Abstract
Being continuously exposed to variable environmental conditions, plants produce phytohormones to react quickly and specifically to these changes. The phytohormone ethylene is produced in response to multiple stresses. While the role of ethylene in defense responses to pathogens is widely recognized, recent studies in arabidopsis and crop species highlight an emerging key role for ethylene in the regulation of organ growth and yield under abiotic stress. Molecular connections between ethylene and growth-regulatory pathways have been uncovered, and altering the expression of ethylene response factors (ERFs) provides a new strategy for targeted ethylene-response engineering. Crops with optimized ethylene responses show improved growth in the field, opening new windows for future crop improvement. This review focuses on how ethylene regulates shoot growth, with an emphasis on leaves.
Keywords: ethylene signaling, ethylene response factors, growth regulation, leaf growth, stress response
Highlights
An increasing number of transcriptome studies in plants exposed to biotic or abiotic stress highlight a role for ethylene under a broad range of stresses.
The role of ethylene under stress is dual: it regulates a defense response, mostly in full-grown leaves, and a growth response in young leaves.
In young leaves, ethylene and the downstream ERFs emerge as central regulators of leaf growth inhibition, orchestrating both cell division and cell expansion.
The knowledge of ethylene-mediated growth inhibition can be successfully implemented in crops to improve plant growth and stress tolerance.
Adapting Plant Growth to the Environment: Why and How?
The sessility of plants is undoubtedly their most disadvantageous feature compared to other living organisms, and implies that their survival can be threatened by environmental perturbations. However, plants have developed fascinating mechanisms enabling rapid detection of changing conditions accompanied by highly complex molecular responses, resulting in remarkable phenotypic plasticity. During the vegetative growth stage, one tightly controlled process is plant growth. Under favorable conditions, root and shoot growth is crucial to enable continuous nutrient uptake and energy production through photosynthesis, respectively. Leaf growth, for example, is controlled by no less than six different cellular mechanisms, including precise orchestration of the switch between cell division, that drives the growth of very young leaf primordia, and cell expansion and differentiation (reviewed in [1]). By contrast, sustaining growth under unfavorable conditions could be detrimental. For example, growth under drought stress would increase the evaporative surface of the plant, rendering the plant even more susceptible. Plants thus constantly evaluate whether the environmental signals are favorable for growth or not, and redirect their resources either for growth or for stress defense.
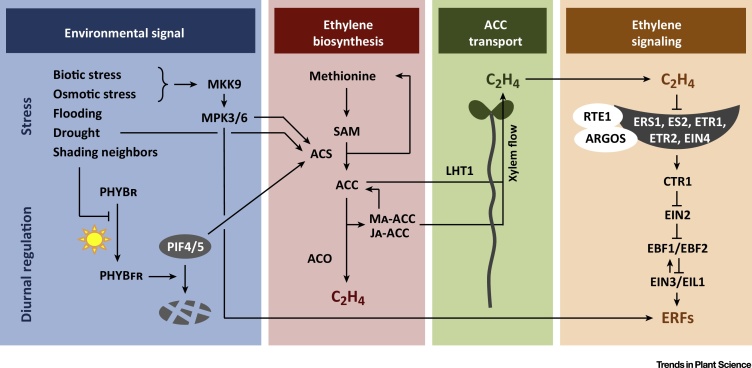
At the physiological level, the integration of environmental signals into proper phenotypic responses is orchestrated by phytohormones. Ethylene, the smallest phytohormone with the simple C2H4 structure, is gaseous and therefore enables plant-to-plant communication. Since its discovery around one century ago, the multiple facets of this hormone as a signaling molecule have fascinated scientists, and this led to the unraveling of its biosynthesis and signaling (Box 1 and Figure 1), and the identification of its various functions: regulation of leaf development, senescence, fruit ripening, stimulation of germination, etc. Importantly, ethylene is produced in response to multiple environmental stresses (Figure 1), both abiotic and biotic, suggesting that it acts as a bridge between a changing environment and developmental adaptation. The abiotic stress conditions that trigger ethylene synthesis include submergence, heat, shade, exposure to heavy metals and high salt, low nutrient availability, and water deficiency 2, 3, 4, 5, 6.
Box 1. Recent Advances in Ethylene Biosynthesis and Signaling.
The ethylene biosynthesis pathway consists of a simple, three-step process: methionine is converted into S-adenosyl methionine (SAM; see Glossary), which is further converted by ACC-synthases (ACS) to ACC, the direct precursor of ethylene (Figure 1). Recycling of methylthioadenosine enables rapid ethylene biosynthesis when necessary [85]. Because the conversion from ACC to ethylene is an exothermic reaction that only requires oxygen, ethylene biosynthesis is regulated at the level of ACS enzymes, which are also under post-translational control: they can be phosphorylated before ubiquitin-mediated protein degradation by, for instance, ETO1 and CUL3 86, 87. ACS induction and activation are responsive to environmental factors that trigger ethylene accumulation. As such, ACS genes are transcriptionally induced by drought [5] and by shade, under the control of PIF4 [58]. ACS2 and ACS6 are post-translationally activated through phosphorylation by a MAPK-phosphorylation cascade involving MKK9 and MPK3/6 [88]. ACC levels are also regulated by conjugation and release from conjugates such as malonyl- or jasmonyl-ACC [89]. The soluble ethylene precursor ACC can be taken up by the amino acid transporter LHT1 and further transported through the plant via the xylem (Figure 1) [90].
In the destination organ, ethylene triggers a signaling cascade initiated by ethylene receptors in the ER and Golgi membrane: ERS1 (ETHYLENE RESPONSE SENSOR 1), ERS2, ETR1 (ETHYLENE RESISTANCE 1), ETR2 and EIN4 (ETHYLENE INSENSITIVE 4). These receptors are active in the absence of ethylene, and their activity can be controlled by complex formation with RTE1 (REVERSION TO ETHYLENE SENSITIVITY) and ARGOS proteins: these are positive regulators of the ethylene receptors, and thus are negative regulators of ethylene sensitivity 11, 91, 92. In the absence of ethylene, active receptors subsequently bind to and thereby activate the CTR1 protein [93]. The levels of the receptors are regulated by ethylene and CTR1: slightly increasing ethylene levels stimulate the transcription of the receptors and stabilization of CTR1, whereas higher ethylene levels push the receptor/CTR1 towards proteasome-mediated degradation [94]. CTR1 is a kinase that represses EIN2, an ER-located membrane protein. When this repression is released in the presence of ethylene, EIN2 is dephosphorylated and cleaved, releasing a C-terminal fragment that either moves to P-bodies or to the nucleus 95, 96. The downstream mode of action of the EIN2 fragment has long been a mystery, but recent studies have shown that it is involved in gene-specific regulation of translation 95, 96. The EIN2 fragment binds to the 3'-untranslated regions (3'-UTRs) of EBF1 and EBF2 transcripts, thereby repressing their translation. EBF1 and EBF2 are two central F-box proteins that target the primary ethylene-responsive TFs EIN3 and EIN3-LIKE 1 (EIL1) for protein degradation in the absence of ethylene 97, 98. In the presence of ethylene, EIN3 and EIL1 induce the expression of numerous secondary transcription factors (TFs), the ERFs [99]. The activity of some ERFs has been reported to be increased by phosphorylation through the MPK3/6-cascade that also regulates ethylene biosynthesis, providing dual-level regulation of the ERF-mediated response 24, 100.
Alt-text: Box 1
Figure 1.
Overview of Ethylene Biosynthesis and Signaling Pathways in Arabidopsis and Environmental Factors That Modulate Ethylene Signaling. Ethylene is synthesized from the amino acid methionine by a three-step pathway (Box 1). Synthesis of the intermediate product 1-aminocyclopropane-1-carboxylic acid (ACC) by ACS enzymes is rate-limiting and controlled by numerous environmental conditions including biotic, osmotic, and drought stress. Ethylene biosynthesis is also diurnally regulated by the red:far-red light ratio. Light-activated PHYB (PHYBFR) binds to and degrades PIF4/5, which can no longer induce ACS transcription. Shading by neighboring plants also influences the PHYB–PIF4/5 pathway. The direct ethylene precursor ACC can be transported through the xylem via the LHT1 transporter or can be conjugated into malonyl-ACC (Ma-ACC) or jasmonyl-ACC (JA-ACC), which are also transported through the xylem. In the destination organ, ethylene targets ethylene receptors, and thus relieves CTR1 inhibition of EIN2. EIN2 activation triggers the stabilization of EIN3 and EIL1, primary transcription factors that further control the expression of the downstream ERFs (Box 1).
Ethylene: An Inhibitor of Leaf Growth
Arabidopsis (Arabidopsis thaliana) plants overproducing ethylene are generally dwarfed, and plant growth is reduced by exposure to ethylene 7, 8, 9. Consequently, when the positive regulators of the ethylene signaling pathway (Box 1 and Figure 1) are mutated, plants are generally found to have larger rosettes with larger leaves in comparison to control plants. Increased growth has, for example, been observed upon mutation the endoplasmic reticulum (ER)- anchored protein EIN2 [10]. Conversely, mutants of negative regulators of ethylene signaling, such as the receptors ETR1 and ERS1 (Box 1), show a growth decrease [9]. Accordingly, overexpression of the negative regulators ARGOS or ARGOS-LIKE (ARL) stimulates leaf growth in arabidopsis 11, 12. Moreover, plant lines in which the ethylene sensitivity is reduced, or treatments reducing sensitivity to ethylene, cause larger leaves. For instance, plants overexpressing NEIP2 or TCTP, genes encoding proteins interacting with the Nicotiana tabacum ethylene receptor, show decreased ethylene sensitivity but improved growth 13, 14. Similarly, Pseudomonas frederiksbergensis, a soil bacterium that reduces plant sensitivity to ethylene, promotes the growth of red pepper plants [15]. Finally, some rhizosphere bacteria that promote plant growth do so by expressing ACC-DEAMINASE, decreasing the levels of 1-aminocyclopropane-1-carboxylic acid (ACC) in plants exposed to stress, and this has a positive effect on growth [16].
Exceptionally, ethylene has been reported to stimulate leaf growth. In the presence of very low ethylene concentrations, Poa alpina and Poa compressa show increased leaf elongation rates [17], and also the primary leaves of sunflower (Helianthus annuus) are enlarged [18]. However, the opposite effect was observed as soon as ethylene levels are increased to concentrations higher than this low growth-promoting optimum. This general negative correlation between ethylene sensitivity and leaf growth has led to the classification of ethylene as a growth-repressing hormone.
Effects of Ethylene on Cell Division
In plants, where growth mainly occurs post-embryonically through well-orchestrated cell divisions, the progression through the cell cycle is tightly governed by more than 70 core cell-cycle proteins (reviewed in [19]). Controlled by endogenous cues and environmental signals, cell-cycle progression and regulation vary depending on the plant organ, and the effect of ethylene is similarly organ-dependent. For instance, during the early development of the apical hook, ethylene participates in stimulating cell divisions, although its contribution is not crucial for curving of the apical hook [20]. Moreover, ethylene and the downstream transcription factors (TFs) ERF018 and ERF109 promote cell division during vasculature development in arabidopsis stems [21]. Thus, in these specific developmental contexts, ethylene can have a positive effect on cell division.
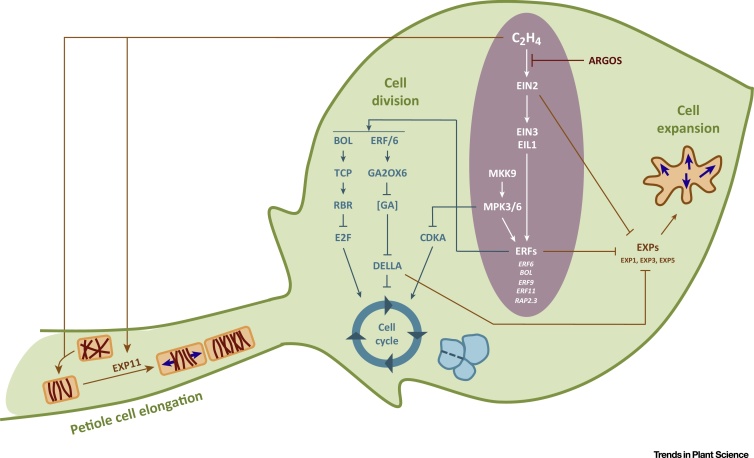
In leaves of plants exposed to environmental stress, ethylene appears to have a negative effect on the cell cycle. When plants are exposed to less than 10 h of osmotic stress, ethylene mediates a temporary and reversible stop of the cell cycle. This is likely to occur through the inactivation of the CDKA by phosphorylation, possibly through the MPK3/6 pathway but independently from EIN3/EIL1 (Figure2) [2]. Moreover, at least four mechanisms in leaves link ethylene to the exit of cell division and a shift to endoreduplication and differentiation. First, accumulation of ethylene and induction of the BOLITA TF (an ERF, Table 1) triggers the activation of type II TEOSINTE BRANCHED 1/CYCLOIDEA/PCF (TCP) genes (Figure 2) [22]. These TCP proteins bind to the promoter of RETINOBLASTOMA RELATED 1 (RBR1), and the encoded protein phosphorylates E2Fa and thus represses the transcription of the E2F target genes, thereby inhibiting progression into the S-phase and cell division. Second, ethylene induces the expression of ERF5 and ERF6, two closely related TFs, in actively growing leaves of plants exposed to stress 2, 23, 24. ERF6 induces the expression of a gibberellic acid (GA)-inactivating enzyme, GA2-OX6, which triggers a reduction in bioactive GA levels and the accumulation of DELLA proteins (Figure 2). The DELLA proteins further repress the expression of the DEL1 and UVI4 genes, causing a premature exit from the cell cycle [25]. A third cell-cycle inhibitory mechanism relies on the downregulation of the CYCLIN genes. Overexpression of ACS8 in poplar leaves results in downregulation of several A- and B-type CYCLIN genes and a CDKA-like gene [26]. Notably, in roots, where ethylene also represses the cell cycle, CYCB1;1 expression is unaffected, but at the protein level CYCB1;1 was degraded in the presence of ethylene, highlighting a post-translational regulatory mechanism [27]. Finally, it should be noted that the CDK-inhibitory genes SIAMESE and SIAMESE-RELATED 8 (SMR8) are direct targets of EIN3 in etiolated seedlings [28], and that, in roots, ethylene has been shown to induce the expression of KRP1/ICK1 and SMR1 [27] (M.D. and P. Genschik, unpublished data). These evidences could provide a fourth mechanism of action that potentially also occurs in leaves as well. Multiple mechanisms thus connect ethylene to cell-cycle inhibition, but the precise regulatory connections have not yet been fully elucidated.
Figure 2.
Molecular Pathways in Arabidopsis Leaves Connecting Ethylene to Cell Division, Cell Expansion, and Petiole Cell Elongation. In actively growing arabidopsis leaves, ethylene regulates cell division through different pathways. The MPK3/6-phosphorylation cascade regulates ethylene biosynthesis and ethylene response factors (ERFs) (simplified view, a complete scheme is given in Figure 1), and inactivates CDKA in a EIN3/EIL1-independent manner. Downstream ERFs inhibit cell division directly through E2F inhibition, and indirectly by inducing DELLA protein stabilization. Positive regulators of ethylene signaling, such as EIN2 or ERFs, negatively affect leaf growth by inhibiting cell expansion. Conversely, negative regulators of ethylene sensitivity, such as ARGOS and ARGOS-LIKE proteins, have a growth-stimulatory effect in leaves. Ethylene also stimulates the elongation of the abaxial petiole cells, causing hyponasty (Box 2).
Table 1.
Overview of ERF Mutant Lines with Shoot Growth Phenotypesa
| Gene name | Origin | Studied plant | Shoot/leaf size | Downstream of ethylene? | EIN3 target [28] | Refs |
|---|---|---|---|---|---|---|
| BOL/DRN-like | A. thaliana | A. thaliana | GOF: reduced | GOF: reduced ACC sensitivity | N | 22, 61 |
| BOL/DRN-like | A. thaliana | N. tabacum | GOF: reduced | NT | N | [22] |
| CaPF1 | Capsicum annuum | Pinus virginiana | GOF: increased | Induced by ethephon | NA | [46] |
| CRF6 | A. thaliana | A. thaliana | GOF: increased | Induced by ACC and ethylene | N | [62] |
| CRF8/ERF070 | A. thaliana | A. thaliana | GOF: reduced | NT | N | [63] |
| DRN/ESR1 | A. thaliana | A. thaliana | GOF: reduced | NT | N | [61] |
| EBE | A. thaliana | A. thaliana | GOF: reduced | NT | N | [64] |
| ERF-1 | A. thaliana | A. thaliana | GOF: reduced | Induced by ACC and ethylene | Y | [65] |
| ERF2 | A. thaliana | A. thaliana | LOF: reduced | Induced by ACC and ethylene | Y | [33] |
| ERF11 | A. thaliana | A. thaliana | GOF: reduced | Induced by ACC and ethylene | Y | 33, 37 |
| ERF14 | A. thaliana | A. thaliana | GOF: reduced | LOF: reduced ethylene signaling | N | [66] |
| ERF15 | A. thaliana | A. thaliana | GOF: reduced | NT | Y | [67] |
| ERF4 | A. thaliana | A. thaliana | GOF: reduced | Induced by ACC | N | [68] |
| ERF6 | A. thaliana | A. thaliana | GOF: reduced | Induced by ACC and ethylene | N | 23, 33 |
| ERF73/HRE1 | A. thaliana | A. thaliana | RNAi: reduced | Induced by ACC | N | [69] |
| ERF8 | A. thaliana | A. thaliana | GOF: reduced LOF: increased |
Induced by ACC and ethylene | Y | 5, 33, 68 |
| ERF9 | A. thaliana | A. thaliana | GOF: reduced | Induced by ACC and ethylene | N | [33] |
| ERF98 | A. thaliana | A. thaliana | GOF: reduced | Induced by ACC and ethylene | N | [33] |
| HhERF2 and PeDREB2a | Hamilodendron halodendron and Populus euphratica | Gossypium hirsutum L. | Double GOF: reduced | NT | NA | [70] |
| HYR | Oryza sativa | Oryza sativa | GOF: increased | NT | NA | [47] |
| JcERF011 | Jatropha curcas L. | A. thaliana | GOF: reduced | NT | NA | [71] |
| LEP | A. thaliana | A. thaliana | GOF: reduced | NT | Y | [72] |
| NtERF3 | N. tabacum | A. thaliana | GOF: reduced | NT | NA | [68] |
| ORA59/ERF59 | A. thaliana | A. thaliana | GOF: reduced GOF: increased |
Induced by ethylene | N | 33, 73 |
| OsEATB | Oryza sativa | Oryza sativa | GOF: reduced | Repressed by ethephon | NA | [34] |
| OsERF1 | Oryza sativa | A. thaliana | GOF: reduced | Induced by ethrel | NA | [74] |
| OsERF48 | Oryza sativa | Oryza sativa | GOF: reduced | NT | NA | [75] |
| pti4 | Lycopersicon esculentum | A. thaliana | GOF: reduced | Induced by ethylene | NA | [76] |
| PvERF001 | Panicum virgatum | Panicum virgatum | GOF: increased | NT | NA | [48] |
| RAP2.12 | A. thaliana | A. thaliana | GOF: reduced | NT | N | [77] |
| RAP2.6 | A. thaliana | A. thaliana | GOF: reduced | Induced by ACC and ethylene | Y | [78] |
| RRTF1 | A. thaliana | A. thaliana | GOF: reduced | Induced by ACC | N | [79] |
| SHN1/WIN1 | A. thaliana | A. thaliana | GOF: reduced | NT | N | [80] |
| SlERF5 | Solanum lycopersicum | Solanum lycopersicum | GOF: reduced | Induced by ACC | NA | [81] |
| SUB1A | Oryza sativa | Oryza sativa | GOF: reduced | Induced by ethylene | NA | [82] |
| TERF1 | Solanum lycopersicum | N. tabacum | GOF: reduced | Induced by ethylene | NA | [83] |
| WXP2 | Medicago truncatula | A. thaliana | GOF: reduced | NT | NA | [84] |
Abbreviations: BOL, BOLITA; CRF, cytokinin response factor; DRN, DORNRÖSCHEN; EATB, ERF protein associated with tillering and panicle branching; EBE, ERF BUD ENHANCER; ERF, ethylene response factor; ESR1, ENHANCER OF SHOOT REGENERATION 1; GOF, gain of function; HRE1, hypoxia responsive ERF 1; LEP, LEAFY PETIOLE; LOF, loss-of-function; NA, not applicable; NT, not tested; ORA59, octadecanoid-responsive AP2/ERF; RAP, RELATED TO APETALA; RRTF1, REDOX RESPONSIVE TRANSCRIPTION FACTOR 1; SUB1A, SUBMERGENCE 1A; WIN/SHN1, WAX INDUCER 1/SHINE 1; WXP, WAX PRODUCTION.
Effects of Ethylene on Cell Expansion
In a simplified view, cellular growth requires both uptake of water and the corresponding extensibility of the cell wall, thus involving two basic actions: relaxation of the cell wall, mediated by cell wall-remodeling enzymes, and the import of water, mainly through a difference in water potential and facilitated by aquaporins. Similarly as for cell division, specific examples are known where ethylene has a positive effect on cell expansion, such as in petioles or in hypocotyls grown in light (Box 2) 29, 30. Ethylene does so by directly acting on microtubule orientation and on genes of the EXPANSIN family (Box 2) [30]. In Sagittaria pygmaea and grape berry, ethylene induces the expression of xyloglucan endotransglycolases/hydrolases (XTHs), also stimulating cell-wall loosening and cell expansion 31, 32.
Box 2. Hyponasty – Growth-Related and Ethylene-Mediated.
In addition to growing, leaves also move up and down to optimize light capture in changing environments. This phenomenon, called hyponasty (up) and epinasty (down), has been observed in multiple plant species but is most pronounced in rosette plants such as arabidopsis. Leaves move in a diurnal way, moving upwards during daytime to reach their most vertical position at dusk [101]. Leaves also move upwards during shade avoidance, light stress, or flooding stress 102, 103. The involvement of ethylene in hyponastic leaf movement under shade or submergence has been known for a long time: ACS genes are induced by stress-responsive TFs (Figure 1) 57, 104, and etr1-1 tobacco mutants as well as the arabidopsis aco5 mutants show reduced hyponastic responses 102, 105. Whether ethylene also regulates the diurnal hyponastic leaf movements under non-stress conditions is still under debate, but recent evidence points in this direction: etr1-1, ein2, and acs2 mutants show reduced leaf-movement amplitudes throughout the day [106].
At the cellular level, hyponasty is established by elongation of the cells on the lower side of the petiole. To enable elongation, cortical microtubules (CMTs), which strengthen the cell wall and inhibit growth in their orientation, are reoriented to enable longitudinal growth. This reorientation is stimulated by ethylene, specifically in the proximal abaxial petiole cells, and coincides with ethylene-mediated transcriptional induction of EXPANSIN11 (Figure 2) [30]. At the molecular level, this is also likely to involve alterations in brassinosteroid and auxin metabolism [104]. Recently, elongation-mediated petiole growth and the involvement of ethylene have been modeled mathematically, also highlighting a role for cell division in this process [107]. The model suggests that the extent of elongation should be greater than what was actually observed, unless the increase in cell elongation is compensated by repression of cell division in the proximal abaxial petiole cells. Experimental validation indeed showed that, in addition to stimulating cell expansion, ethylene also moderates the level of hyponasty by negatively acting on the cell cycle of petiole cells [107].
Alt-text: Box 2
However, similarly to cell division, the influence of ethylene on cell growth in leaves is almost exclusively negative. Overproduction of ethylene or overexpression of proteins of the signaling cascade results in smaller leaves because of restricted cell expansion, as illustrated by overexpression of ACS8 [26], EIN2 [10], BOLITA [22], ERF6 [23], and multiple other ERF-encoding genes, discussed in the next section. By contrast, mutants with a reduced ethylene sensitivity have an increased leaf size resulting from enhanced cell expansion, as demonstrated in ein2 [10] and in lines overexpressing negative regulators of ethylene signaling of the ARGOS family [12]. Molecularly, the connection between the proteins downstream of ethylene signaling and effectors of cell expansion is not entirely clear, but the data point toward convergence at the level of EXPANSINs. EXP3 and EXP5 are downregulated in plants overexpressing EIN2 and are upregulated in ein2 knockout plants, and expression of EXP1 and EXP5 is strongly repressed in dwarfed BOLITA gain-of-function plants (Figure 2) 10, 22. Alternatively, in the ERF6-mediated growth-inhibitory pathway, the inhibition of cell expansion might be regulated by DELLA proteins that are stabilized by ERF6 overexpression. Numerous molecular mechanisms connect DELLAs to the inhibition of cell expansion (reviewed in [25]), but a notable mechanism is the DELLA-mediated degradation of the PIF4 and PIF5 proteins, which activate genes involved in cell-wall remodeling. In conclusion, at least two parallel pathways are likely to repress cell expansion upon ethylene accumulation in leaves.
Exploring ERFs as Growth-Regulating TFs
In arabidopsis, 65 ERFs have been identified and, to our knowledge, 31 have been studied at the phenotypic level. Of these, 22 show a growth phenotype when overexpressed or knocked down (Table 1), which provides multiple additional connections between ethylene and growth-regulating pathways.
Interestingly, a large number of ethylene-responsive ERFs, including ERF-1, ERF2, ERF5, ERF6, ERF8, ERF9, ERF11, ERF59, ERF98, and RAP2.6L (Table 1), have been shown to be part of a transcriptional network that regulates leaf growth inhibition upon mild osmotic stress [33]. In this network, which also contains TFs outside the ERF family, all the TFs are densely connected and regulate each other’s transcription, rendering the network-mediated growth regulation particularly complex. The network is mainly composed of inhibitors of leaf growth, including ERF6, ERF8, ERF9, ERF11, and ERF98, but also contains some growth-promoting ERFs, such as ERF2, ERF59, and RAP2.6L. Most ERFs of the network are transcriptional activators that are induced quickly upon stress to activate the response, whereas two repressing ERFs, ERF8 and ERF9, are induced later to avoid overactivation and enable fine-tuning of the stress response. This network is transcriptionally induced upon osmotic stress, but most likely also acts under other abiotic stress conditions such as salt or drought stress. ERF8, for example, is a strong inhibitor of cell division and leaf growth, and is an important factor in the drought stress response 5, 23, 33.
Several ERFs, including the core components of this growth-regulatory network, have been shown to directly or indirectly regulate the expression of GA biosynthesis or degradation enzymes in growing leaves. ERF6, ERF9, ERF11, and ERF98 regulate the transcription of GA2-OX6, encoding a GA degradation enzyme, upon osmotic stress, possibly resulting in growth inhibition when these ERFs are overexpressed 23, 33. Rice plants overexpressing OsEATB (an ERF) display a dwarfed phenotype because of decreased GA levels resulting from downregulation of the GA biosynthesis gene ENT-COPALYL DIPHOSPHATE SYNTHASE 2 (OsCPS2), although the GA20-OX2 gene was upregulated, and SLR1, encoding a DELLA protein, was downregulated [34]. Moreover, the dwarfed phenotype of the ERF6-overexpression line and of dwarfy, an ACS8-overexpression poplar line in which several ERFs are upregulated, could be rescued by increasing GA levels 23, 26.
Connections between ethylene, ERFs, and the growth-regulating GA/DELLA pathway have also been established for stem growth. Ethylene accumulation under flooding conditions induces the expression of SNORKEL1 and SNORKEL2, which promote internode elongation possibly through GA [35]. These two ERFs have a contrasting function with SUB1A, another ERF induced by ethylene, that inhibits elongation upon submergence [36]. This example illustrates the diverse modes of action of different ERFs in the same environmental context. In addition, the opposite holds true: one ERF can exert different growth-related functions in different biological contexts. As such, the growth-regulatory capacities of ERF11 depend on the organ: ERF11 overexpression induces GA2-OX6 in leaves and hence inhibits leaf growth 33, 37, whereas in the internodes it results in downregulation of GA2-OX6 and increased elongation [38]. In the latter, ERF11 was also found to interact with the DELLA protein RGA, counteracting its growth-repressing function [38]. These observations show that ERFs are connected with each other and with the GA signaling pathway, as has been also reported for other hormones [39], resulting in complex crosstalk that regulates shoot growth.
Ethylene and Crop Yield
Over the past decade research on the genetics of ethylene biosynthesis has successfully been translated from arabidopsis to crops, and from the laboratory to the field. For example, a Zea mays ACS6 gene loss-of-function mutant shows slower leaf senescence and maintains photosynthesis for a longer period when exposed to drought stress [40]. In accordance, ACS6 RNAi lines, which have reduced ethylene biosynthesis and sensitivity, show a significant increase in grain yield when exposed to drought stress in the field [41].
In addition to the targeted alteration of ethylene biosynthesis, modified expression of ethylene signaling genes appears to have promising applications for improving field crops, the most notable example being maize plants with increased expression levels of ARGOS. Overexpression of some genes of the ARGOS family, including ARGOS, ARL, AtOSR1, and AtOSR2, stimulates cell division, cell expansion, and thereby leaf size in arabidopsis 42, 43. ARGOS proteins interact with the ethylene receptors and negatively regulate ethylene responses 11, 12. The same ethylene insensitivity and growth advantage were observed in arabidopsis plants overexpressing the maize orthologs ZmARGOS1 and ZmARGOS8. Most interestingly, CRISPR/Cas9-engineered variants of maize with increased ZmARGOS8 expression levels show a higher grain yield under drought stress conditions, and also a mild but significant increase in plant height under well-watered conditions, albeit without grain yield advantage [44]. Conversely, inducing strong constitutive ethylene responses can be disadvantageous. For example, overexpression of MHZ7, the rice EIN2 homolog, results in the field in shorter plants and reduced yield [45].
Genetic alteration of upstream members of the ethylene signaling pathway results in pleiotropic phenotypes, both desirable and undesirable, because ethylene has wide-ranging molecular functions in almost all plant organs 10, 27. To avoid these undesirable effects, more downstream players, such as the ERFs, could be more attractive candidates to target because they are more specific in terms of molecular function or expression pattern. Overexpression of specific ERFs has therefore successfully resulted in increased shoot biomass and yield in the field. For example, overexpression of a pepper ethylene-responsive ERF, CaPF1, in Virginia pine resulted in increased tolerance to a range of stresses and in enhanced shoot growth as a result of a larger number of cells [46]. In addition, the overexpression line of HIGHER YIELD RICE (HYR), a rice ERF, has increased shoot biomass and grain yield under normal and drought conditions [47]. Finally, increased biomass was observed in switchgrass overexpressing PvERF001 [48]. To avoid unwanted side effects, the use of spatially or temporally regulated promoters to control ERF expression could also provide success in the field. For example, increased drought tolerance without growth penalty under normal conditions was obtained with rice plants overexpressing OsERF71 specifically in the roots [49].
Because ERFs are known to have dual roles in regulating both growth and stress-tolerance mechanisms, attempts have been made to increase defense mechanisms by altering the expression of ERF genes. Plants with improved defense mechanisms upon stress will survive or grow better following stress, and are expected to produce more yield at the end of the season. Sl-ERF.B.3 transcripts accumulate upon ethylene treatment, and antisense transgenic tomato plants had improved tolerance to cold stress, without growth penalty, and even a slight tendency towards enhanced height [50]. Using CRISPR/Cas9, some rice variants with deletions in OsERF922 have been generated, and these showed enhanced resistance to Magnaporthe oryzae, but no difference with the wild type for several yield-related traits [51]. In several cases, ERF overexpression resulted in increased tolerance to stress without affecting growth under normal conditions. AP37 and AP59 overexpression gave increased tolerance to severe drought stress and high-salinity stress in rice, without affecting growth [52]. Overexpression of OsERF109 or OsERF3, that are both induced by ethylene, reduced drought tolerance without causing any growth defect under normal conditions 53, 54. However, in a series of studies, ERF overexpression improved stress tolerance, but with a negative effect on plant growth. This was observed for rice OsERF1 and OsERF48 (Table 1). This shows that, although numerous crops with altered ethylene sensitivity or ERF expression have successfully made it to the field, undesired effects are still unavoidable. The highly complex regulatory connections observed between growth-promoting and growth-repressing ERFs [33] might explain such undesired and unpredictable phenotypic effects. In this respect, unraveling the networks in which ERFs are involved will be crucial for understanding the growth-regulatory pathways in shoots, and will enable new advances in targeted engineering of ethylene-mediated shoot growth.
Does Ethylene Regulate Leaf Growth Dynamics?
Ethylene levels are not only increased by adverse environmental conditions but also vary in growth-favorable conditions, for example throughout a day/night cycle. Diurnal oscillations of ethylene levels have been observed in several plants species including sorghum [55], the potato subspecies Andigena [56], and arabidopsis [57]. In general, ethylene levels are low at dawn, increase during the first half of the day, peak between midday and evening, and decrease again during the evening, with the peak slightly shifting depending on the species 55, 56, 57. Interestingly, these oscillations are maintained when plants are transferred to continuous light or dark, pointing to endogenously controlled regulation [57]. In arabidopsis, the fluctuating levels of ethylene in seedlings result from oscillating expression patterns mainly of ACS8, but also of ACS5 and ACS9 [57]. ACS8 is most likely a target of the circadian clock because the ACS8 promoter contains an element typically found in clock-regulated genes. Accordingly, ethylene oscillations are altered in the arabidopsis clock mutant toc1-1 and in a CCA1 overexpressor [57]. Moreover, the ACS8 gene is also under the control of the dark-stabilized PIF4 and, accordingly, phyB (PHYTOCHROME B) mutants show higher ACS8 transcript levels in leaves (Figure 1) 58, 59. Finally, in sorghum, the amplitude of diurnal ethylene oscillations can be influenced by light or shade treatments, as well as by the presence of PHYB [55]. These observations thus suggest that ethylene oscillations are controlled both by a clock-entrained mechanism and a light/dark-regulated response.
This tightly controlled regulation of ethylene fluctuations, even when plants are not exposed to stress conditions, supports the hypothesis that ethylene could also act under normal conditions to exert its growth-regulatory function. Indeed, leaf growth dynamics also vary according to the time of the day under control of the circadian clock [60]. The diurnal oscillations of ethylene levels and growth dynamics have both been thoroughly studied independently, but whether oscillating ethylene levels regulate these leaf growth dynamics has, surprisingly, never been investigated. Based on both the oscillation patterns and the anti-correlation between ethylene and leaf growth, largely illustrated in this review, it may be speculated that ethylene could regulate diurnal leaf growth dynamics. Arabidopsis leaf growth has a maximal rate at dawn [60], which corresponds to the moment when ethylene levels are the lowest [57]. Subsequently, until the afternoon, ethylene levels increase, and leaf growth decreases correspondingly. During the night, leaf growth rates increase, while ethylene levels remain low and invariable. This hypothetical model shows some inconsistencies, particularly regarding growth in the evening when both ethylene and growth rates are low. To further investigate this model it will be crucial to measure ethylene levels specifically in young and actively growing leaves instead of in whole seedlings or hypocotyls as it has been done until now. Validation of this model could subsequently be obtained by detailed measurements of leaf growth dynamics in ethylene-overproducing lines or mutants. Such detailed investigations would enrich our basic knowledge of the role of ethylene in leaf growth, which is currently restricted to biotic and abiotic stress conditions.
Concluding Remarks and Future Perspectives
Over the past decade, studies have provided multiple molecular connections between ethylene and growth, cell division, and cell expansion. The effects of ethylene accumulation on cell division and cell expansion can be either positive or negative, depending on the environmental context and the organ. In leaves, the effect of ethylene on cellular processes that mediate growth is almost exclusively negative, with the exception of several ERFs, illustrated in this paper, that appear to have positive effects. Although the inhibitory effect of ethylene on shoot growth has been observed in multiple studies, including studies on field-grown crop species, the precise molecular pathways connecting ethylene to growth inhibition are far less understood. Ethylene accumulation in leaves causes rapid inhibition of cell division and cell expansion, either through DELLA-mediated mechanisms or through more direct connections with core cell-cycle or EXPANSIN genes, respectively. These connections, however, are still vague, and strong evidence for direct regulatory links is still missing (see Outstanding Questions). It is likely that the ERF TFs play a major role in these regulatory pathways. Identification of their direct target genes would be helpful and would improve our understanding of their sometimes contradictory roles in shoot growth. Given the emerging importance of ethylene-mediated growth inhibition of plants exposed to environmental stresses, unraveling the molecular connections with the effector genes of cell division and cell expansion would be highly valuable for engineering of crops with less-pronounced growth inhibition in adverse conditions. Although impressive results were already obtained in the field by engineering ethylene sensitivity or signaling, undesired effects are still observed, and progress can still be made in uncoupling the defense-inducing and growth-inhibitory mechanisms by targeted engineering using new genome-editing techniques.
Outstanding Questions.
What are the precise molecular mechanisms underlying ethylene-mediated inhibition of the cell cycle in leaves? Are the cell-cycle genes directly targeted by EIN3/EIL1 and/or by ERFs in leaves?
Ethylene-engineered crops in the field score high success rates in terms of biomass and grain yield production, but what is the molecular origin of this advantage? Understanding the molecular reasons would enable future engineering in a more targeted manner.
Is ethylene a pure stress-responsive hormone, or does it also have a function under growth-favorable conditions, for example in controlling the diurnal dynamics of leaf growth?
Acknowledgments
We thank Drs Annick Bleys and Hannes Claeys for helpful suggestions for improving the manuscript. This work was supported by Ghent University [Bijzonder Onderzoeksfonds Methusalem Project BOF08/01M00408] and by the European Research Council (ERC) under the 7th Framework Programme (FP7/2007–2013) of the EU under ERC grant agreement 339341-AMAIZE. L.V.d.B. is a predoctoral fellow of the Research Foundation Flanders (FWO 131013).
Glossary
- S-Adenosyl methionine (SAM)
a conjugated form of the amino acid methionine. It is an intermediate product of ethylene biosynthesis and the precursor of ACC.
- 1-Aminocyclopropane-1-carboxylic acid (ACC)
the precursor of ethylene. It is stable and can be transported throughout the plant.
- CDKA
A-TYPE CYCLIN-DEPENDENT KINASE, a key regulator of the cell cycle that is important at both the G1–S and G2–M phase transitions.
- CTR1
CONSTITUTIVE TRIPLE RESPONSE 1. When mutated, CTR1 confers the constitutive triple response, a signature phenotype of ethylene-treated, dark-grown seedlings characterized, in comparison to wild-type etiolated seedlings, by a less-elongated, thickened hypocotyl, and a curling apical hook.
- CYCLINs
key proteins controlling the different steps of the cell cycle by associating with the CYCLIN-DEPENDENT KINASES (CDKs). Their cyclic expression and subsequent protein degradation ensures unidirectional progression through the cell cycle.
- DEL1 and UVI4
proteins that control the shift between the mitotic cell cycle and endoreduplication.
- Endoreduplication
a variant of the plant cell cycle in which the S-phase (DNA synthesis) still takes place but not the M-phase (mitosis), resulting in doubling of the amount of DNA per cell. In many plants, endoreduplication coincides with cell expansion and differentiation.
- EXPANSIN
an enzyme responsible for loosening the cell wall.
- KRP1/ICK1 and SMR1
CDK-inhibitory proteins that bind to and inhibit CDKs, and thus repress cell-cycle progression.
- LHT1
LYSINE HISTIDINE TRANSPORTER has been shown to also mediate ACC transport.
References
- 1.Gonzalez N. Leaf size control: complex coordination of cell division and expansion. Trends Plant Sci. 2012;17:332–340. doi: 10.1016/j.tplants.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 2.Skirycz A. Pause-and-stop: the effects of osmotic stress on cell proliferation during early leaf development in Arabidopsis and a role for ethylene signaling in cell cycle arrest. Plant Cell. 2011;23:1876–1888. doi: 10.1105/tpc.111.084160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thao N.P. Role of ethylene and its cross talk with other signaling molecules in plant responses to heavy metal stress. Plant Physiol. 2015;169:73–84. doi: 10.1104/pp.15.00663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang M. The regulatory roles of ethylene and reactive oxygen species (ROS) in plant salt stress responses. Plant Mol. Biol. 2016;91:651–659. doi: 10.1007/s11103-016-0488-1. [DOI] [PubMed] [Google Scholar]
- 5.Dubois M. Time of day determines Arabidopsis transcriptome and growth dynamics under mild drought. Plant Cell Environ. 2017;40:180–189. doi: 10.1111/pce.12809. [DOI] [PubMed] [Google Scholar]
- 6.Savada R.P. Heat stress differentially modifies ethylene biosynthesis and signaling in pea floral and fruit tissues. Plant Mol. Biol. 2017;95:313–331. doi: 10.1007/s11103-017-0653-1. [DOI] [PubMed] [Google Scholar]
- 7.Burg S.P., Burg E.A. Ethylene formation in pea seedlings; its relation to the inhibition of bud growth caused by indole-3-acetic acid. Plant Physiol. 1968;43:1069–1074. doi: 10.1104/pp.43.7.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vogel J.P. Recessive and dominant mutations in the ethylene biosynthetic gene ACS5 of Arabidopsis confer cytokinin insensitivity and ethylene overproduction, respectively. Proc. Natl. Acad. Sci. U. S. A. 1998;95:4766–4771. doi: 10.1073/pnas.95.8.4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qu X. A strong constitutive ethylene-response phenotype conferred on Arabidopsis plants containing null mutations in the ethylene receptors ETR1 and ERS1. BMC Plant Biol. 2007;7:3. doi: 10.1186/1471-2229-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng G. The Arabidopsis EIN2 restricts organ growth by retarding cell expansion. Plant Signal. Behav. 2015;10 doi: 10.1080/15592324.2015.1017169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rai M.I. The ARGOS gene family functions in a negative feedback loop to desensitize plants to ethylene. BMC Plant Biol. 2015;15:157. doi: 10.1186/s12870-015-0554-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi J. Overexpression of ARGOS genes modifies plant sensitivity to ethylene, leading to improved drought tolerance in both Arabidopsis and maize. Plant Physiol. 2015;169:266–282. doi: 10.1104/pp.15.00780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao Y.-R. Tobacco ankyrin protein NEIP2 interacts with ethylene receptor NTHK1 and regulates plant growth and stress responses. Plant Cell Physiol. 2015;56:803–818. doi: 10.1093/pcp/pcv009. [DOI] [PubMed] [Google Scholar]
- 14.Tao J.-J. Tobacco translationally controlled tumor protein interacts with ethylene receptor tobacco histidine kinase1 and enhances plant growth through promotion of cell proliferation. Plant Physiol. 2015;169:96–114. doi: 10.1104/pp.15.00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chatterjee P. Beneficial soil bacterium Pseudomonas frederiksbergensis OS261 augments salt tolerance and promotes red pepper plant growth. Front. Plant Sci. 2017;8:705. doi: 10.3389/fpls.2017.00705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen L. The rhizobacterium Variovorax paradoxus 5C-2, containing ACC deaminase, promotes growth and development of Arabidopsis thaliana via an ethylene-dependent pathway. J. Exp. Bot. 2013;64:1565–1573. doi: 10.1093/jxb/ert031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fiorani F. Ethylene emission and responsiveness to applied ethylene vary among Poa species that inherently differ in leaf elongation rates. Plant Physiol. 2002;129:1382–1390. doi: 10.1104/pp.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee S.H., Reid D.M. The role of endogenous ethylene in the expansion of Helianthus annuus leaves. Can. J. Bot. 1997;75:501–508. doi: 10.1139/b97-054. [DOI] [PubMed] [Google Scholar]
- 19.Polyn S. Cell cycle entry, maintenance, and exit during plant development. Curr. Opin. Plant Biol. 2015;23:1–7. doi: 10.1016/j.pbi.2014.09.012. [DOI] [PubMed] [Google Scholar]
- 20.Raz V., Koornneef M. Cell division activity during apical hook development. Plant Physiol. 2001;125:219–226. doi: 10.1104/pp.125.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Etchells J.P. Plant vascular cell division is maintained by an interaction between PXY and ethylene signalling. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marsch-Martinez N. BOLITA, an Arabidopsis AP2/ERF-like transcription factor that affects cell expansion and proliferation/differentiation pathways. Plant Mol. Biol. 2006;62:825–843. doi: 10.1007/s11103-006-9059-1. [DOI] [PubMed] [Google Scholar]
- 23.Dubois M. ETHYLENE RESPONSE FACTOR6 acts as a central regulator of leaf growth under water-limiting conditions in Arabidopsis. Plant Physiol. 2013;162:319–332. doi: 10.1104/pp.113.216341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meng X. Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell. 2013;25:1126–1142. doi: 10.1105/tpc.112.109074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Claeys H. Gibberellins and DELLAs: central nodes in growth regulatory networks. Trends Plant Sci. 2014;19:231–239. doi: 10.1016/j.tplants.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 26.Plett J.M. Heterologous over-expression of ACC SYNTHASE8 (ACS8) in Populus tremula × P. alba clone 717-1B4 results in elevated levels of ethylene and induces stem dwarfism and reduced leaf size through separate genetic pathways. Front. Plant Sci. 2014;5:514. doi: 10.3389/fpls.2014.00514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Street I.H. Ethylene inhibits cell proliferation of the Arabidopsis root meristem. Plant Physiol. 2015;169:338–350. doi: 10.1104/pp.15.00415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang K.N. Temporal transcriptional response to ethylene gas drives growth hormone cross-regulation in Arabidopsis. eLife. 2013;2 doi: 10.7554/eLife.00675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smalle J. Ethylene can stimulate Arabidopsis hypocotyl elongation in the light. Proc. Natl. Acad. Sci. U. S. A. 1997;94:2756–2761. doi: 10.1073/pnas.94.6.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Polko J.K. Ethylene-induced differential petiole growth in Arabidopsis thaliana involves local microtubule reorientation and cell expansion. New Phytol. 2012;193:339–348. doi: 10.1111/j.1469-8137.2011.03920.x. [DOI] [PubMed] [Google Scholar]
- 31.Ookawara R. Expression of α-expansin and xyloglucan endotransglucosylase/hydrolase genes associated with shoot elongation enhanced by anoxia, ethylene and carbon dioxide in arrowhead (Sagittaria pygmaea Miq.) tubers. Ann. Bot. 2005;96:693–702. doi: 10.1093/aob/mci221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chervin C. Stimulation of the grape berry expansion by ethylene and effects on related gene transcripts, over the ripening phase. Physiol. Plant. 2008;134:534–546. doi: 10.1111/j.1399-3054.2008.01158.x. [DOI] [PubMed] [Google Scholar]
- 33.Van den Broeck L. From network to phenotype: the dynamic wiring of an Arabidopsis transcriptional network induced by osmotic stress. Mol. Syst. Biol. 2017;13:961. doi: 10.15252/msb.20177840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qi W. Rice ethylene-response AP2/ERF factor OsEATB restricts internode elongation by down-regulating a gibberellin biosynthetic gene. Plant Physiol. 2011;157:216–228. doi: 10.1104/pp.111.179945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hattori Y. The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature. 2009;460:1026–1030. doi: 10.1038/nature08258. [DOI] [PubMed] [Google Scholar]
- 36.Fukao T. A variable cluster of ethylene response factor-like genes regulates metabolic and developmental acclimation responses to submergence in rice. Plant Cell. 2006;18:2021–2034. doi: 10.1105/tpc.106.043000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dubois M. The ETHYLENE RESPONSE FACTORS ERF6 and ERF11 antagonistically regulate mannitol-induced growth inhibition in Arabidopsis. Plant Physiol. 2015;169:166–179. doi: 10.1104/pp.15.00335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou X. The ERF11 transcription factor promotes internode elongation by activating gibberellin biosynthesis and signaling. Plant Physiol. 2016;171:2760–2770. doi: 10.1104/pp.16.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Müller M., Munné-Bosch S. Ethylene response factors: a key regulatory hub in hormone and stress signaling. Plant Physiol. 2015;169:32–41. doi: 10.1104/pp.15.00677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Young T.E. ACC synthase expression regulates leaf performance and drought tolerance in maize. Plant J. 2004;40:813–825. doi: 10.1111/j.1365-313X.2004.02255.x. [DOI] [PubMed] [Google Scholar]
- 41.Habben J.E. Transgenic alteration of ethylene biosynthesis increases grain yield in maize under field drought-stress conditions. Plant Biotechnol. J. 2014;12:685–693. doi: 10.1111/pbi.12172. [DOI] [PubMed] [Google Scholar]
- 42.Wang B. Expression of a rice OsARGOS gene in Arabidopsis promotes cell division and expansion and increases organ size. J. Genet. Genomics. 2009;36:31–40. doi: 10.1016/S1673-8527(09)60004-7. [DOI] [PubMed] [Google Scholar]
- 43.Feng G. Arabidopsis ORGAN SIZE RELATED1 regulates organ growth and final organ size in orchestration with ARGOS and ARL. New Phytol. 2011;191:635–646. doi: 10.1111/j.1469-8137.2011.03710.x. [DOI] [PubMed] [Google Scholar]
- 44.Shi J. ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 2017;15:207–216. doi: 10.1111/pbi.12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma B. Identification of rice ethylene-response mutants and characterization of MHZ7/OsEIN2 in distinct ethylene response and yield trait regulation. Mol. Plant. 2013;6:1830–1848. doi: 10.1093/mp/sst087. [DOI] [PubMed] [Google Scholar]
- 46.Tang W. Overexpression of the pepper transcription factor CaPF1 in transgenic Virginia pine (Pinus Virginiana Mill.) confers multiple stress tolerance and enhances organ growth. Plant Mol. Biol. 2005;59:603–617. doi: 10.1007/s11103-005-0451-z. [DOI] [PubMed] [Google Scholar]
- 47.Ambavaram M.M.R. Coordinated regulation of photosynthesis in rice increases yield and tolerance to environmental stress. Nat. Commun. 2014;5:5302. doi: 10.1038/ncomms6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wuddineh W.A. Identification and molecular characterization of the switchgrass AP2/ERF transcription factor superfamily, and overexpression of PvERF001 for improvement of biomass characteristics for biofuel. Front. Bioeng. Biotechnol. 2015;3:101. doi: 10.3389/fbioe.2015.00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee D.-K. Rice OsERF71-mediated root modification affects shoot drought tolerance. Plant Signal. Behav. 2017;12 doi: 10.1080/15592324.2016.1268311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klay I. Ethylene response factor Sl-ERF.B.3 is responsive to abiotic stresses and mediates salt and cold stress response regulation in tomato. Sci. World J. 2014;2014:167681. doi: 10.1155/2014/167681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang F. Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922. PLoS One. 2016;11 doi: 10.1371/journal.pone.0154027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oh S.-J. Overexpression of the transcription factor AP37 in rice improves grain yield under drought conditions. Plant Physiol. 2009;150:1368–1379. doi: 10.1104/pp.109.137554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wan L. Transcriptional activation of OsDERF1 in OsERF3 and OsAP 2-39 negatively modulates ethylene synthesis and drought tolerance in rice. PLoS One. 2011;6 doi: 10.1371/journal.pone.0025216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu Y. The ethylene response factor OsERF109 negatively affects ethylene biosynthesis and drought tolerance in rice. Protoplasma. 2017;254:401–408. doi: 10.1007/s00709-016-0960-4. [DOI] [PubMed] [Google Scholar]
- 55.Finlayson S.A. The mechanism of rhythmic ethylene production in sorghum. The role of phytochrome B and simulated shading. Plant Physiol. 1999;119:1083–1089. doi: 10.1104/pp.119.3.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chincinska I. Photoperiodic regulation of the sucrose transporter StSUT4 affects the expression of circadian-regulated genes and ethylene production. Front. Plant Sci. 2013;4:26. doi: 10.3389/fpls.2013.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thain S.C. Circadian rhythms of ethylene emission in Arabidopsis. Plant Physiol. 2004;136:3751–3761. doi: 10.1104/pp.104.042523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nomoto Y. Circadian clock- and PIF4-controlled plant growth: a coincidence mechanism directly integrates a hormone signaling network into the photoperiodic control of plant architectures in Arabidopsis thaliana. Plant Cell Physiol. 2012;53:1950–1964. doi: 10.1093/pcp/pcs137. [DOI] [PubMed] [Google Scholar]
- 59.Vandenbussche F. Ethylene and auxin control the Arabidopsis response to decreased light intensity. Plant Physiol. 2003;133:517–527. doi: 10.1104/pp.103.022665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ruts T. Diel patterns of leaf and root growth: endogenous rhythmicity or environmental response? J. Exp. Bot. 2012;63:3339–3351. doi: 10.1093/jxb/err334. [DOI] [PubMed] [Google Scholar]
- 61.Kirch T. The DORNRÖSCHEN/ENHANCER OF SHOOT REGENERATION1 gene of Arabidopsis acts in the control of meristem cell fate and lateral organ development. Plant Cell. 2003;15:694–705. doi: 10.1105/tpc.009480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zwack P.J. Cytokinin response factor 6 negatively regulates leaf senescence and is induced in response to cytokinin and numerous abiotic stresses. Plant Cell Physiol. 2013;54:971–981. doi: 10.1093/pcp/pct049. [DOI] [PubMed] [Google Scholar]
- 63.Ramaiah M. ETHYLENE RESPONSE FACTOR070 regulates root development and phosphate starvation-mediated responses. Plant Physiol. 2014;164:1484–1498. doi: 10.1104/pp.113.231183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mehrnia M. EBE, an AP2/ERF transcription factor highly expressed in proliferating cells, affects shoot architecture in Arabidopsis. Plant Physiol. 2013;162:842–857. doi: 10.1104/pp.113.214049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lorenzo O. ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell. 2003;15:165–178. doi: 10.1105/tpc.007468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oñate-Sánchez L. AtERF14, a member of the ERF family of transcription factors, plays a nonredundant role in plant defense. Plant Physiol. 2007;143:400–409. doi: 10.1104/pp.106.086637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee S.-b. AtERF15 is a positive regulator of ABA response. Plant Cell Rep. 2015;34:71–81. doi: 10.1007/s00299-014-1688-2. [DOI] [PubMed] [Google Scholar]
- 68.Koyama T. A regulatory cascade involving class II ETHYLENE RESPONSE FACTOR transcriptional repressors operates in the progression of leaf senescence. Plant Physiol. 2013;162:991–1005. doi: 10.1104/pp.113.218115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang C.-Y. The AP2/ERF transcription factor AtERF73/HRE1 modulates ethylene responses during hypoxia in Arabidopsis. Plant Physiol. 2011;156:202–212. doi: 10.1104/pp.111.172486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li J.B. Simultaneous overexpression of the HhERF2 and PeDREB2a genes enhanced tolerances to salt and drought in transgenic cotton. Protein Pept. Lett. 2016;23:450–458. doi: 10.2174/0929866523666160314153212. [DOI] [PubMed] [Google Scholar]
- 71.Tang Y. Genome-wide analysis of the AP2/ERF gene family in physic nut and overexpression of the JcERF011 gene in rice increased its sensitivity to salinity stress. PLoS One. 2016;11 doi: 10.1371/journal.pone.0150879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ward J.M. A new role for the Arabidopsis AP2 transcription factor, LEAFY PETIOLE, in gibberellin-induced germination is revealed by the misexpression of a homologous gene, SOB2/DRN-LIKE. Plant Cell. 2006;18:29–39. doi: 10.1105/tpc.105.036707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pré M. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 2008;147:1347–1357. doi: 10.1104/pp.108.117523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hu Y. Overexpression of OsERF1, a novel rice ERF gene, up-regulates ethylene-responsive genes expression besides affects growth and development in Arabidopsis. J. Plant Physiol. 2008;165:1717–1725. doi: 10.1016/j.jplph.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 75.Jung H. Overexpression of OsERF48 causes regulation of OsCML16, a calmodulin-like protein gene that enhances root growth and drought tolerance. Plant Biotechnol. J. 2017;15:1295–1308. doi: 10.1111/pbi.12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu K. Functional analysis of tomato Pti4 in Arabidopsis. Plant Physiol. 2002;128:30–37. [PMC free article] [PubMed] [Google Scholar]
- 77.Paul M.V. Oxygen sensing via the ethylene response transcription factor RAP2.12 affects plant metabolism and performance under both normoxia and hypoxia. Plant Physiol. 2016;172:141–153. doi: 10.1104/pp.16.00460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Krishnaswamy S. Functional characterization of four APETALA2-family genes (RAP2.6, RAP2.6L, DREB19 and DREB26) in Arabidopsis. Plant Mol. Biol. 2011;75:107–127. doi: 10.1007/s11103-010-9711-7. [DOI] [PubMed] [Google Scholar]
- 79.Matsuo M. High REDOX RESPONSIVE TRANSCRIPTION FACTOR1 levels result in accumulation of reactive oxygen species in Arabidopsis thaliana shoots and roots. Mol. Plant. 2015;8:1253–1273. doi: 10.1016/j.molp.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 80.Aharoni A. The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell. 2004;16:2463–2480. doi: 10.1105/tpc.104.022897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pan Y. An ethylene response factor (ERF5) promoting adaptation to drought and salt tolerance in tomato. Plant Cell Rep. 2012;31:349–360. doi: 10.1007/s00299-011-1170-3. [DOI] [PubMed] [Google Scholar]
- 82.Xu K. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature. 2006;442:705–708. doi: 10.1038/nature04920. [DOI] [PubMed] [Google Scholar]
- 83.Huang Z. Tomato TERF1 modulates ethylene response and enhances osmotic stress tolerance by activating expression of downstream genes. FEBS Lett. 2004;573:110–116. doi: 10.1016/j.febslet.2004.07.064. [DOI] [PubMed] [Google Scholar]
- 84.Zhang J.-Y. Heterologous expression of two Medicago truncatula putative ERF transcription factor genes, WXP1 and WXP2, in Arabidopsis led to increased leaf wax accumulation and improved drought tolerance, but differential response in freezing tolerance. Plant Mol. Biol. 2007;64:265–278. doi: 10.1007/s11103-007-9150-2. [DOI] [PubMed] [Google Scholar]
- 85.Sauter M. Methionine salvage and S-adenosylmethionine: essential links between sulfur, ethylene and polyamine biosynthesis. Biochem. J. 2013;451:145–154. doi: 10.1042/BJ20121744. [DOI] [PubMed] [Google Scholar]
- 86.Thomann A. Arabidopsis CULLIN3 genes regulate primary root growth and patterning by ethylene-dependent and -independent mechanisms. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yoon G.M. New insights into the protein turnover regulation in ethylene biosynthesis. Mol. Cells. 2015;38:597–603. doi: 10.14348/molcells.2015.0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xu J., Zhang S. Regulation of ethylene biosynthesis and signaling by protein kinases and phosphatases. Mol. Plant. 2014;7:939–942. doi: 10.1093/mp/ssu059. [DOI] [PubMed] [Google Scholar]
- 89.Van de Poel B., Van Der Straeten D. 1-Aminocyclopropane-1-carboxylic acid (ACC) in plants: more than just the precursor of ethylene! Front. Plant Sci. 2014;5:640. doi: 10.3389/fpls.2014.00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shin K. Genetic identification of ACC-RESISTANT2 reveals involvement of LYSINE HISTIDINE TRANSPORTER1 in the uptake of 1-aminocyclopropane-1-carboxylic acid in Arabidopsis thaliana. Plant Cell Physiol. 2015;56:572–582. doi: 10.1093/pcp/pcu201. [DOI] [PubMed] [Google Scholar]
- 91.Resnick J.S. REVERSION-TO-ETHYLENE SENSITIVITY1, a conserved gene that regulates ethylene receptor function in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 2006;103:7917–7922. doi: 10.1073/pnas.0602239103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shi J. Maize and Arabidopsis ARGOS proteins interact with ethylene receptor signaling complex, supporting a regulatory role for ARGOS in ethylene signal transduction. Plant Physiol. 2016;171:2783–2797. doi: 10.1104/pp.16.00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lacey R.F., Binder B.M. How plants sense ethylene gas – the ethylene receptors. J. Inorg. Biochem. 2014;133:58–62. doi: 10.1016/j.jinorgbio.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 94.Shakeel S.N. Ethylene regulates levels of ethylene receptor/CTR1 signaling complexes in Arabidopsis thaliana. J. Biol. Chem. 2015;290:12415–12424. doi: 10.1074/jbc.M115.652503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li W. EIN2-directed translational regulation of ethylene signaling in Arabidopsis. Cell. 2015;163:670–683. doi: 10.1016/j.cell.2015.09.037. [DOI] [PubMed] [Google Scholar]
- 96.Merchante C. Gene-specific translation regulation mediated by the hormone-signaling molecule EIN2. Cell. 2015;163:684–697. doi: 10.1016/j.cell.2015.09.036. [DOI] [PubMed] [Google Scholar]
- 97.Guo H., Ecker J.R. Plant responses to ethylene gas are mediated by SCFEBF1/EBF2-dependent proteolysis of EIN3 transcription factor. Cell. 2003;115:667–677. doi: 10.1016/s0092-8674(03)00969-3. [DOI] [PubMed] [Google Scholar]
- 98.Potuschak T. EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell. 2003;115:679–689. doi: 10.1016/s0092-8674(03)00968-1. [DOI] [PubMed] [Google Scholar]
- 99.Nakano T. Identification of genes of the plant-specific transcription-factor families cooperatively regulated by ethylene and jasmonate in Arabidopsis thaliana. J. Plant Res. 2006;119:407–413. doi: 10.1007/s10265-006-0287-x. [DOI] [PubMed] [Google Scholar]
- 100.Yoo S.-D., Sheen J. MAPK signaling in plant hormone ethylene signal transduction. Plant Signal. Behav. 2008;3:848–849. doi: 10.4161/psb.3.10.5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dornbusch T. Differentially phased leaf growth and movements in Arabidopsis depend on coordinated circadian and light regulation. Plant Cell. 2014;26:3911–3921. doi: 10.1105/tpc.114.129031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pierik R. Canopy studies on ethylene-insensitive tobacco identify ethylene as a novel element in blue light and plant-plant signalling. Plant J. 2004;38:310–319. doi: 10.1111/j.1365-313X.2004.02044.x. [DOI] [PubMed] [Google Scholar]
- 103.Sasidharan R. Signal dynamics and interactions during flooding stress. Plant Physiol. 2017 doi: 10.1104/pp.17.01232. Published online November 2, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Stamm P., Kumar P.P. The phytohormone signal network regulating elongation growth during shade avoidance. J. Exp. Bot. 2010;61:2889–2903. doi: 10.1093/jxb/erq147. [DOI] [PubMed] [Google Scholar]
- 105.Rauf M. NAC transcription factor SPEEDY HYPONASTIC GROWTH regulates flooding-induced leaf movement in Arabidopsis. Plant Cell. 2013;25:4941–4955. doi: 10.1105/tpc.113.117861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bours R. Antiphase light and temperature cycles affect PHYTOCHROME B-controlled ethylene sensitivity and biosynthesis, limiting leaf movement and growth of Arabidopsis. Plant Physiol. 2013;163:882–895. doi: 10.1104/pp.113.221648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Polko J.K. Ethylene-mediated regulation of A2-type CYCLINs modulates hyponastic growth in Arabidopsis. Plant Physiol. 2015;169:194–208. doi: 10.1104/pp.15.00343. [DOI] [PMC free article] [PubMed] [Google Scholar]