatin riboside O-glucoside | 2.3 ± 0.3 | 11.7 ± 3.1 | 5 |
| Dihydrozeatin 7-glucoside | 2.8 ± 1.6 | 15.1 ± 7.2 | 5 |
| Dihydrozeatin 9-glucoside | 9.7 ± 9.1 | 13.9 ± 0.0 | 1 |
Mutant calli and wild-type seedlings (wt, s) were grown in vitro on solidified hormone-free MS medium. Cytokinin concentrations were determined by micro liquid chromatography with column switch configuration coupled to positive ion electrospray tandem mass spectrometry using multiple reactant monitoring as described in “Materials and Methods.” The data are the means ± se of two biological independent replications.
Data from a single sample.
Increase in c1 callus compared with wild-type seedlings.
Analysis of Cytokinin Oxidase Activity
The presence of higher cytokinin metabolite concentrations in line c1 could be due to increased cytokinin synthesis and/or decreased catabolism. Cytokinin oxidase is the key enzyme of cytokinin degradation in plants (Armstrong, 1994). Table IV shows that c1 has approximately 5% of wild-type cytokinin oxidase activity. None of the other tested lines (r1, c2, s1, s4, and s6) had significantly altered cytokinin oxidase activities compared with wild-type callus (Table IV; data not shown).
Table IV.
Cytokinin oxidase activity in mutant lines
| Plant Material | Cytokinin Oxidase Activity |
|---|---|
| pmol mg−1 protein h−1 | |
| Seedling (WT) | 11.4 ± 1.7 |
| Callus | |
| WT | 4.9a |
| c1 | 0.6 ± 0.3 |
| c2 | 4.5 ± 2.5 |
| s1 | 4.5 ± 0.5 |
| r1 | 4.8 ± 0.7 |
Cytokinin oxidase assays were based on the rate of conversion of [2,8-3H]iP to adenine. Substrate and product were separated by thin-layer chromatography. Radioactivity was measured using a liquid scintillation counter. Other details are described in “Materials and Methods.” Wild-type (WT) seedlings and mutant calli were grown on solidified, hormone-free MS medium. WT callus was grown on 1 mg/L naphthylacetic acid and 0.1 mg/L iP. The values represent the means ± se of three independent samples.
Data from a single replicate.
Steady-State mRNA Levels of IAA1 and IAA2
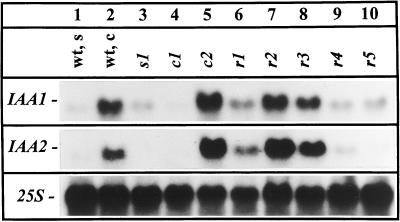
We used the auxin primary response genes IAA1 and IAA2 (Abel et al., 1995) as marker genes to test for altered auxin signaling in the mutants. Figure 3 shows that r1, r2, r3, and c2 contained approximately 3- to 5-fold elevated steady-state mRNA levels of the IAA1 and IAA2 genes. In contrast, the s lines and line c1 contained no detectable or only low levels of these transcripts (Fig. 5 and data not shown). Lines r3 and c2 do not have an elevated auxin content, which indicates altered auxin signal transduction.
Figure 3.
Northern-blot analyses of IAA1 and IAA2 steady-state mRNA levels in wild-type seedlings (wt, s), wild-type calli (wt, c), and mutant calli. Total RNA (50 μg) was separated in a denaturing formaldehyde gel and, after blot transfer to a nylon filter, hybridized with 32P-labeled specific cDNA probes. Hybridization with a 25S rDNA probe served as a control for loading.
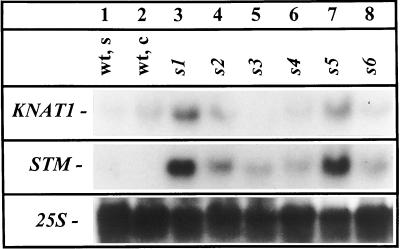
Figure 5.
Northern-blot analysis KNAT1 and STM steady-state mRNA levels in wild-type seedlings (wt, s), wild-type calli (wt, c), and mutant calli. Total RNA (50 μg) was separated in a denaturing formaldehyde gel and, after blot transfer to a nylon filter, hybridized with 32P-labeled specific cDNA probes. Hybridization with a 25S rDNA probe served as a control for loading.
Steady-State mRNA Level CKI1
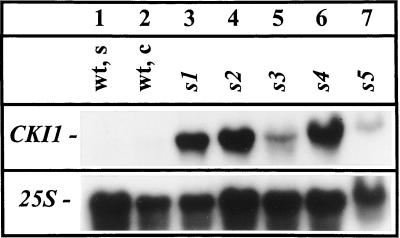
The shooty callus mutant lines showed cytokinin-autonomous proliferation and shoot formation similar to Arabidopsis calli overexpressing the CKI1 gene (Kakimoto, 1996). Figure 4 shows that lines s1, s2, and s4 contained at least a 5-fold-increased steady-state mRNA level of CKI1 compared with wild-type control tissues. Lines s3 and s5 contained elevated levels of CKI1 mRNA, but the increase was not as large (Fig. 4).
Figure 4.
Northern-blot analysis of CKI1 steady-state mRNA levels in wild-type seedlings (wt, s), wild-type calli (wt, c), and mutant calli. Total RNA (50 μg) was separated in a denaturing formaldehyde gel and, after blot transfer to a nylon filter, hybridized with 32P-labeled specific cDNA probes. Hybridization with a 25S rDNA probe served as a control for loading.
Steady-State mRNA Levels of KNAT1 and STM
The transgenic overproduction of cytokinins, KNAT1, or its maize homolog, KN1, are sufficient to induce ectopic shoot formation in tobacco and Arabidopsis, indicating that they might act on a common pathway (Sinha et al., 1993; Hewelt et al., 1994; Chuck et al., 1996). Therefore, deregulated expression of homeobox genes could be linked to the phenotypic traits of the shooty callus mutant class. Figure 5 shows that the steady mRNA levels of STM was 3- to 4-fold increased in all shooty callus lines (especially in lines s1, s2, and s5) compared with wild-type seedlings and calli. KNAT1 mRNA accumulated also in these lines. In contrast, lines s3, s4, and s6 contained KNAT1 mRNA levels comparable to wild-type and seedlings and calli (Fig. 5). Both genes were poorly expressed in the rooty callus lines (data not shown).
DISCUSSION
We have isolated three mutant classes that exhibited loss of control of cell division activity after germination and could grow as a callus on hormone-free medium. These mutant tissue lines resemble phenotypically somatic mutants isolated previously after γ-radiation of Arabidopsis seeds and seedlings (Persinger and Town, 1991; Campell and Town, 1992). The lines described here were analyzed for differences in cell cycle gene expression and, in particular, for differences in auxin and cytokinin content or signaling to detect an eventual correlation between the mutant phenotype and changes of these parameters.
All mutant lines, in particular s6 and r3, had a higher growth rate than wild-type callus grown on auxin- and cytokinin-containing medium (Table I). The steady-state mRNA level of cell cycle genes was not strongly altered in the mutant lines, unlike the deregulation that occurs in animal tumors (Hunter, 1997). In particular, the human cyclin D1 gene, and perhaps other D-type cyclins, are putative proto-oncogenes possibly activated by deregulated transcription (Motokura and Arnold, 1993). In plants, the correlation between overexpression of genes encoding cell cycle regulators and differentiation is less clear. Cdc2 and CycB1 overexpression in Arabidopsis neither altered development nor caused neoplasia (Hemerly et al., 1995; Doerner et al., 1996). In contrast, ectopic and enhanced expression of CycD3 perturbed plant growth and differentiation (Riou-Khamlichi et al., 1999).
Most mutant lines showed a ratio of the S-phase marker H4 and Cdc2a similar to wild type. The Cdc2a/H4 ratio can be taken as a measure of the ratio of division-competent cells to cells that are actually dividing. A decrease in the Cdc2a/H4 ratio indicates a higher percentage of dividing cells. Indeed, the Cdc2/H4 mRNA ratios were lowest in the fast-growing mutant lines s6 and r3 (Table I), indicating that cell division, and not just cell expansion, contributes to the faster growth. The fast-growing mutant lines s6, c2, and r3 also have the highest CycD3 steady-state levels (Table I). This could indicate that CycD3 is limiting for growth in wild type and the other mutant lines and that deregulated CycD3 expression might be causally linked to the rapid growth.
The rooty callus mutants mimic an auxin phenotype, and the shooty callus mutants a cytokinin phenotype in the absence of exogenous hormones. This led us to study hormone content and signal transduction in the mutant lines. In the following sections the results are discussed separately for rooty and shooty callus lines.
The rooty Mutant Lines
In three of the rooty callus lines (r1, r2, and r6), we detected a significantly elevated content of free and conjugated IAA (Table II). Two mutants that overproduce auxin have been previously reported in Arabidopsis. One mutant overproliferating lateral roots was independently isolated several times and called sur1 (Boerjan et al., 1995), rty (King et al., 1995), hls3 (Lehman et al., 1996), and alf1 (Celenza et al., 1995). The RTY/SUR1 gene encodes a protein similar to Tyr aminotransferases possibly implicated in auxin synthesis (Golparaj et al., 1996). sur1 mutants produce calli that grow on hormone-free medium (Boerjan et al., 1995; King et al., 1995; Delarue et al., 1998). Recently, a second auxin-overproducing mutant, sur2, has been identified in Arabidopsis (Delarue et al., 1998). However, sur2 explants are unable to sustain auxin-autonomous growth. It is possible that the auxin-overproducing rooty callus harbor mutated alleles of the RTY/SUR1 gene.
Lines r3 and c2 had normal auxin contents but increased IAA1 and IAA2 transcript levels. This indicates that auxin signal transduction is affected in these mutant lines. Possibly, a loss-of-function mutation of a repressor of the auxin response results in a constitutive auxin response. Similar mutants have yet to be described. Two mutants, age1 and age2, that show enhanced auxin sensitivity of an IAA gene promoter or ectopic promoter activity in the absence of exogenous IAA were isolated previously (Oono et al., 1998). In contrast to the lines described here, these mutants did not show altered tissue differentiation.
Interestingly, r4 and r5 have neither an altered auxin content nor an altered transcript level of the IAA1 and IAA2 genes (Fig. 3). However, r4 and r5 have enhanced steady-state mRNA levels of another auxin response gene, IAA9 (M. Frank and T. Schmülling, unpublished results). This suggests either that r4 and r5 are mutated in a different auxin response pathway than the other rooty callus mutants, or they are mutated in a gene that regulates functions downstream of IAA1 and IAA2. In comparison, undifferentiated or root-forming Arabidopsis mutant lines obtained by γ-radiation did neither contain increased auxin concentrations, which supports the notion that this phenotype can be induced without increased auxin (Campell and Town, 1991). In conclusion, these results indicate that the screening procedure has the potential to identify new loci of the auxin response pathway.
The shooty Mutant Lines
None of the shooty callus lines had a significantly altered auxin or cytokinin content. Only in line c1 did we detect strongly elevated cytokinin concentrations (Table III). The strong reduction of cytokinin oxidase activity in this line (Table IV) could affect c1 hormone homeostasis. The accumulation of cytokinin conjugates might be a detoxification mechanism that operates to avoid the accumulation of toxic concentrations of biologically active cytokinins. The results presented here suggest that a reduction in cytokinin oxidase activity may result in accumulation of conjugates and unorganized growth of tissue. A cytokinin oxidase gene has been cloned from maize (Houba-Hérin et al., 1999; Morris et al., 1999), and several homologous candidate genes are identifiable by comparison with the genomic sequence in Arabidopsis (K. Lemcke, T. Werner, and T. Schmülling, unpublished results). Therefore, loss-of-function studies become feasible for this enzyme. Experiments to test the influence of reduced cytokinin oxidase activity on differentiation processes are in progress.
The steady-state mRNA level of the CKI1 His kinase gene is enhanced in all shooty callus mutant lines (Fig. 4). CKI1 is likely to be involved in cytokinin signaling (Kakimoto, 1996). Transgenic overexpressors of the CKI1 gene share phenotypic characteristics such as cytokinin-independent proliferation and shoot formation with the shooty callus lines. Mutations in negative regulatory elements of CKI1 could be the reason for the enhanced CKI1 transcript levels and the constitutive cytokinin response phenotype of the shooty callus lines. However, the present data do not exclude the possibility that the higher gene expression levels are merely a consequence of the mutant's morphology and not a cause.
Simlarily, we observed in the shooty callus lines an increase in the steady-state mRNA levels of the shoot-meristem-specifying class I homeobox gene STM (Fig. 5). We hypothesize that lines overexpressing both CKI1 and STM are mutated in a pathway that links CKI1 and STM functionally. The differences in relative transcript abundance could be due to different types of mutations and/or to overproliferation of different tissues. Likewise, differences in the up-regulation of KNAT1 in the different mutant lines and in comparison with STM accumulation (Fig. 5) could reflect a different contribution of the original KNAT1 expression domain to the callus tissue. KNAT1 transcript is localized in the wild type primarily in the peripheral zone of the vegetative shoot apical meristem (Lincoln et al., 1994), while STM is expressed in a central region (Endrizzi et al., 1996; Long et al., 1996).
In this context it is noteworthy that the cytokinin-overproducing amp1 mutant and transgenic cytokinin overproducers also have increased steady-state mRNA levels of STM and KNAT1. It was therefore hypothesized that cytokinins participate in the regulation of homeobox gene expression (Rupp et al., 1999). The fact that STM- and KNAT1-overexpressing shooty callus lines do not have an altered cytokinin content is compatible with the hypothesis that cytokinins act upstream of KNAT1 and STM.
Enhanced cytokinin signaling by CKI1 in the shooty callus lines might be causally related to enhanced shoot meristem activity through deregulation of shoot-meristem-specifying homeobox genes. This raises the possibility that cytokinins, CKI1, and these homeobox genes act on the same pathway. Further experimental proof is required to support this hypothetical model.
CONCLUSIONS
Our experimental results indicate that the phenotype of all rooty callus lines and of line c2 is linked to increased auxin content or signaling. In all shooty callus lines and in line c1, we found increased cytokinin content or indications of enhanced cytokinin signaling. Thus, the analyses of hormone content and hormone sensitivity have so far confirmed our operational concept. This supports the idea that tightly regulated auxin and cytokinin metabolism and signaling are a prerequisite for coordinated growth and development.
This pilot study demonstrates that screening for such mutants and analysis of known hormonal parameters provides a useful approach to identify new factors involved in auxin and cytokinin metabolism and/or signal transduction. The phenotypic differences between different shooty callus lines and differences in the expression of marker genes suggests that mutations of different genes have occurred in the various lines. One new genetic locus has been identified in the progeny of line s1 and we have been able to identify similar mutants from mutagenized single lines that map to different chromosomal locations. We are currently analyzing the early events during the de-differentiation process caused by these mutations (M. Frank, I. Lorenz-Meyer, A. Guivarch, D. Chriqui, and T. Schmülling, unpublished results).
ACKNOWLEDGMENTS
We thank Franka Schermutzki for help with seed sterilization and plating, and S. Öden for skillful technical assistance. Plasmids harboring part of the KNAT1 and STM cDNAs were a generous gift of Dr. Thomas Laux. The CycD3 gene probe was kindly provided by Dr. James A. H. Murray, and the IAA1 and IAA2 gene probes were kindly provided by Drs. Steffen Abel and Anastasios Theologis. We also acknowledge proofreading by Catherine Scott-Taggart.
Footnotes
This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB 446), the Volkswagenstiftung, and International Association for the Promotion of Cooperation with Scientists from the New Independent States of the former Soviet Union (INTAS) to T.S. M.F. received a stipend from the Studienstiftung des deutschen Volkes. E.P. and H.V.O. are, respectively, a postdoctoral fellow and the research director of the Fund for Scientific Research Flanders.
LITERATURE CITED
- Abel S, Nguyen MD, Theologis A. The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J Mol Biol. 1995;251:533–549. doi: 10.1006/jmbi.1995.0454. [DOI] [PubMed] [Google Scholar]
- Armstrong J. Cytokinin oxidase and the regulation of cytokinin degradation. In: Mok DWS, Mok MC, editors. Cytokinins: Chemistry, Activity and Function. Boca Raton, FL: CRC Press; 1994. pp. 139–154. [Google Scholar]
- Bialek K, Cohen JD. Quantitation of indoleacetic acid conjugates in bean seeds by direct tissue hydrolysis. Plant Physiol. 1989;90:398–400. doi: 10.1104/pp.90.2.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieleski WJ. The problem of halting enzyme action when extracting plant tissues. Anal Biochem. 1964;9:431–442. doi: 10.1016/0003-2697(64)90204-0. [DOI] [PubMed] [Google Scholar]
- Boerjan W, Cervera MT, Delarue M, Beeckman T, Dewitte W, Bellini C, Caboche M, Van Onckelen H, Van Montagu M, Inzé D. superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell. 1995;7:1405–1419. doi: 10.1105/tpc.7.9.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzovkina IS, Kneshke I, Lutova LA. The model of tumorigenesis in vitro in inbred and hybrids of radish. Genetika. 1993;29:1002–1008. [Google Scholar]
- Campell BR, Town CD. Physiology of hormone autonomous tissue lines derived from radiation-induced tumors of Arabidiopsis thaliana. Plant Physiol. 1991;97:1166–1173. doi: 10.1104/pp.97.3.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campell BR, Town CD. Characterization of overexpressed cDNAs isolated from a hormone-autonomous, radiation-induced tumor tissue line of Arabidopsis thaliana. Plant Physiol. 1992;100:2018–2023. doi: 10.1104/pp.100.4.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardarelli M, Mariotti D, Pomponi M, Spano L, Capone I, Costantino P. Agrobacterium rhizogenesT-DNA genes capable of inducing hairy root phenotype. Mol Gen Genet. 1987;209:475–480. doi: 10.1007/BF00331152. [DOI] [PubMed] [Google Scholar]
- Celenza JL, Grisafi PL, Fink GGR. A pathway for lateral root formation in Arabidopsis thaliana. Genes Dev. 1995;9:2131–2142. doi: 10.1101/gad.9.17.2131. [DOI] [PubMed] [Google Scholar]
- Chatfield JM, Armstrong DJ. Regulation of cytokinin oxidase activity in callus tissues of Phaseolus vulgarisL. cv Great Northern. Plant Physiol. 1986;80:493–499. doi: 10.1104/pp.80.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck C, Lincoln C, Hake S. KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell. 1996;8:1277–1289. doi: 10.1105/tpc.8.8.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delarue M, Prinsen E, Van Onckelen H, Caboche M, Bellini C. Sur2 mutations of Arabidopsis thalianadefine a new locus involved in the control of auxin homeostasis. Plant J. 1998;14:603–611. doi: 10.1046/j.1365-313x.1998.00163.x. [DOI] [PubMed] [Google Scholar]
- Dockx J, Kock P, Willemsen V, Weisbeek P, Smeekens S, Scheres B (1996) The role of the homeobox gene KNAT2 in Arabidopsis development. Presented at the 7th International Conference on Arabidopsis Research, Norwich, UK, poster 196
- Doerner P, Jorgensen JE, You R, Stepphuhn J, Lamb C. Control of root growth and development by cyclin expression. Nature. 1996;380:520–523. doi: 10.1038/380520a0. [DOI] [PubMed] [Google Scholar]
- Endrizzi K, Moussian B, Haecker A, Levin JZ, Laux T. The SHOOT MERISTEMLESS gene is required for maintenance of undifferentiated cells in Arabidopsis shoot and floral meristems and acts at a different regulatory level than the meristem genes WUSCHEL and ZWILLE. Plant J. 1996;10:967–979. doi: 10.1046/j.1365-313x.1996.10060967.x. [DOI] [PubMed] [Google Scholar]
- Epstein E, Cohen JD. Microscale preparation of pentafluorobenzyl esters: electron-capture gas chromatographic detection of indole-3-acetic acid from plants. J Chromatogr. 1981;209:413–420. [Google Scholar]
- Faiss M, Zalubílova J, Strnad M, Schmülling T. Conditional transgenic expression of the iptgene indicates a function for cytokinins in paracrine signaling in whole tobacco plants. Plant J. 1997;12:401–415. doi: 10.1046/j.1365-313x.1997.12020401.x. [DOI] [PubMed] [Google Scholar]
- Golparaj M, Tseng TS, Olszewski N. The ROOTY gene of Arabidopsisencodes a protein with high similarity to aminotransferases (abstract no. 469) Plant Physiol. 1996;111:S-114. [Google Scholar]
- Hemerly A, Engler JdA, Bergounioux C, Van Montagu M, Engler G, Inzé D, Ferreira P. Dominant negative mutants of the Cdc2 kinase uncouple cell division from iterative plant development. EMBO J. 1995;14:3925–3936. doi: 10.1002/j.1460-2075.1995.tb00064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemerly AS, Ferreira P, de Almeida Engler J, Van Montagu M, Engler G, Inzé D. cdc2aexpression in Arabidopsis is linked with competence for cell division. Plant Cell. 1993;5:1711–1723. doi: 10.1105/tpc.5.12.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewelt A, Prinsen E, Schell J, Van Onckelen H, Schmülling T. Promoter tagging with a promoterless iptgene leads to cytokinin-induced phenotypic variability in transgenic tobacco plants: implications of gene dosage effects. Plant J. 1994;6:879–891. doi: 10.1046/j.1365-313x.1994.6060879.x. [DOI] [PubMed] [Google Scholar]
- Hooykaas PJJ, den Dulk-Ras H, Schilperoort RA. The Agrobacterium tumefaciens T-DNA gene 6b is an oncgene. Plant Mol Biol. 1988;11:791–794. doi: 10.1007/BF00019519. [DOI] [PubMed] [Google Scholar]
- Houba-Hérin N, Pethe C, d'Alayer J, Laloue M. Cytokinin oxidase from Zea mays: purification, cDNA cloning and expression in moss protoplasts. Plant J. 1999;17:615–626. doi: 10.1046/j.1365-313x.1999.00408.x. [DOI] [PubMed] [Google Scholar]
- Hunter T. Oncoprotein networks. Cell. 1997;88:333–346. doi: 10.1016/s0092-8674(00)81872-3. [DOI] [PubMed] [Google Scholar]
- Ichikawa T, Syono K. Tobacco genetic tumors. Plant Cell Physiol. 1991;32:1123–1128. [Google Scholar]
- Kakimoto T. CKI1, a histidine kinase homolog implicated in cytokinin signal transduction. Science. 1996;274:982–985. doi: 10.1126/science.274.5289.982. [DOI] [PubMed] [Google Scholar]
- King JJ, Stimart DP, Fisher RH, Bleecker AB. A mutation altering auxin homeostasis and plant morphology in Arabiopsis. Plant Cell. 1995;7:2023–2037. doi: 10.1105/tpc.7.12.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman A, Black R, Ecker J. HOOKLESS1, an ethylene response gene, is required for differential cell elongation in the Arabidopsishypocotyl. Cell. 1996;85:183–194. doi: 10.1016/s0092-8674(00)81095-8. [DOI] [PubMed] [Google Scholar]
- Lincoln C, Long J, Yamaguchi J, Serikawa K, Hake S. A knotted1-like homeobox gene in Arabidopsisis expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell. 1994;6:1859–1876. doi: 10.1105/tpc.6.12.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature. 1996;379:66–69. doi: 10.1038/379066a0. [DOI] [PubMed] [Google Scholar]
- Lutova LA, Buzovkina IS, Smirnova OA, Tikhodeyev ON, Shishkova SO, Trifonova IM. Genetic control of in vitro differentiation processes in radish. In Vitro Cell Dev Biol Plant. 1997;33:269–274. [Google Scholar]
- Maurel C, Barbier-Brygoo H, Spena A, Tempe J, Guern J. Single rol genes from Agrobacterium rhizogenes TL-DNA alter some of the cellular responses to auxin in Nicotiana tabacum. Plant Physiol. 1991;97:212–216. doi: 10.1104/pp.97.1.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer AD, Aebi R, Meins F. Tobacco plants carrying a tms locus of Ti-plasmid origin and the Hl-1allele are tumor prone. Differentiation. 1997;61:213–221. doi: 10.1046/j.1432-0436.1997.6140213.x. [DOI] [PubMed] [Google Scholar]
- Morris RO. Genes specifying auxin and cytokinin biosynthesis in prokaryotes. In: Davies PJ, editor. Plant Hormones: Physiology, Biochemistry and Molecular Biology. Kluwer Academic Publishers. The Netherlands: Dordrecht; 1995. pp. 318–339. [Google Scholar]
- Morris RO, Bilyeu KD, Laskey JG, Cheikh NN. Isolation of a gene encoding a glycosylated cytokinin oxidase from maize. Biochem Biophys Res Commun. 1999;255:328–333. doi: 10.1006/bbrc.1999.0199. [DOI] [PubMed] [Google Scholar]
- Motokura T, Arnold A. Cyclin D and oncogenesis. Curr Opin Gene Dev. 1993;3:5–10. doi: 10.1016/s0959-437x(05)80334-x. [DOI] [PubMed] [Google Scholar]
- Motyka V, Faiss M, Strnad M, Kamínek M, Schmülling T. Changes in cytokinin content and cytokinin oxidase activity in response to derepression of iptgene transcription in transgenic tobacco calli and plants. Plant Physiol. 1996;112:1035–1043. doi: 10.1104/pp.112.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motyka V, Kamínek M. Cytokinin oxidase from auxin- and cytokinin-dependent callus cultures of tobacco (Nicotiana tabacumL.) J Plant Growth Regul. 1994;13:1–9. [Google Scholar]
- Murashige T, Skoog F. A revised medium for plant growth and bioassays with tobacco tissue culture. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Oono Y, Chen-Qianhong G, Overvoorde PJ, Koehler C, Theologis A. age mutants of Arabidopsisexhibit altered auxin-regulated gene expression. Plant Cell. 1998;10:1649–1662. doi: 10.1105/tpc.10.10.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otten L, Schmidt J. A T-DNA from the Agrobacterium tumefacienslimited-host-range strain AB2/73 contains a single oncogene. Mol Plant-Microbe Interact. 1998;11:335–342. doi: 10.1094/MPMI.1998.11.5.335. [DOI] [PubMed] [Google Scholar]
- Persinger SM, Town CD. Isolation and characterization of hormone-autonomous tumors of Arabidopsis thaliana. J Exp Bot. 1991;42:1363–1370. [Google Scholar]
- Prinsen E, Redig P, Strnad M, Galis I, Van Dongen W, Van Onckelen H. Quantifying phytohormones in transformed plants. In: Gartland KMA, Davey MR, editors. Methods in Molecular Biology. Vol. 44. Totowa, NJ: Humana Press; 1995a. pp. 245–262. [DOI] [PubMed] [Google Scholar]
- Prinsen E, Redig P, Van Dongen W, Esmans EL, Van Onckelen H. Quantitative analysis of cytokinins by electrospray tandem mass spectrometry. Rapid Commun Mass Spectrom. 1995b;9:948–953. [Google Scholar]
- Prinsen E, Van Dongen W, Esmans EL, Van Onckelen H. Micro and capillary LC-MS/MS: a new dimension in phytohormone research. J Chromatogr A. 1998;826/1:25–37. [Google Scholar]
- Riou-Khamlichi C, Huntley R, Jaqmard A, Murray JAH. Cytokinin activation of Arabidopsiscell division through a D-type cyclin. Science. 1999;283:1541–1544. doi: 10.1126/science.283.5407.1541. [DOI] [PubMed] [Google Scholar]
- Rupp HM, Frank M, Werner T, Strnad M, Schmülling T. Increased steady state mRNA levels of the STM and KNAT1 homeobox genes in cytokinin overproducing Arabidopsis thalianaindicate a role for cytokinins in the shoot apical meristem. Plant J. 1999;18:557–563. doi: 10.1046/j.1365-313x.1999.00472.x. [DOI] [PubMed] [Google Scholar]
- Schmülling T, Fladung M, Grossmann K, Schell J. Hormonal content and sensitivity of transgenic tobacco and potato plants expressing single rol genes of Agrobacterium rhizogenesT-DNA. Plant J. 1993;3:371–382. [Google Scholar]
- Sinha NR, Williams RE, Hake S. Overexpression of the maize homeobox gene, KNOTTED-1, causes a switch from determinate to indeterminate cell fates. Genes Dev. 1993;7:787–795. doi: 10.1101/gad.7.5.787. [DOI] [PubMed] [Google Scholar]
- Skoog F, Miller CO. Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp Soc Exp Biol. 1957;11:118–131. [PubMed] [Google Scholar]
- Smith HH, Kao KN, Combatti NC. Interspecific hybridization by protoplast fusion in Nicotiana. J Hered. 1976;67:123–128. [Google Scholar]
- Verwoerd TC, Dekker MM, Hoekema A. A small-scale procedure for the rapid isolation of plant RNA. Nucleic Acid Res. 1989;17:2362. doi: 10.1093/nar/17.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]