Abstract
Background
Synergistic combination antimicrobial therapy may provide new options for treatment of MDR infections. However, comprehensive in vitro synergy data are limited and facile methods to perform synergy testing in a clinically actionable time frame are unavailable.
Objectives
To systematically investigate a broad range of antibiotic combinations for evidence of synergistic activity against a collection of carbapenem-resistant Enterobacteriaceae (CRE) isolates.
Methods
We made use of an automated method for chequerboard array synergy testing based on inkjet printer technology in the HP D300 digital dispenser to test 56 pairwise antimicrobial combinations of meropenem, aztreonam, cefepime, colistin, gentamicin, levofloxacin, chloramphenicol, fosfomycin, trimethoprim/sulfamethoxazole, minocycline and rifampicin, as well as the double carbapenem combination of meropenem and ertapenem.
Results
In a screening procedure, we tested these combinations against four CRE strains and identified nine antibiotic combinations that showed potential clinically relevant synergy. In confirmatory testing using 10 CRE strains, six combinations demonstrated clinically relevant synergy with both antimicrobials at the minimum fractional inhibitory concentration (FICI-MIN) in the susceptible or intermediate range in at least one trial. These included two novel combinations: minocycline plus colistin and minocycline plus meropenem. In 80% of strains at least one combination demonstrated clinically relevant synergy, but the combinations that demonstrated synergy varied from strain to strain.
Conclusions
This work establishes the foundation for future systematic, broad-range investigations into antibiotic synergy for CRE, emphasizes the need for individualized synergy testing and demonstrates the utility of inkjet printer-based technology for the performance of automated antimicrobial synergy assays.
Introduction
The spread of carbapenem-resistant Enterobacteriaceae (CRE) and other MDR pathogens constitutes a worldwide public health crisis.1 Antimicrobial synergy may offer the ability to treat pathogens otherwise resistant to all available or acceptable therapies. Antimicrobial synergy is a phenomenon in which two agents exert greater than additive activity when used together. Specifically, the MICs of two agents that are in the resistant range individually may be moved into the susceptible range when used in combination, thereby providing additional treatment options. Furthermore, synergistic combinations may provide therapeutic benefit even if susceptibility to both agents alone already exists by establishing more favourable drug exposure–MIC relationships. However, the synergy field has been limited by the laborious technical requirements for standard synergy testing, whether performed using chequerboard or time–kill methods, as a consequence of which such testing has generally only been applied retrospectively to limited numbers of drug combinations and bacterial isolates.2
For CRE, clinical data supporting the use of antibiotic combinations exist primarily in the form of small, retrospective studies that do not incorporate in vitro synergy data.3,4 Furthermore, in vitro investigations of synergistic activity against CRE have employed variable antimicrobials and methods, and the findings amongst studies have at times been contradictory,5–8 perhaps because results for individual strains are not generalizable to other CRE. As a result of these limitations, evidence-based guidelines for use of combination therapy for CRE have proven impossible to formulate.
Our laboratory previously pioneered the development of an automated method for antimicrobial susceptibility testing based on inkjet printer technology in the HP D300 digital dispenser.9–11 This commercially available inkjet printing technology dispenses droplets that can be varied in size from 11 pL to 10 μL, allowing the exact amount of antimicrobial needed to be dispensed into a microwell from an antimicrobial stock solution. Therefore, multiple chequerboard synergy grids can be set up in seconds using inexpensive, commercially available supplies. The only human pipetting required is the loading of the two selected antimicrobial stock solutions into dispensehead cassette wells that are equivalent to ink chambers in a standard printer. Here, we have adapted this technology to facilitate combinatorial chequerboard synergy testing and, as proof of principle, performed a comprehensive examination of antimicrobial combinations against a collection of CRE clinical isolates.
Materials and methods
Bacterial strains
The 10 de-identified CRE clinical isolates used in the study were collected at our institution under institutional review board-approved protocols and were sequenced through the CRE genome initiative at the Broad Institute (Cambridge, MA, USA). All contained a Klebsiella pneumoniae carbapenemase (blaKPC) gene and were colony purified, minimally passaged and stored at −80°C prior to use in this study. Escherichia coli ATCC 25922 was obtained from the ATCC (Manassas, VA, USA).
Antimicrobial agents
Antimicrobials were obtained from the following suppliers: Sigma–Aldrich, St Louis, MO, USA (levofloxacin, chloramphenicol, fosfomycin, gentamicin); Alfa Aesar, Tewksbury, MA, USA (gentamicin); Ark Pharm, Libertyville, IL, USA (meropenem); MP Biomedicals, Santa Ana, CA, USA (aztreonam); Research Products International, Mount Prospect, IL, USA (trimethoprim, ertapenem); Chem-Impex International, Wood Dale, IL, USA (sulfamethoxazole, minocycline, cefepime); Santa Cruz Biotechnology, Santa Cruz, CA, USA and Alfa Aesar (colistin); and Fisher Scientific, Pittsburgh, PA, USA (rifampicin). Antibiotic stock solutions used in reference broth microdilution (BMD) testing were dissolved according to CLSI guidelines,12 with the exception of trimethoprim and sulfamethoxazole, which were dissolved in DMSO (Sigma–Aldrich) as they were not soluble in CLSI-recommended solvents at the concentrations required. Antibiotic stock solutions used for the HP D300 digital dispensing method (DDM) were dissolved in either sterile water according to CLSI guidelines12 with the addition of 0.3% polysorbate 20 (P-20; Sigma–Aldrich) or in DMSO (chloramphenicol, trimethoprim, sulfamethoxazole and rifampicin). The DMSO concentration ranged from 0.0008% to 0.968%, below the CLSI-recommended maximum concentration of 1%.12 P-20 is required for proper fluid handling of aqueous antimicrobial stock solutions by the D300 instrument as part of the DDM used for setting up chequerboard arrays. The final concentrations of surfactant in microdilution wells ranged from 3.1 × 10−7% to 0.015%. Of note, a different surfactant, polysorbate 80 (P-80), at a concentration of 0.002%, has been noted to lower colistin MICs for organisms with colistin MICs of <2 mg/L in standard BMD assays.13,14 However, in our assays, P-20 was only introduced in assay wells at concentrations ≥0.002% when colistin concentrations were ≥64 mg/L (with the exception of colistin at ≥2 mg/L in combination with aztreonam and levofloxacin at ≥256 and ≥128 mg/L, respectively) and therefore was considered unlikely to interfere with assays. Our laboratory previously demonstrated that P-20 at all concentrations tested (up to 0.0015%) had no effect on DDM results in comparison with reference BMD.11 Therefore P-20 should have no discernible effect on MIC values, as supported by the high essential agreement between DDM and BMD presented in the Results section. Antimicrobials were stored as aliquots at −20°C and discarded after a single use. All antibiotic stock solutions were quality control (QC) tested with E. coli ATCC 25922 prior to experiments and were used only if they produced an MIC result within the accepted CLSI QC range.12
MIC determination for individual antimicrobials
Reference BMD testing was performed according to CLSI guidelines using the direct colony suspension method.15 Serial 2-fold dilutions of each antimicrobial were prepared at double concentrations in 50 μL volumes of CAMHB (BD Diagnostics, Franklin Lakes, NJ, USA) in 96-well plates (Evergreen Scientific, Los Angeles, CA, USA), which were stored at −80°C until use. Each plate contained negative control wells to assess for contamination of broth or reagents, and positive control wells to verify bacterial growth. A representative plate from each lot was QC tested with E. coli ATCC 25922 prior to use of that lot for clinical strain testing. Maximum antimicrobial concentrations were at least one 2-fold dilution above the resistance breakpoint for Enterobacteriaceae; in the case of rifampicin, for which there are no interpretive criteria for Enterobacteriaceae, concentrations up to two 2-fold dilutions above the maximum expected MIC for E. coli ATCC 25922 were included.12
Bacterial inocula were prepared by suspending and diluting colonies in CAMHB to an OD600 of 0.0006, which corresponds to ∼1 × 106 cfu/mL for E. coli ATCC 25922. Fifty microlitres of the bacterial suspension was added to each well, bringing the bacteria to a final concentration of ∼5 × 105 cfu/mL. Panels were incubated at 37°C in ambient air for 16–20 h. The MIC was defined as the lowest concentration of antimicrobial resulting in complete inhibition of growth as determined visually. BMD MICs were determined in duplicate for each strain and antibiotic. If the two results were discrepant, the higher MIC was considered the final BMD MIC.
For fosfomycin, agar dilution reference testing was performed instead, as recommended by CLSI.12,15 Agar dilution plates were prepared by adding one part of fosfomycin stock solution at 10 times the final concentration to nine parts of molten Bacto agar (Becton, Dickinson and Company, Sparks, MD, USA) containing non-cation-adjusted Mueller–Hinton broth (Becton, Dickinson and Company) and glucose-6-phosphate (G6P; Sigma–Aldrich; final concentration 25 mg/L). At the time of use, bacterial inocula were adjusted to an OD600 of 0.01, which corresponds to ∼1–2 × 107 cfu/mL for E. coli ATCC 25922. Two microlitres of this bacterial suspension was spotted on the surface of each agar plate, with each spot containing ∼1 × 104 cfu. QC testing of E. coli ATCC 25922 was performed in parallel.
DDM MIC testing was performed with the HP D300 digital dispenser (HP, Inc., Palo Alto, CA, USA) as previously described by our laboratory.11 Immediately prior to addition of bacterial suspensions, antimicrobial stock solutions were dispensed by the D300 into empty, flat-bottomed, untreated 384-well polystyrene plates (Greiner Bio-One, Monroe, NC, USA) in volumes ranging from 0.0521 to 323 nL to produce the final desired doubling dilution concentrations with maximum final concentrations at least one 2-fold dilution above the resistance breakpoint for each antibiotic.
Bacterial inocula were adjusted to an OD600 of 0.0003 in CAMHB, which corresponds to ∼5 × 105 cfu/mL for E. coli ATCC 25922, and 50 μL of this bacterial suspension was added to each well using a multichannel pipette. For fosfomycin testing, the bacterial suspension was supplemented with 25 mg/L G6P. Plates were incubated at 37°C in ambient air for 16–20 h. After incubation, bacterial growth was quantified by measurement of OD600 using an Epoch (BioTek, Winooski, VT, USA) or Spark 10M microplate reader (Tecan, Morrisville, NC, USA). An OD600 reading of ≥0.08 (approximately twice typical background readings in wells containing broth alone) was considered indicative of bacterial growth (as also appreciable by visual assessment).
Chequerboard array testing
To create chequerboard arrays, serial 2-fold dilutions of antimicrobial pairings were dispensed in orthogonal titrations by the D300, i.e. two-dimensional DDM. Titrations consisted of up to seven doubling dilutions for each antibiotic. When an isolate’s MIC was below the resistance breakpoint, the maximum concentration tested was two doubling dilutions above the MIC. When the MIC was at or above the resistance breakpoint, the maximum concentration tested was at least one doubling dilution above the resistance breakpoint. Inoculum addition, incubation and growth determination were performed as described for single antimicrobial DDM.
For wells in which growth was inhibited, a fractional inhibitory concentration (FIC) for each antimicrobial was calculated by dividing the concentration of the antibiotic in the well by the MIC of the antibiotic when tested alone.16 The FIC index (FICI) was determined by summing the FICs of the two antimicrobials in each inhibited well. The lowest FICI value in each chequerboard array (FICI-MIN) was used to determine whether the combination was synergistic, as described below. When a ‘skipped well’ occurred (i.e. inhibition of bacterial growth at a given FICI, but growth at the next highest FICI), the higher FICI was considered the FICI-MIN in order to avoid false-positive synergy interpretations.
CLSI-recommended interpretive breakpoints for Enterobacteriaceae were used for all categorical interpretations12 with the exception of colistin, for which EUCAST breakpoints were used,17 and rifampicin, for which formal interpretive criteria are not available and for which an MIC of ≥4 mg/L was considered resistant in accordance with previous investigations in Acinetobacter species.18,19
Data analysis
Statistical analysis was performed using R software v3.1 (R Foundation, Vienna, Austria) and Microsoft Excel 2013 (Microsoft Corporation, Redmond, WA, USA). The χ2 test was used for comparison of proportions.
Results
Precision and accuracy
Overall, 1585 MIC values were collected during the two stages of DDM synergy testing described below; 78.7% were on-scale (i.e. they fell within the range of MICs included in the antibiotic titration). Modal DDM MICs were calculated by taking the mode of all DDM MIC measurements obtained for a given antibiotic–strain combination, inclusive of initial MIC titrations and single-antibiotic rows/columns in synergy arrays (Table 1). Among on-scale MIC results for which the modal DDM MIC was also on-scale, 96.8% (1141/1179) were within plus or minus one 2-fold dilution of the modal DDM MICs, indicating high intra-method precision as observed previously.9,11 Among on-scale DDM MIC results for which the reference BMD MIC result determined prior to synergy experiments was also on-scale, 91.3% (1025/1123) were within plus or minus one 2-fold dilution of the BMD result. Among all DDM MIC results from assays in which dilution ranges spanned the susceptible, intermediate and resistant breakpoints, 89.1% (1408/1580) were in categorical agreement with BMD results. Among all DDM MIC results, the minor (either BMD or DDM result intermediate and the other susceptible or resistant), major (DDM resistant and BMD susceptible) and very major (DDM susceptible and BMD resistant) error rates were 10%, 0.13% and 0.44%, respectively. Consistent with our previous results,9,11 these data showed DDM MIC measurements to be both robustly precise and accurate by generally accepted standards,20 in this case for a highly antimicrobial-resistant CRE strain set with many MICs lying on a breakpoint, which inherently increases rates of minor categorical disagreement. Our results thereby supported use of the underlying technology in two-dimensional chequerboard testing of CRE.
Table 1.
Characteristics and MICs of carbapenem-resistant bacterial isolates examined in synergy experiments
| Strain characteristics |
DDM modal MICs, mg/L |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BIDMC strain | species | KPC type | MEM | ETP | ATM | FEP | CST | MIN | GEN | CHL | LVX | SXT | FOF | RIFa |
| 4 | KPN | 3 | 32 | 32 | >64 | 64 | 0.25 | 8 | 32 | >256 | 128 | 4/76 | 128 | 32 |
| 5 | KPN | 3 | 8 | 16 | >128 | 16 | 0.5 | 16 | 32 | >256 | >64 | >8/152 | 8 | 32 |
| 10 | KPN | 3 | 4 | 16 | >512 | 16 | 0.25 | 4 | 32 | >64 | 16 | >32/608 | 64 | 32 |
| 12A | KPN | 3 | 8 | 32 | >512 | 16 | 0.25 | 4 | 1 | 64 | 32 | >8/152 | 8 | 32 |
| 18A | KPN | 2 | 32 | 64 | >128 | >64 | >32 | 2 | 64 | >256 | 64 | >8/152 | 4 | 16 |
| 6 | ECO | 2 | 2 | 8 | 256 | 8 | 0.25 | 1 | >32 | 8 | 16 | >32/608 | 0.5 | 16 |
| 9 | ECO | 2 | 4 | 8 | >512 | 32 | 0.25 | 1 | >32 | 8 | 16 | >32/608 | 0.5 | 16 |
| 15 | ECO | 2 | 4 | 32 | 512 | 4 | 0.25 | 1 | 64 | 8 | 32 | >8/152 | 0.5 | 32 |
| 17A | ECO | 2 | 2 | 8 | 256 | 4 | 0.25 | 1 | >32 | 8 | 16 | >32/608 | 0.5 | 32 |
| 20A | ECO | 3 | 1 | 4 | 256 | 8 | 0.25 | 4 | 0.5 | 16 | 32 | >32/608 | 1 | 16 |
ECO, E. coli; KPN, K. pneumoniae; ATM, aztreonam; CHL, chloramphenicol; CST, colistin; ETP, ertapenem; FEP, cefepime; FOF, fosfomycin; GEN, gentamicin; LVX, levofloxacin; MEM, meropenem; MIN, minocycline; RIF, rifampicin; SXT, trimethoprim/sulfamethoxazole.
Dark grey shading indicates an MIC classified by CLSI or EUCAST (for colistin) as resistant, light grey shading indicates an MIC classified by CLSI or EUCAST (for colistin) as intermediate (susceptible dose-dependent for cefepime) and no shading indicates an MIC classified by CLSI or EUCAST (for colistin) as susceptible.
There are no established Enterobacteriaceae interpretive criteria for rifampicin. Rifampicin MICs ≥4 mg/L are classified as resistant as discussed in the text.
Synergy testing screen
All two-drug combinations of meropenem, aztreonam, cefepime, colistin, gentamicin, levofloxacin, chloramphenicol, fosfomycin, trimethoprim/sulfamethoxazole, minocycline and rifampicin, as well as the double carbapenem combination of meropenem and ertapenem, were initially tested in duplicate against four bacterial screening strains (BIDMC 4, BIDMC 5, BIDMC 12A and BIDMC 15). Trials were repeated with a new inoculum if they were uninterpretable due to multiple skipped wells or if the MIC of either of the individual drugs was more than one 2-fold dilution above or below the MIC determined by DDM in advance of the synergy experiments. If multiple skipped wells recurred on repeat testing, the combination was not further assayed against that strain.
In total, 521 trials were performed, which included 448 trials comprising 56 antibiotic combinations assayed in duplicate against four bacterial strains. Forty-nine of 521 trials (9.4%) were uninterpretable due to multiple skipped wells. Nearly all trials with multiple skipped wells (46/49; 94%) included cefepime and/or fosfomycin and the rates of multiple skipped wells were significantly higher among trials containing cefepime (29/110, 26%) and fosfomycin (21/94; 22.3%) than among trials containing neither of these antibiotics (4/327; 1.2%, P < 0.001).
For each of the 448 trials used for data analysis, the FICI-MIN was calculated as described in the Materials and methods section and the concentration of each antibiotic at the FICI-MIN was categorized as susceptible, intermediate or resistant. Trials for which the FICI-MIN was ≤0.75 and the concentrations of both antibiotics at the FICI-MIN were within the susceptible or intermediate category were considered to show potential clinically relevant synergy. The FICI-MIN cut-off of ≤0.75, which is higher than the traditionally accepted cut-off of ≤0.5 for synergy,21 was chosen for screening in order to increase sensitivity for detection of combinations that might show synergy against bacterial strains other than those used at the screening stage.
Overall, 206/448 trials (46%) had an FICI-MIN of ≤0.75 and 51/448 (11%) met criteria for potential clinically relevant synergy as listed for each combination in Table S1 (available as Supplementary data at JAC Online). No trials demonstrated antagonism (FICI-MIN >4.0).21
Spectrum of activity evaluation
The nine antibiotic combinations that met criteria for potential clinically relevant synergy in two or more trials in the screening stage were selected for activity spectrum evaluation. These were minocycline and colistin; colistin and rifampicin; gentamicin and colistin; minocycline and gentamicin; levofloxacin and colistin; meropenem and colistin; minocycline and meropenem; chloramphenicol and colistin; and chloramphenicol and meropenem. Combinations containing fosfomycin and/or cefepime were excluded based on the high rates of skipped wells. Each selected combination was tested in duplicate on separate days against the 10 clinical KPC-producing CRE strains (5 K. pneumoniae and 5 E. coli) listed in Table 1, including the original 4 used in the synergy testing screen.
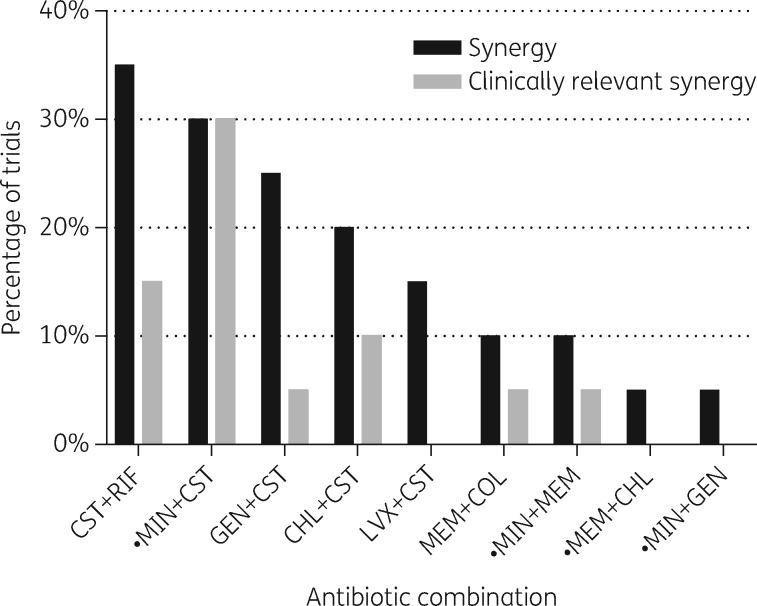
A trial was classified as demonstrating synergy if it had an FICI-MIN of ≤0.5,21 and clinically relevant synergy if concentrations of both antibiotics at the FICI-MIN were also within the susceptible or intermediate category. Overall, 31/180 trials (17.2%) demonstrated synergy and 14/180 trials (7.8%) demonstrated clinically relevant synergy. The percentage of trials that demonstrated synergy and clinically relevant synergy varied among the antibiotic combinations, with the combination of colistin plus minocycline demonstrating the highest rate of clinically relevant synergy at 30% (Figure 1). For 8 of the 10 strains (80%), combinations were identified that demonstrated clinically relevant synergy in at least one trial, but these combinations varied among strains (Table 2). The results of all 180 trials are detailed in Table S2.
Figure 1.
Combinatorial activity spectrum. Percentage of trials of indicated antimicrobial combinations demonstrating synergy (FICI-MIN ≤0.5) and clinically relevant synergy (FICs of both antibiotics at the FICI-MIN within the susceptible or intermediate category) against a collection of five K. pneumoniae and five E. coli CRE strains. CHL, chloramphenicol; CST, colistin; GEN, gentamicin; LVX, levofloxacin; MEM, meropenem; MIN, minocycline; RIF, rifampicin. Filled circles identify combinations for which synergy testing against CRE has not previously been reported.
Table 2.
Antibiotic combinations demonstrating clinically relevant synergy against a CRE strain set
 |
Grey shading indicates a combination that demonstrated clinically relevant synergy in at least one trial for the designated isolate.
ECO, E. coli; KPN, K. pneumoniae; CHL, chloramphenicol; CST, colistin; GEN, gentamicin; LVX, levofloxacin; MEM, meropenem; MIN, minocycline; RIF, rifampicin.
A filled circle indicates a combination for which synergy testing against CRE has not previously been reported.
In 120/180 trials (67%) at least one of the antibiotics had an MIC in the resistant range for the isolate being tested. Clinically relevant synergy was demonstrated in 8/120 (7%) of these cases. In other words, the concentrations of the antibiotic(s) with resistant MICs were brought into the intermediate or susceptible range. These 8 trials represent 57% of the 14 total trials with clinically relevant synergy. In the other six trials, both antibiotics had MICs in the susceptible or intermediate range individually. Notably, for strain BIDMC 18A, which is highly resistant to colistin (MIC >128 mg/L), the combinations of colistin plus minocycline and colistin plus rifampicin resulted in reduction of inhibitory colistin concentrations into the susceptible range (1–2 mg/L) and similarly dramatic reduction in minocycline and rifampicin inhibitory concentrations (0.5 and 1 mg/L), respectively.
Discussion
Through the application of an inkjet printer-based automated chequerboard method, we were able to rapidly test 56 antimicrobial combinations, including many that have not been evaluated in the literature to date, against a screening set of CRE strains. We identified six combinations that showed clinically relevant synergy in one or more trials in activity spectrum evaluation. Among these were two combinations that have frequently been reported as demonstrating synergy against CRE (colistin plus rifampicin8,22,23 and a carbapenem plus colistin8,24,25).
Three of our findings merit further discussion. First, we identified two novel antibiotic combinations demonstrating clinically relevant synergy whose efficacy against CRE has not previously been described (Figure 1; minocycline plus colistin and minocycline plus meropenem). Both of these combinations include minocycline, a tetracycline antibiotic that has recently attracted attention as a therapeutic option for CRE and other resistant Gram-negative bacteria.26,27 Minocycline has several potential advantages over the tetracycline derivative tigecycline, a glycylcycline antibiotic that has been used to treat CRE both alone and in combination.27,28 Unlike tigecycline, minocycline is available in both oral and intravenous (iv) forms, allowing easier outpatient therapy in patients with less severe infections or those for whom a longer course of therapy is desired after completion of an initial iv antibiotic course. Minocycline also has a generally favourable side-effect profile,29 while tigecycline is associated with significant rates of nausea and vomiting.30 Furthermore, unlike tigecycline, which has limited urinary excretion,31 raising concerns about its utility for treatment of urinary tract infection (UTI),30,32 minocycline has an FDA-approved indication for UTI,33 which is one of the most common manifestations of CRE infection.1,34 Minocycline is also potentially a preferable agent for treatment of bloodstream infections, as it reaches higher serum concentrations than tigecycline.31,33
Second, while some double carbapenem combinations have previously been shown to demonstrate in vitro synergy against CRE,35 we did not observe clinically relevant synergy for the combination of meropenem and ertapenem in the screening stage. This may have been due to the high ertapenem MICs of the strains, which ranged from 8 to 64 mg/L. A previous investigation of the combination of meropenem and ertapenem similarly showed high rates of synergy, but at concentrations above those clinically achievable.36 Therefore, data suggest that meropenem/ertapenem combinations may not provide reliable benefit.
Third, significant heterogeneity in synergistic activity was observed against different strains, a finding that is consistent with prior studies, but not generally emphasized.5,8,35,37,38 Even combinations with the highest rates of synergistic activity showed clinically relevant synergy in no more than one-third of trials. Importantly, this finding underscores the need for individualized synergy testing to determine which combinations will be effective for a given patient’s isolate. Studies such as ours, which identify those combinations that are most likely to be synergistic, can be used to select high-yield combinations for clinical testing.
In our study, we classified combinations according to whether or not they demonstrated clinically relevant synergy. We considered combinations in which inhibitory concentrations were lowered into the intermediate range clinically relevant because there is increasing interest in the use of higher doses of antibiotics, particularly β-lactams, to treat organisms with MICs classified as intermediate.39–41 The concept of clinically relevant synergy has not been consistently applied in the literature to date, with some studies reporting only synergistic combinations in which concentrations fall within a clinically achievable range based on pharmacodynamic parameters,5 while others present only limited data, if any, on the final concentrations of antibiotics in synergistic combinations.8,37 Furthermore, the clinical significance of synergy in combinations in which an isolate is already susceptible to each antibiotic individually is less well established and warrants further investigation. It is possible, for example, that dosing of some of the most toxic antibiotics, including colistin and aminoglycosides, could be reduced, thus potentially decreasing the risk of toxicity.
In determining which combinations met criteria for synergy in the activity spectrum evaluation, we used the standard FICI-MIN cut-off of ≤0.5. A conservative cut-off of this type corrects for the well-known plus or minus one 2-fold variability inherent in MIC testing.20,42 This variability is inevitably increased when two antibiotics are assayed simultaneously. However, it is plausible that FICI-MIN values that repeatedly fall just above the 0.5 cut-off are truly synergistic. Notably, the ease with which DDM chequerboard synergy testing can be performed could allow combinations to be routinely tested in duplicate or triplicate, thereby increasing confidence in the FICI-MIN result.
We noted a markedly higher rate of skipped wells in synergy arrays containing fosfomycin and/or cefepime. Frequent skipped wells and inconsistent MIC results have been noted when fosfomycin is tested by BMD, due in part to particular sensitivity to the inoculum effect,43 so this finding was not entirely unexpected. It is possible that skipped wells with cefepime were due in part to the inoculum effect, which is pronounced for later-generation cephalosporin antibiotics such as cefepime when tested against β-lactamase-producing Enterobacteriaceae.44
Our study has certain limitations. First, chequerboard array testing does not provide bactericidal or time–kill data and, while there are insufficient data on the predictive clinical value of different synergy testing methods to determine whether one is superior to another,45 it is possible that time–kill synergy assays, which provide data on bactericidal activity over time, may more accurately reflect in vivo synergy. However, time–kill studies are significantly more labour intensive than chequerboard studies and the time–kill method is not practical for a large screen such as we performed here. We expect to test promising combinations by time–kill assay as a follow-up step. Second, without accepted interpretive breakpoints for rifampicin, we used a resistance cut-off based on previous investigations of rifampicin for use in Acinetobacter species. Given the lack of pharmacodynamic data on the use of rifampicin against Enterobacteriaceae, it is uncertain how accurately this cut-off reflects clinical efficacy. Third, more generally, pharmacokinetic/pharmacodynamic studies in animal infection models and future clinical trials will be essential to establish linkage between in vitro synergy results and treatment benefit.
Importantly, we demonstrated the utility of a new technology to support systematic testing of a wide range of antibiotic combinations against a collection of CREs. The DDM synergy method essentially eliminates cumulative error in serial dilution and dramatically reduces the time required to perform a chequerboard array. Using DDM, a chequerboard array can be set up in ∼2 min, which includes pipetting of the two stock antimicrobial solutions (one for each antibiotic) into the D300 cassette channel and dispensing of antimicrobials by the D300 using pre-programmed protocols. In contrast, a synergy array prepared manually according to the protocol published by the American Society for Microbiology46 involves the use of eight different stock solutions (up to five per antibiotic depending on the concentration range to be tested), preparation of 18 intermediate antimicrobial concentrations, dispensing of each of these intermediate concentrations into an individual row or column of a microtitre plate and addition of extra liquid medium to reach final appropriate volumes, a process requiring 30 min at minimum.
We identified novel synergistic combinations and have also illustrated the variability of synergistic activity against different CRE strains. Future studies that expand synergy testing against a comprehensive, diverse collection of CREs will be needed to provide the foundation for a more definitive understanding of the antibiotic combinations that are most frequently synergistic against specific types of CRE, which will in turn serve as guidance for empirical combination therapy and for focused testing of patient isolates. This study provides proof-of-concept support for the ability of DDM technology to perform synergy analysis within an actionable time frame and to serve as a powerful and versatile general approach for synergy chequerboard exploration using prospectively or retrospectively collected isolates.
Supplementary Material
Acknowledgments
Funding
This work was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health (T32HD055148 to T. B.-K.) and the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (R21AI119114, R21AI112694 to J. E. K.).
Transparency declarations
The HP D300 digital dispenser and associated consumables were provided by Tecan (Morrisville, NC, USA). J. E. K. also received an honorarium and travel support from Tecan. Tecan had no role in study design, data collection/interpretation, manuscript preparation or decision to publish.
Disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Supplementary data
Tables S1 and S2 are available as Supplementary data at JAC Online.
References
- 1. Temkin E, Adler A, Lerner A. et al. Carbapenem-resistant Enterobacteriaceae: biology, epidemiology, and management. Ann NY Acad Sci 2014; 1323: 22–42. [DOI] [PubMed] [Google Scholar]
- 2. Zusman O, Avni T, Leibovici L. et al. Systematic review and meta-analysis of in vitro synergy of polymyxins and carbapenems. Antimicrob Agents Chemother 2013; 57: 5104–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Paul M, Carmeli Y, Durante-Mangoni E. et al. Combination therapy for carbapenem-resistant Gram-negative bacteria. J Antimicrob Chemother 2014; 69: 2305.. [DOI] [PubMed] [Google Scholar]
- 4. Rafailidis PI, Falagas ME.. Options for treating carbapenem-resistant Enterobacteriaceae. Curr Opin Infect Dis 2014; 27: 479–83. [DOI] [PubMed] [Google Scholar]
- 5. Clock SA, Tabibi S, Alba L. et al. In vitro activity of doripenem alone and in multi-agent combinations against extensively drug-resistant Acinetobacter baumannii and Klebsiella pneumoniae. Diagn Microbiol Infect Dis 2013; 76: 343–6. [DOI] [PubMed] [Google Scholar]
- 6. Hirsch EB, Guo B, Chang KT. et al. Assessment of antimicrobial combinations for Klebsiella pneumoniae carbapenemase-producing K. pneumoniae. J Infect Dis 2013; 207: 786–93. [DOI] [PubMed] [Google Scholar]
- 7. Tängdén T, Hickman RA, Forsberg P. et al. Evaluation of double- and triple-antibiotic combinations for VIM- and NDM-producing Klebsiella pneumoniae by in vitro time-kill experiments. Antimicrob Agents Chemother 2014; 58: 1757–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tascini C, Tagliaferri E, Giani T. et al. Synergistic activity of colistin plus rifampin against colistin-resistant KPC-producing Klebsiella pneumoniae. Antimicrob Agents Chemother 2013; 57: 3990–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Smith KP, Brennan-Krohn T, Weir S. et al. Improved accuracy of cefepime susceptibility testing for ESBL-producing Enterobacteriaceae using an on-demand digital dispensing method. J Clin Microbiol 2016; 55: 470–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Smith KP, Kirby JE.. How inkjet printing technology can defeat multidrug-resistant pathogens. Future Microbiol 2016; 11: 1375–7. [DOI] [PubMed] [Google Scholar]
- 11. Smith KP, Kirby JE.. Verification of an automated, digital dispensing platform for at-will broth microdilution-based antimicrobial susceptibility testing. J Clin Microbiol 2016; 54: 2288–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Sixth Informational Supplement M100-S26. CLSI, Wayne, PA, USA, 2016. [Google Scholar]
- 13. Hindler JA, Humphries RM.. Colistin MIC variability by method for contemporary clinical isolates of multidrug-resistant Gram-negative bacilli. J Clin Microbiol 2013; 51: 1678–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sutherland CA, Nicolau DP.. To add or not to add polysorbate 80: impact on colistin MICs for clinical strains of Enterobacteriaceae and Pseudomonas aeruginosa and quality controls. J Clin Microbiol 2014; 52: 3810.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically—Tenth Edition: Approved Standard M07-A10. CLSI, Wayne, PA, USA, 2015. [Google Scholar]
- 16. Elion GB, Singer S, Hitchings GH.. Antagonists of nucleic acid derivatives. VIII. Synergism in combinations of biochemically related antimetabolites. J Biol Chem 1954; 208: 477–88. [PubMed] [Google Scholar]
- 17. EUCAST. Clinical Breakpoints http://www.eucast.org/clinical_breakpoints/.
- 18. Henwood CJ, Gatward T, Warner M. et al. Antibiotic resistance among clinical isolates of Acinetobacter in the UK, and in vitro evaluation of tigecycline (GAR-936). J Antimicrob Chemother 2002; 49: 479–87. [DOI] [PubMed] [Google Scholar]
- 19. Tan TY, Ng LS, Poh K.. Susceptibility testing of unconventional antibiotics against multiresistant Acinetobacter spp. by agar dilution and Vitek 2. Diagn Microbiol Infect Dis 2007; 58: 357–61. [DOI] [PubMed] [Google Scholar]
- 20. Clark R, Lewinski M, Loeffelholz M. et al. Cumitech 31A, Verification and Validation of Procedures in the Clinical Microbiology Laboratory. Washington, DC: ASM Press, 2009. [Google Scholar]
- 21. Odds FC. Synergy, antagonism, and what the chequerboard puts between them. J Antimicrob Chemother 2003; 52: 1.. [DOI] [PubMed] [Google Scholar]
- 22. Bratu S, Tolaney P, Karumudi U. et al. Carbapenemase-producing Klebsiella pneumoniae in Brooklyn, NY: molecular epidemiology and in vitro activity of polymyxin B and other agents. J Antimicrob Chemother 2005; 56: 128–32. [DOI] [PubMed] [Google Scholar]
- 23. Elemam A, Rahimian J, Doymaz M.. In vitro evaluation of antibiotic synergy for polymyxin B-resistant carbapenemase-producing Klebsiella pneumoniae. J Clin Microbiol 2010; 48: 3558–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jernigan MG, Press EG, Nguyen MH. et al. The combination of doripenem and colistin is bactericidal and synergistic against colistin-resistant, carbapenemase-producing Klebsiella pneumoniae. Antimicrob Agents Chemother 2012; 56: 3395–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pankey GA, Ashcraft DS.. Detection of synergy using the combination of polymyxin B with either meropenem or rifampin against carbapenemase-producing Klebsiella pneumoniae. Diagn Microbiol Infect Dis 2011; 70: 561–4. [DOI] [PubMed] [Google Scholar]
- 26. Pogue JM, Neelakanta A, Mynatt RP. et al. Carbapenem-resistance in Gram-negative bacilli and intravenous minocycline: an antimicrobial stewardship approach at the Detroit Medical Center. Clin Infect Dis 2014; 59: S388–93. [DOI] [PubMed] [Google Scholar]
- 27. Thaden JT, Pogue JM, Kaye KS.. Role of newer and re-emerging older agents in the treatment of infections caused by carbapenem-resistant Enterobacteriaceae. Virulence 2017; 8: 403–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ni W, Han Y, Liu J. et al. Tigecycline treatment for carbapenem-resistant Enterobacteriaceae infections: a systematic review and meta-analysis. Medicine (Baltimore) 2016; 95: e3126.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ritchie DJ, Garavaglia-Wilson A.. A review of intravenous minocycline for treatment of multidrug-resistant Acinetobacter infections. Clin Infect Dis 2014; 59: S374–80. [DOI] [PubMed] [Google Scholar]
- 30. Doan TL, Fung HB, Mehta D. et al. Tigecycline: a glycylcycline antimicrobial agent. Clin Ther 2006; 28: 1079–106. [DOI] [PubMed] [Google Scholar]
- 31. Wyeth Pharmaceuticals. Tygacil® (Tigecycline) for Injection, Package Insert. Philadelphia, PA, USA, 2010. [Google Scholar]
- 32. Muralidharan G, Micalizzi M, Speth J. et al. Pharmacokinetics of tigecycline after single and multiple doses in healthy subjects. Antimicrob Agents Chemother 2005; 49: 220–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. The Medicines Company. MINOCIN® (Minocycline) for Injection, Package Insert. Parsippany, NJ, USA, 2015. [Google Scholar]
- 34. Guh AY, Bulens SN, Mu Y. et al. Epidemiology of carbapenem-resistant Enterobacteriaceae in 7 US communities, 2012-2013. JAMA 2015; 314: 1479–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Poirel L, Kieffer N, Nordmann P.. In vitro evaluation of dual carbapenem combinations against carbapenemase-producing Enterobacteriaceae. J Antimicrob Chemother 2016; 71: 156–61. [DOI] [PubMed] [Google Scholar]
- 36. Oliva A, Gizzi F, Mascellino MT. et al. Bactericidal and synergistic activity of double-carbapenem regimen for infections caused by carbapenemase-producing Klebsiella pneumoniae. Clin Microbiol Infect 2016; 22: 147–53. [DOI] [PubMed] [Google Scholar]
- 37. Berçot B, Poirel L, Dortet L. et al. In vitro evaluation of antibiotic synergy for NDM-1-producing Enterobacteriaceae. J Antimicrob Chemother 2011; 66: 2295–7. [DOI] [PubMed] [Google Scholar]
- 38. Albiero J, Sy SKB, Mazucheli J. et al. Pharmacodynamic evaluation of the potential clinical utility of fosfomycin and meropenem in combination therapy against KPC-2-producing Klebsiella pneumoniae. Antimicrob Agents Chemother 2016; 60: 4128–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Morrill HJ, Pogue JM, Kaye KS. et al. Treatment options for carbapenem-resistant Enterobacteriaceae infections. Open Forum Infect Dis 2015; 2: ofv050.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Daikos GL, Markogiannakis A.. Carbapenemase-producing Klebsiella pneumoniae: (when) might we still consider treating with carbapenems? Clin Microbiol Infect 2011; 17: 1135–41. [DOI] [PubMed] [Google Scholar]
- 41. Alves MD, Ribeiro VB, Tessari JP. et al. Effect of cefepime dose on mortality of patients with Gram-negative bacterial bloodstream infections: a prospective cohort study. J Antimicrob Chemother 2014; 69: 1681–7. [DOI] [PubMed] [Google Scholar]
- 42. US FDA. Class II Special Controls Guidance Document: Antimicrobial Susceptibility Test (AST) Systems. Rockville, MD: US FDA, 2009. [Google Scholar]
- 43. Fuchs PC, Barry AL, Brown SD.. Susceptibility testing quality control studies with fosfomycin tromethamine. Eur J Clin Microbiol Infect Dis 1997; 16: 538–40. [DOI] [PubMed] [Google Scholar]
- 44. Queenan AM, Foleno B, Gownley C. et al. Effects of inoculum and β-lactamase activity in AmpC- and extended-spectrum β-lactamase (ESBL)-producing Escherichia coli and Klebsiella pneumoniae clinical isolates tested by using NCCLS ESBL methodology. J Clin Microbiol 2004; 42: 269–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Doern CD. When does 2 plus 2 equal 5? A review of antimicrobial synergy testing. J Clin Microbiol 2014; 52: 4124–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Humphries RM, Testing: broth microdilution checkerboard and broth macrodilution methods In: Leber AL, ed. Clinical Microbiology Procedures Handbook. Washington, DC: American Society of Microbiology, 2016; 5.16.1–5.8.23. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.