Abstract
Objectives: The performance of cadexomer iodine was determined against microbial populations from chronic non-healing diabetic foot ulcers (DFUs) complicated by biofilm in vivo, using molecular, microscopy and zymography methods.
Methods: Chronic non-healing DFUs due to suspected biofilm involvement were eligible for enrolment. DNA sequencing and real-time quantitative PCR was used to determine the microbial load and diversity of tissue punch biopsies obtained pre- and post-treatment. Scanning electron microscopy and/or fluorescence in situ hybridization confirmed the presence or absence of biofilm. Zymography was used to determine levels of wound proteases.
Results: Seventeen participants were recruited over a 6 month period. Scanning electron microscopy and or fluorescence in situ hybridization confirmed the presence of biofilm in all samples. Eleven participants exhibited log10 reductions in microbial load after treatment (range 1–2 log10) in comparison with six patients who experienced <1 log10 reduction (P = 0.04). Samples were tested for levels of wound proteases pre- and post-treatment. Reductions in the microbial load correlated to reductions in wound proteases pre- and post-treatment (P = 0.03).
Conclusions: To the best of our knowledge, this study represents the first in vivo evidence, employing a range of molecular and microscopy techniques, of the ability of cadexomer iodine to reduce the microbial load of chronic non-healing DFUs complicated by biofilm. Further analyses correlating log reductions to optimal duration of therapy and improvements in clinical parameters of wound healing in a larger cohort are required.
Introduction
In a person with diabetes, foot ulceration leaves a physical break in the protective barrier of the skin. Factors including a retarded host immune response and pathogen-related dynamics (such as virulence or pathogenicity) may predispose the diabetic foot ulcer (DFU) to further planktonic microbial replication and invasion. This may result in damage to host tissues and an inflammatory response that is characterized as a clinical infection.1
Increasing evidence into the role of microorganisms involved in DFUs (and other wound aetiologies) has identified that single free-floating microorganisms (planktonic) that are responsible for acute infections, may not necessarily represent the ecology of microorganisms present in chronic non-healing wounds. Instead, the focus has orientated towards the concept of biofilms, which differ markedly in both their physiology and activity. The exact mechanisms of biofilm impairment on the healing processes of wounds are not clear, but general consensus suggests that biofilms maintain an elevated inflammatory state within the wound.2
Microorganisms in biofilm exhibit enhanced tolerances to chemical, biological and host attack compared with those in planktonic forms. In vitro biofilm models have demonstrated that microbial biofilms can withstand antimicrobial concentrations 100–1000 times higher than that of their planktonic counterparts.3–6 This may go towards explaining why some wounds fail to heal with standard care and why chronic infections persist.7In vitro models assessing the effectiveness of many antimicrobials used in wound-related products have identified that these treatments often have variable and poor results against microbial cells in biofilm phenotypes.8–10
One potential explanation for this treatment failure is because the wound care treatments do not target or are ineffective against biofilm. Cadexomer iodine, however, has demonstrated superior efficacy against microbial biofilms when tested in vitro and in animal models, when compared against other topical antimicrobials used in wound care dressings.8–10
Therefore, as a pilot study massively parallel DNA sequencing, real-time quantitative PCR (qPCR), microscopy techniques [scanning electron microscopy (SEM) and fluorescence in situ hybridization (FISH)] and gel zymography were used to identify if the topical antimicrobial cadexomer iodine could reduce the microbial load of chronic non-healing DFUs complicated by biofilm in vivo. Additional interests were to explore the effects of cadexomer iodine on the microbial communities pre- and post-treatment, and determine the levels of wound proteases [matrix metalloproteinase (MMP)-2 and MMP-9]. It was not the intention of this pilot study to correlate any effects of microbial loads or diversity to wound healing outcomes.
Materials and methods
Ethics
Ethics approval for this study was granted by the South West Sydney Local Health District Research and Ethics Committee (HREC/14/LPOOL/487, SSA/14/LPOOL/489).
Exclusion criteria
Patients with clinical signs of infection as per the IDSA guidelines,1 known osteomyelitis that was associated with the DFU, or patients who had received any topical or systemic antimicrobial therapy 2 weeks prior to enrolment, were not eligible. The reason for excluding patients with clinical infection was the assumption that these wounds would be driven predominantly by planktonic microorganisms.11 Participants with general contraindications to the use of cadexomer iodine as per the manufacturer’s guidelines were also excluded.12
Subjects and sample collection
Individuals were eligible for the study if they had a chronic non-healing DFU defined as a wound of >6 weeks duration failing to respond to standard care.13 Cadexomer iodine was applied every second day over a 7 day treatment period (total of three applications). Sharp debridement of DFUs was withheld over the 7 day treatment period given this was likely to impact the results significantly.14 A tissue biopsy was obtained from the wound edge for each participant before and after treatment. All tissue samples were frozen at −80°C until completion of the last patient and processed in bulk so as to reduce any bias. Additionally, we collected broad demographics and wound metrics. Primary endpoint was a reduction in microbial load 7 days post-treatment. Secondary analysis included the exploration of community richness and diversity of DFUs pre- and post-treatment, visual changes to biofilm structures and alteration to levels of wound proteases (MMP-2 and MMP-9).
Cadexomer iodine (Iodosorb®, Smith & Nephew)
Iodosorb® ointment is designed as a carrier system enabling the delivery of iodine, which can penetrate the cell wall of microorganisms and disrupt protein and nucleic acid structure and synthesis.15 Iodosorb® consists of small polysaccharide beads (cadexomer) containing 0.9% iodine, which, in the presence of wound exudate, causes the polysaccharide beads to swell allowing a slow sustained release of iodine into the wound.
Specimen collection and storage
A 3 mm (width)×10 mm (depth) tissue punch biopsy was obtained from the edge of each DFU after cleansing the wound with 0.9% NaCl. Tissue biopsy samples were obtained for all participants at baseline and day 7. For SEM, an additional 2 mm (width) × 10 mm (depth) was necessary. Following removal, tissue samples were rinsed vigorously in a PBS bath to remove any coagulated blood and to reduce the number of planktonic microorganisms. Samples were cut transversely into two 1.5 mm pieces for DNA sequencing and FISH. DNA samples were immediately placed into RNAlater® (Ambion, Inc.) for 24 h at 4°C and then frozen at −80°C until DNA extraction. Tissue samples for FISH were immediately fixed in 4% paraformaldehyde overnight at 4°C, then transferred into PBS and frozen at −80°C. Tissue samples for SEM were immediately fixed in 3% glutaraldehyde overnight at 4°C, then transferred into 0.1% PBS and frozen at −80°C. All tissue samples remained at −80°C until study completion to reduce any bias and were processed in bulk.
Tissue processing workflow
DNA extraction
Five to ten milligrams of human chronic DFU biopsy samples was defrosted on ice prior to DNA extraction. Genomic DNA was extracted using a Mo Bio PowerBiofilm DNA isolation kit (Mo Bio cat. no. 24000–50) following the manufacturer’s instructions.
Next-generation DNA sequencing to determine bacterial diversity
Massively Parallel DNA sequencing was carried out by a commercial laboratory (Australian Centre for Ecogenomics) targeting the V4 region of the 16S rRNA gene using eubacterial universal primers 515F (TCGTCGG CAGCGT CAGATGTGTATAAGAGACAGCCAGCAGCYGCGGTAAN) and 806R (GTCTCG TGGGCTCGGAGATGTGTATAAGAGACAG GGACTA CHVGGGTWTCTAAT).
Preparation of the 16S library was performed as described at the Australian Centre for Ecogenomics using the workflow outlined by Illumina. In the first stage, PCR products of ∼466 bp were amplified according to the specified workflow with an alteration in polymerase used to substitute Q5 Hot Start High-Fidelity 2X Master Mix (New England Biolabs, Ipswich, MA, USA) in standard PCR conditions. Resulting PCR amplicons were purified using Agencourt AMPure XP beads (Beckman Coulter, Brea, CA, USA). Purified DNA was indexed with unique 8 bp barcodes using the Illumina Nextera XT 384 sample Index Kit A-D (Illumina FC-131–1002) in standard PCR conditions with Q5 Hot Start High-Fidelity 2X Master Mix. Indexed amplicons were pooled together in equimolar concentrations and sequenced on the MiSeq Sequencing System (Illumina) using paired end sequencing with V3 300 bp chemistry.
Sequence analysis and quality control
Reads in FASTQ format were imported to CLC genomics workbench version 8.5.1 using the microbial genome finishing module (CLC bio; Qiagen, Aarhus, Denmark), for sequence quality control and analysis. Workflows for sequence quality control and operational taxonomic units (OTUs) clustering were based on previously reported wound microbiome analysis.16 OTUs were defined as molecular proxies for describing organisms based on their phylogenetic relationships to other organisms, and were reported at either the genera or species level identification where possible.
Briefly, after sequence and quality control measures, reads were assigned to OTUs using SILVA17 at 99% similarity at the genus level and species level where possible. OTUs were aligned using MUSCLE18 to reconstruct a phylogenetic tree, allowing the estimation of the alpha diversity pre- and post-treatment for each DFU. This included both community richness (rarefaction) and community diversity (Shannon–Weaver Index). Rarefaction curves allow the estimation of the number of unique microbial taxa within a sample and the Shannon–Weaver Index is a measure of diversity that includes the number of unique microbial taxa and their relative evenness within each sample. Thus, a higher Shannon–Weaver Index correlates to a greater diversity.
qPCR to determine microbial load in DFU biofilms
We utilized qPCR to determine microbial load in DFU biofilms, as previously reported.19,20 To quantify the total amount of bacterial DNA/mg of tissue in each wound tissue sample we obtained the copy number of the 16S rRNA gene (copies/μL), which was normalized against the human 18S rRNA gene (copies/μL) in each chronic wound sample.
Characterization and visualization of DFU biofilm
The presence or absence of biofilms in chronic non-healing DFUs was confirmed through SEM or FISH. For the purpose of this study, we used definitions promoted by an expert group to characterize biofilm as being (i) microbial aggregates surrounded by a self-produced or host-derived matrix adhering to natural or artificial surfaces in the host, or (ii) aggregates associated with, but not directly adherent to, the surface.21
FISH
Two to three millimetres of DFU tissue was embedded in optimal cutting temperature (OCT) embedding matrix (Tissue-Plus™ O.C.T Compound, Thermo Fisher Scientific, Waltham, MA, USA) and frozen at −80°C overnight. DFU tissues were sectioned to a thickness of 6 μm and mounted on SuperFrost Plus slides (Thermo Fisher Scientific). For visualization of microorganisms and biofilm, confocal laser scanning microscopy was combined with FISH. The hybridization process used was previously described by Thurnheer et al.22 Briefly, two different probes were utilized for in situ hybridization: Fluor 488-labelled universal probe23 (final concentration 5 ng/μL); and Cy3-labelled Staphylococcus spp.-specific probe (final concentration 5 ng/μL).24 All images were examined under confocal laser scanning microscopy (Carl Zeiss Ltd, Herefordshire, UK). All images were processed using ZEISS ZEN Imaging Software (black edition).
SEM and image interpretation
In vivo microbial biofilms associated with DFU tissue were sampled at 5–200 μm for optimal visualization through SEM.25 Two to 3 mm of DFU tissue was fixed in 3% glutaraldehyde, followed by three washes of 0.1 M PBS prior to serial ethanol dehydration and hexamethyldisilazane (Polysciences, Inc., Warrington, PA, USA), as described previously.20 Dried samples were coated with 20 nm gold film and examined using SEM. Each sample was scored based on the amount of bacteria/biofilm observed using an arbitrary five-point scale as previously reported:26 score 0 = no bacteria observed; 1 = single individual cells; 2 = small microcolonies (∼10 cells); 3 = large microcolonies (∼100 cells); 4 = continuous film; and 5 = thick continuous film.
Determination of wound proteases
Wound fluids were collected from DFUs pre- and post-treatment and stored at −80°C for quantification of MMPs (MMP-2 and total MMP-9) by gel band zymography, as previously described.27
Statistics
Wound metrics and microbiome data were analysed through Statistical Package for Social Sciences Version 23 (IBM, Armonk, NJ, USA). Wilcoxon signed-rank tests for paired samples of non-parametric data were performed on pre- and post-treatment microbial log10 reductions. χ2 was used to correlate OTUs and Shannon–Weaver indices to log10 reduction. One-way ANOVA was used to estimate variances between MMP levels before and after treatment. Logistic regression was employed to correlate microbial reduction to MMP levels. CLC genomics workbench version 8.5.1 in combination with the microbial genome-finishing module (CLC bio; Qiagen) was used to analyse DNA sequence data. For all comparisons and modelling, the level of significance was set at P < 0.05. Data are given as mean, standard deviation (±) and 95% CI.
Results
A total of 17 patients with chronic non-healing DFUs were enrolled. One ulcer from each patient was biopsied before and after treatment. Patient demographics and wound metrics are shown in Table 1. Microbial load was determined via qPCR for all 17 DFU samples pre- and post-treatment. However, three samples were removed from exploring community richness (rarefaction) and diversity (Shannon–Weaver Index) due to a low number of 16S rRNA gene sequence reads, leaving 12 samples available for analysis. In total (for these 12 samples), 703 346 high-quality DNA sequences were generated (before = 384 772 and after = 318 574), with a median of 31 452 per sample level data (range 1137–61 820). The clustering of OTUs identified 1976 unique taxa from which low abundance OTUs were removed (<0.1%), leaving 368 OTUs for further analysis. Only eight wound fluid samples were available for analysis. Protein concentrations in the remaining samples were too low for analysis.
Table 1.
Patient demographics and wound metrics at baseline
| Patient demographics | |
| male | 11 (65%) |
| female | 6 (35%) |
| type 1 diabetes | 2 (12%) |
| type 2 diabetes | 15 (88%) |
| mean age in years | 66 (±13.6) |
| Wound metrics | |
| location of ulcer | |
| plantar metatarsal head | 8 |
| calcaneum | 3 |
| dorsal foot | 2 |
| ankle | 2 |
| hallux | 2 |
| duration of ulcer in weeks | |
| mean duration of ulcer at baseline | 25 (±20.7, range 6–72) |
| University of Texas wound classification | |
| 1A | 10 (59%) |
| 1C | 1 (5.8%) |
| 2A | 5 (29.4%) |
| 2C | 1 (5.8%) |
| size of ulcer | |
| mean DFU size at baseline | 3.7 × 2.7 cm (L × W) |
Confirmation of the presence or absence of biofilms in each DFU
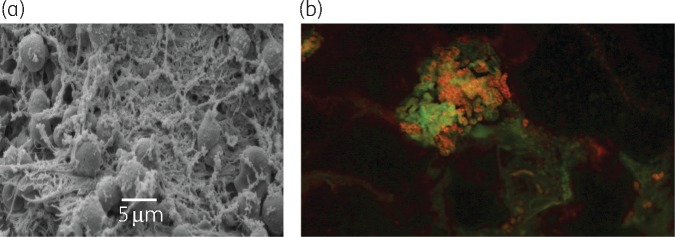
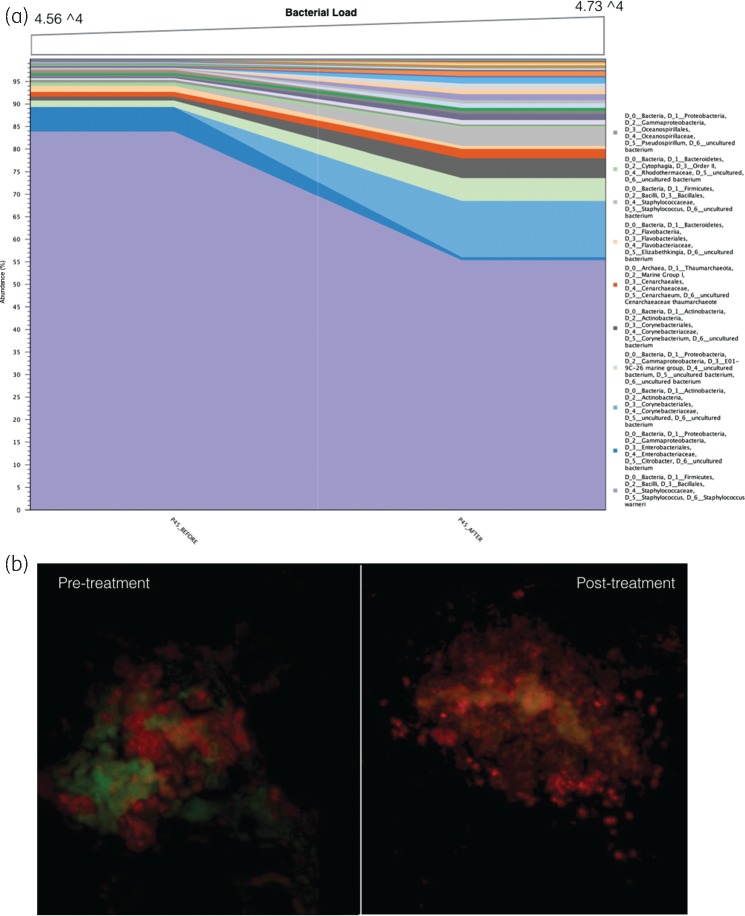
The presence of biofilm was visualized and confirmed in all 17 participants using SEM, FISH or both (Figure 1a and b). Biofilm architecture was graded (via SEM) using an arbitrary sliding scale previously reported.26 The median value of DFU biofilm architecture reduced between pre- and post-treatment samples; pre-treatment median was 4 (large microcolonies ∼100 cells and a continuous film/matrix) and the post-treatment median was 3 (large microcolonies ∼100 cells).
Figure 1.
(a) SEM from a sample pre-treatment. Microbial aggregates with self-produced or host-derived matrix that we describe as a stringy-like glue appearance. SEM imaged at 5 μm. (b) FISH image depicts microbial aggregates of mixed species biofilm [Fluor 488-labelled universal bacterial probe (red) and Cy3-labelled Staphylococcus spp.-specific probe].
Reduction in microbial load of chronic non-healing DFUs complicated by biofilm
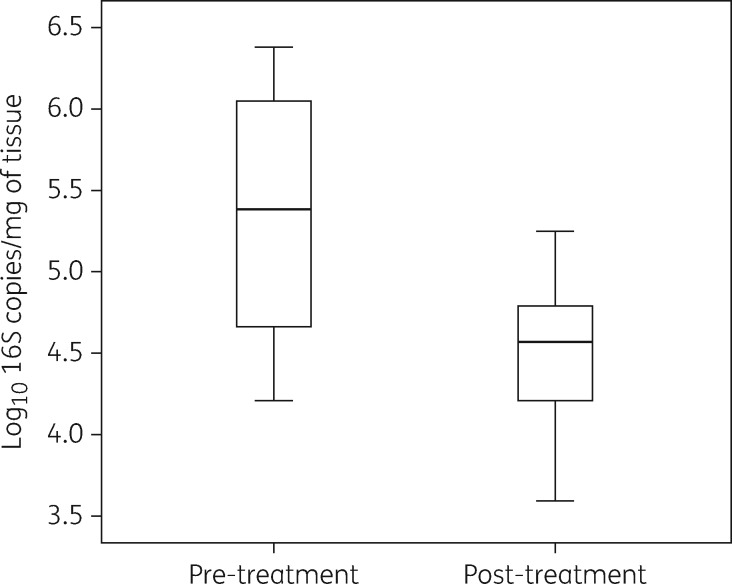
The application of cadexomer iodine resulted in 11 samples achieving up to and greater than a 1 log10 reduction (mean microbial load pre-treatment = 5.92 log10 16S copies/mg of tissue versus 4.56 log10 16S copies/mg of tissue, P = 0.02, 95% CI 3.43 to 4.61 log10) (Figure 2). Six samples had no change or increases in log10 (mean microbial load pre-treatment = 5.22 log10 16S copies/mg of tissue versus 5.20 log10 16S copies/mg of tissue).
Figure 2.
Effect of cadexomer iodine pre- and post-treatment. Boxplots show median and IQR, with whiskers showing the range of log10 values of all 17 patients.
Analysis of community richness and diversity of chronic non-healing DFUs treated with cadexomer iodine
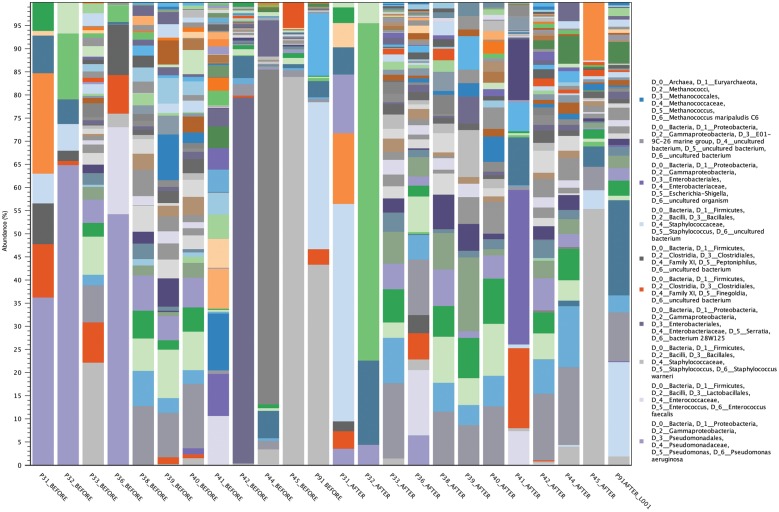
The most abundant OTUs (based on the number of DNA sequences represented when pooling samples) are noted in Figure 3. The richness and diversity of chronic non-healing DFUs pre- and post-treatment are also reported at the individual sample level of microorganisms contributing to >5% of abundance in rank order (Table 2). Post-treatment identified diversity shifts with increasing environmental contaminants. These microorganisms are identifiable only by molecular methods (in the majority), and included in the Proteobacteria-E01-9C-26 Marine Group, Proteobacteria-ARKDMS-49, Archaea-Cenarchaeum, Elizabethkingia spp., Bacteroidetes-Rhodothermacae and Proteobacteria-Rhodothalssium. Furthermore, some of these microorganisms are known extremophiles, existing in hostile, niche environments.
Figure 3.
Most frequent microorganisms based on the pooling of all DNA sequences and the total number of DNA sequences attributed to each microorganism. This graph also exemplifies the contrast between low- and high-frequency taxa at the genus/species level. High-frequency taxa consist of a few predominant microorganisms (typically between 1 and 9 microorganisms; samples 31, 32, 36, 42, 44, 45 and 91) and low-frequency taxa consist of multiple microorganisms (typically 10-fold in comparison with high-frequency taxa; >30–90 microorganisms; samples 33, 38, 39, 40 and 41).
Table 2.
Pre- and post-treatment community diversity in microorganisms contributing to >5% abundance of each individual sample
| Genera | Samples | Average abundance (%) | Aerotolerance |
|---|---|---|---|
| Pre-treatment | |||
| Pseudomonas spp. | 5 | 58.5 | aerobe |
| E01-9C-26 Marine | 4 | 11.2 | unknown |
| Staphylococcus spp. | 4 | 58 | facultative |
| Rhodothermaceae spp. | 3 | 5 | unknown |
| Finegoldia spp. | 3 | 7.8 | anaerobe |
| Elizabethkingia meningoseptica | 3 | 6.3 | aerobe |
| Cornyebacterium spp. | 3 | 5.3 | aerobe |
| Peptoniphilus spp. | 2 | 9.5 | anaerobe |
| Ananerococcus spp. | 2 | 9.2 | anaerobe |
| Proteus penneri | 2 | 20.5 | facultative |
| Post-treatment | |||
| E01-9C-26 Marine | 8 | 10 | unknown |
| ARKDMS49 | 7 | 5.7 | unknown |
| Cenarchaeum | 5 | 5.1 | unknown |
| Cyanobacteria—subsection I | 5 | 7.4 | unknown |
| Rhodothermaceae spp. | 4 | 5.8 | unknown |
| Rhodothalassium spp. | 3 | 7 | unknown |
| Cornyebacterium spp. | 3 | 11 | aerobe |
| Elizabethkingia meningoseptica | 3 | 6.6 | aerobe |
| Staphylococcus spp. | 3 | 42.8 | facultative |
| Proteus penneri | 2 | 39 | facultative |
The taxonomic richness and diversity of pre- and post-treatment pooled samples identified no differences between mean OTUs (pre-treatment = 115, SD = 69.3 versus post-treatment = 112, SD = 74.3, P = 0.53) and mean Shannon–Weaver indices (pre-treatment = 2.8, SD 1.2 versus post-treatment = 3, SD = 1.4, P = 0.58). When exploring this at the individual sample level there was large heterogeneity between pre- and post-treatment OTUs and Shannon–Weaver indices. To ascertain if the number of unique OTUs and their relative abundance were correlated to reductions in log10 values, a χ2 test was performed. Reductions in log10 did not correlate to reductions in either OTUs (P = 0.85) or Shannon–Weaver indices (P = 0.72).
MMP levels pre- and post-treatment
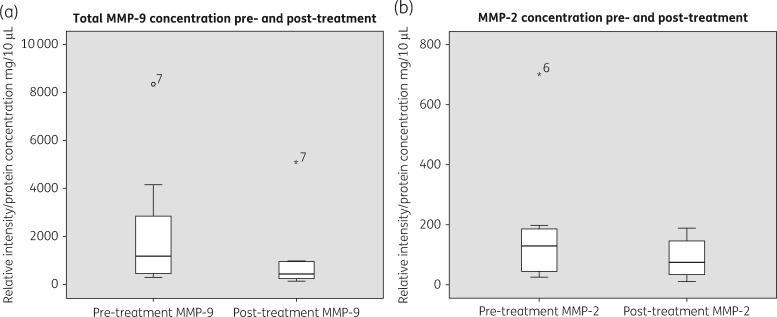
The mean total MMP-9 and MMP-2 levels in eight samples reduced following 7 days application of cadexomer iodine (Figure 4). However, only total MMP-9 reached statistical significance (mean total MMP-9 before = 2202 mg/10 μL versus mean total MMP-9 after = 1065 mg/10 μL, 95% CI −14.5 to 2287, P = 0.05). Mean MMP-2 before = 181.9 versus mean MMP-2 after = 89 (95% CI −61.3 to 246.7, P = 0.197). In general, any reductions in the levels of MMPs were correlated to reductions in the microbial load (P = 0.03).
Figure 4.
(a) Total MMP-9 values pre- and post-treatment. (b) MMP-2 values pre- and post-treatment. Boxplots show the median and IQR, with whiskers showing the range apart from outliers (>1.5×IQR).
Discussion
Using a combination of DNA sequencing, qPCR, SEM and FISH, the ecology of wounds and presence of microbial biofilms was explored. To the best of our knowledge, we are the first to employ these suites of molecular and microscopy techniques to show that cadexomer iodine can reduce the microbial load of chronic non-healing DFUs complicated by biofilm. We also show that in reducing microbial load, concomitant reductions in wound proteases are also achieved.
Massively parallel DNA sequencing allowed the exploration of chronic non-healing DFU microbiome, and provided useful insights that these wounds support complex polymicrobial communities. Molecular methods also demonstrated that cadexomer iodine had a broad level of antimicrobial activity in reducing both facultative anaerobes such as Staphylococcus spp., Serratia spp., Pseudomonas spp. and obligate anaerobes including Clostridiales family XI.
Both facultative and obligate anaerobes were detected together in chronic non-healing DFU samples positive for biofilm presence. Microelectrode and transcriptomic analysis of in vitro biofilms have identified that the interior of biofilms house niche areas of altered pH levels, oxygen and nutrient depletion.3,28 Oxygen in particular is noted for its depletion at the substratum layers of biofilms and in the centre of microcolonies, explaining why wounds complicated by biofilm can harbour such diverse microorganisms. This may indirectly support the action of cadexomer iodine against biofilms, as it suggests penetration to deeper areas that house obligate anaerobes is possible, as identified by 16S rRNA of pre- and post-treatment reductions in obligate anaerobes.
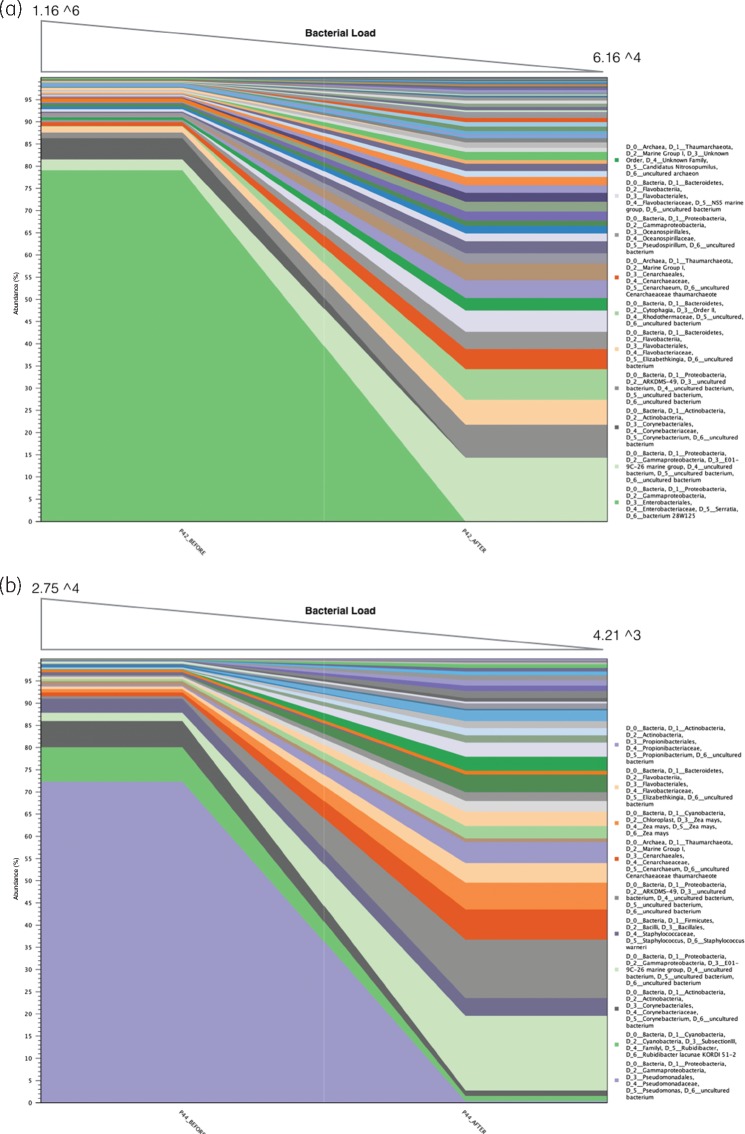
Using a combined molecular and microscopy approach can provide an extended understanding of the effects of antimicrobials on wounds. This approach for example provided insights into how cadexomer iodine affected microbial populations. Using sample level data, Serratia spp. were identified as contributing to 75% of abundance pre-treatment and 0% post-treatment in sample 42 (Figure 5a). This also correlated with a >1 log10 reduction in microbial load as determined by qPCR. Similarly, for sample 44, P. aeruginosa contributed to 88% of abundance pre-treatment and 0.3% post-treatment, with a 0.76 log10 reduction in microbial load (Figure 5b). This suggests that cadexomer iodine was effective in reducing the abundant microorganisms in these cases.
Figure 5.
Pre- and post-treatment community diversity identified through 16S rRNA sequencing. (a) In the left-hand side, sample 42 identifies Serratia spp. contributing to 75% abundance pre-treatment, and in the right-hand side, this reduces to 0% post-treatment. Top of the graph identified the reduction in microbial load by over 1 log10 reduction determined through qPCR (99% reduction in microorganisms). Therefore, the left-hand pane is a reflection of the remaining 1% of microorganisms. (b) Sample 44 identified Pseudomonas spp. contributing to 88% abundance pre-treatment and 0.3% post-treatment with a 0.83 log10 reduction.
In contrast, there was no log10 reduction in microbial load for sample 45 (pre-treatment = 4.565 and post-treatment = 4.735). While community profiling revealed a minor reduction in Staphylococcus spp. (Figure 6a), this was countered with a concomitant increase in other microorganisms. In this case, FISH was able to demonstrate the lack of action against the bulk of microorganisms in sample 45, with pre- and post-treatment images depicting the presence of microbial aggregates as biofilm in chronic non-healing DFU tissue samples (Figure 6b).
Figure 6.
(a) Sample 45 identifies a small reduction in S. aureus. This is countered by a concomitant increase in other microorganisms allowing an increase in community diversity, but not overall microbial load. (b) This lack of action is also identified through FISH. Red colour represents a universal probe (Fluor 488) and the green represents a species-specific probe for S. aureus (Cy3). Some reduction in S. aureus aggregates post-treatment is noted, but the biofilm remains in bulk.
In five patients, no log10 reduction was noted with the use of cadexomer iodine. Considering the accepted notion that biofilms are tolerant to antimicrobials, and combining the potential attributes of extremophile (and non-extremophile) microorganisms, this may explain why some wound microorganisms were tolerant to treatment. Post-treatment microbiome analysis identified that these microorganisms increased in abundance from previously low numbers, whereas skin flora microorganisms such as Pseudomonas spp. decreased in abundance following treatment (indicating cadexomer iodine were effective). This diversity shift may have occurred as nutrient availability increased or where mutual benefit arose.29
Study limitations
The primary aim of this study was to ascertain the effects of cadexomer iodine on microbial populations associated with the presence of biofilm, however without the controlled conditions afforded by in vitro biofilm models we are not able to fully rule out that planktonic microorganisms contaminated samples. Our rationale to provide a strong argument that planktonic microorganisms were of a negligible proportion of microbial cells, with the bulk of cells being biofilm, is supported by our visualization techniques using SEM and FISH. These methods identified significant aggregates of microbial cells with extracellular polymeric substance (EPS). Furthermore, we vigorously rinsed all tissue samples during preparation as a method to reduce planktonic microorganisms not adhered to tissue. Alternative methods to reduce planktonic microorganisms from contaminating biofilm models have only been alluded to from ex vivo animal explants,10 with no reports of this approach on human tissue.
To measure total microbial load, qPCR was utilized as the technique of choice, with previous wound-related PCR studies adopting this methodology.30,31 We acknowledge with this approach the inability of qPCR (based on 16S rRNA gene) to differentiate live or dead bacteria. Other commonly employed techniques such as molecular viability testing (pre-RNA analysis or RT-qPCR) are able to detect live or dead microorganisms. The problem with using these techniques for our diabetic ulcer samples is the predominance of biofilm phenotype cells present in the tissue. Pre-RNA rapidly responds to nutrition stimulation and target metabolism, not actual microbial load. Given that biofilm cells have low metabolism in comparison with planktonic cells, this may lead to variations in the 16S pre-RNA level between the biofilm and planktonic cells and different growth conditions.32 The log reductions noted in this study therefore represent the minimal response and we acknowledge that some of the bacteria detected by qPCR could be dead, resulting in a lower calculated efficacy for cadexomer iodine.
Lastly, only eight samples with adequate protein concentration were available for analysis. In the majority, obtaining enough wound fluid post-treatment was difficult, which may be associated with reduced wound inflammation with reductions in microbial loads. Alternatively, the time of dipstick applications were undertaken during 30 min appointments and this may not have been long enough to allow the wounds to leak adequately onto the three dipsticks required.
Acknowledgements
We would like to acknowledge the support of South West Sydney LHD who presented the lead author with an early career research award, allowing the undertaking of this project as part of a PhD thesis.
Funding
This work was supported by a research grant awarded by Smith & Nephew that contributed to the analysis of tissue samples using DNA sequencing and microscopy techniques.
Transparency declarations
M. M. received a consultancy fee from Smith & Nephew to undertake work not associated with this study, but pertaining to producing educational material on biofilms in chronic wounds. All other authors: none to declare.
References
- 1. Lipsky BA, Berendt AR, Cornia PB. et al. Infectious Diseases Society of America clinical practice guideline for the diagnosis and treatment of diabetic foot infections. Clin Infect Dis 2012; 54: e132–73. [DOI] [PubMed] [Google Scholar]
- 2. Wolcott RD. Biofilms and chronic wound inflammation. J Wound Care 2008; 17: 333–41. [DOI] [PubMed] [Google Scholar]
- 3. Walters MC, Roe F, Bugnicourt A. et al. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob Agents Chemother 2003; 47: 317–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Anwar H, Costerton JW.. Enhanced activity of combination of tobramycin and piperacillin for eradication of sessile biofilm cells of Pseudomonas aeruginosa. Antimicrob Agents Chemother 1990; 34: 1666–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Machado I, Graça J, Lopes H. et al. Antimicrobial pressure of ciprofloxacin and gentamicin on biofilm development by an endoscope-isolated Pseudomonas aeruginosa. ISRN Biotechnol 2012; 2013: 178646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stewart PS, Costerton JW.. Antibiotic resistance of bacteria in biofilms. Lancet 2001; 358: 135–8. [DOI] [PubMed] [Google Scholar]
- 7. Bjarnsholt T, Kirketerp-Møller K, Jensen PØ. et al. Why chronic wounds will not heal: a novel hypothesis. Wound Repair Regen 2008; 16: 2–10. [DOI] [PubMed] [Google Scholar]
- 8. Fitzgerald DJ, Renick PJ, Forrest EC. et al. Cadexomer iodine provides superior efficacy against bacterial wound biofilms in vitro and in vivo. Wound Repair Regen 2016; doi:10.1111/wrr.12497. [DOI] [PubMed] [Google Scholar]
- 9. Hill KE, Malic S, McKee R. et al. An in vitro model of chronic wound biofilms to test wound dressings and assess antimicrobial susceptibilities. J Antimicrob Chemother 2010; 65: 1195–206. [DOI] [PubMed] [Google Scholar]
- 10. Phillips PL, Yang Q, Davis S. et al. Antimicrobial dressing efficacy against mature Pseudomonas aeruginosa biofilm on porcine skin explants. Int Wound J 2015; 12: 469–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wolcott R, Costerton JW, Raoult D. et al. The polymicrobial nature of biofilm infection. Clin Microbiol Infect 2013; 19: 107–12. [DOI] [PubMed] [Google Scholar]
- 12. Schwartz JA, Lantis JC, Gendics C. et al. A prospective, non comparative, multicenter study to investigate the effect of cadexomer iodine on bioburden load and other wound characteristics in diabetic foot ulcers. Int Wound J 2013; 10: 193–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Frykberg RG, Banks J.. Challenges in the treatment of chronic wounds. Adv Wound Care (New Rochelle) 2015; 4: 560–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wolcott RD, Rumbaugh KP, James G. et al. Biofilm maturity studies indicate sharp debridement opens a time-dependent therapeutic window. J Wound Care 2010; 19: 320–8. [DOI] [PubMed] [Google Scholar]
- 15. Edwards-Jones V. Essential Microbiology for Wound Care. Oxford University Press, 2016. [Google Scholar]
- 16. Gardner SE, Hillis SL, Heilmann K. et al. The neuropathic diabetic foot ulcer microbiome is associated with clinical factors. Diabetes 2013; 62: 923–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Quast C, Pruesse E, Yilmaz P. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 2013; 41: D590–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 2004; 5: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hu H, Jacombs A, Vickery K. et al. Chronic biofilm infection in breast implants is associated with an increased T-cell lymphocytic infiltrate: implications for breast implant-associated lymphoma. Plast Reconstr Surg 2015; 135: 319–29. [DOI] [PubMed] [Google Scholar]
- 20. Jacombs A, Allan J, Hu H. et al. Prevention of biofilm-induced capsular contracture with antibiotic-impregnated mesh in a porcine model. Aesthet Surg J 2012; 32: 886–91. [DOI] [PubMed] [Google Scholar]
- 21. Høiby N, Bjarnsholt T, Moser C. et al. ESCMID guideline for the diagnosis and treatment of biofilm infections 2014. Clin Microbiol Infect 2015; 21 Suppl 1: S1–25. [DOI] [PubMed] [Google Scholar]
- 22. Thurnheer T, Gmür R, Guggenheim B.. Multiplex FISH analysis of a six-species bacterial biofilm. J Microbiol Methods 2004; 56: 37–47. [DOI] [PubMed] [Google Scholar]
- 23. Amann RI, Binder BJ, Olson RJ. et al. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol 1990; 56: 1919–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Trebesius K, Leitritz L, Adler K. et al. Culture independent and rapid identification of bacterial pathogens in necrotising fasciitis and streptococcal toxic shock syndrome by fluorescence in situ hybridisation. Med Microbiol Immunol 2000; 188: 169–75. [DOI] [PubMed] [Google Scholar]
- 25. Bjarnsholt T, Alhede M, Alhede M. et al. The in vivo biofilm. Trends Microbiol 2013; 21: 466–74. [DOI] [PubMed] [Google Scholar]
- 26. Han A, Zenilman JM, Melendez JH. et al. The importance of a multifaceted approach to characterizing the microbial flora of chronic wounds. Wound Repair Regen 2011; 19: 532–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Min D, Lyons JG, Jia J. et al. 2-Methoxy-2,4-diphenyl-3(2H)-furanone-labeled gelatin zymography and reverse zymography: a rapid real-time method for quantification of matrix metalloproteinases-2 and -9 and tissue inhibitors of metalloproteinases. Electrophoresis 2006; 27: 357–64. [DOI] [PubMed] [Google Scholar]
- 28. James GA, Zhao AG, Usui M. et al. Microsensor and transcriptomic signatures of oxygen depletion in biofilms associated with chronic wounds. Wound Repair Regen 2016; 24: 373–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hibbing ME, Fuqua C, Parsek MR. et al. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol 2010; 8: 15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gentili V, Gianesini S, Balboni PG. et al. Panbacterial real-time PCR to evaluate bacterial burden in chronic wounds treated with Cutimed™ Sorbact™. Eur J Clin Microbiol Infect Dis 2012; 31: 1523–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Redel H, Gao Z, Li H. et al. Quantitation and composition of cutaneous microbiota in diabetic and nondiabetic men. J Infect Dis 2013; 207: 1105–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Weigel KM, Jones KL, Do JS. et al. Molecular viability testing of bacterial pathogens from a complex human sample matrix. PLoS One 2013; 8: e54886. [DOI] [PMC free article] [PubMed] [Google Scholar]