Abstract
Objectives
To characterize NAI-107 and related lantibiotics for their in vitro activity against Gram-negative pathogens, alone or in combination with polymyxin, and against non-dividing cells or biofilms of Staphylococcus aureus. NAI-107 was also evaluated for its propensity to select or induce self-resistance in Gram-positive bacteria.
Methods
We used MIC determinations and chequerboard experiments to establish the antibacterial activity of the examined compounds against target microorganisms. Time–kill assays were used to evaluate killing of exponential and stationary-phase cells. The effects on biofilms (growth inhibition and biofilm eradication) were evaluated using biofilm-coated pegs. The frequency of spontaneous resistant mutants was evaluated by either direct plating or by continuous sub-culturing at 0.5 × MIC levels, followed by population analysis profiles.
Results
The results showed that NAI-107 and its brominated variant are highly active against Neisseria gonorrhoeae and some other fastidious Gram-negative pathogens. Furthermore, all compounds strongly synergized with polymyxin against Acinetobacter baumannii, Escherichia coli, Klebsiella pneumoniae and Pseudomonas aeruginosa, and showed bactericidal activity. Surprisingly, NAI-107 alone was bactericidal against non-dividing A. baumannii cells. Against S. aureus, NAI-107 and related lantibiotics showed strong bactericidal activity against dividing and non-dividing cells. Activity was also observed against S. aureus biofilms. As expected for a lipid II binder, no significant resistance to NAI-107 was observed by direct plating or serial passages.
Conclusions
Overall, the results of the current work, along with previously published results on the efficacy of NAI-107 in experimental models of infection, indicate that this lantibiotic represents a promising option in addressing the serious threat of antibiotic resistance.
Introduction
The increasing incidence of MDR pathogens has led to the dire prediction that humanity will soon enter the post-antibiotic era, when today's routine surgical procedures will become high-risk endeavours.1,2 This worrisome scenario is exacerbated by the paucity of new antibiotics reaching the market, and in particular of new chemical classes suitable for systemic administration and not affected by prevailing resistance mechanisms. Indeed, since the beginning of the century most antibacterial drugs introduced into human use or under advanced clinical development represent improved analogues of marketed compounds.3
Of particular concern are infections caused by the ESKAPE pathogens, which include Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter spp.4 In particular, infections by MRSA are associated with community and hospital infections, and up to 80% of all MRSA infections in the USA have been ascribed to the MRSA clone USA300, which is also highly resistant to other antibiotics.5 Given the limited options available to treat ESKAPE pathogens, it is important to develop new antibiotics that are not affected by prevailing resistance mechanisms, while at the same time devising strategies that minimize the spread of antibiotic resistance.
One class of antibiotics that has been receiving increasing attention is represented by the lantibiotics. These compounds, which belong to the growing family of ribosomally synthesized and post-translationally modified peptides, are characterized by the presence of (methyl)-lanthionine bridges that confer rigidity and stability on the peptide.6 The prototype lantibiotic is nisin, which has been used for decades as a food preservative.
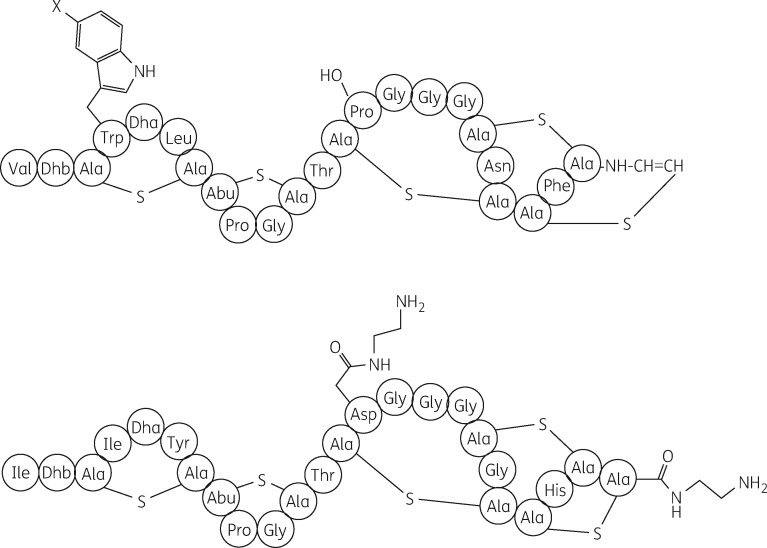
One of the most potent lantibiotics is NAI-107 (Figure 1), a chlorine-containing compound active against MDR Gram-positive pathogens, including MRSA, glycopeptide-intermediate S. aureus (GISA), VRE and penicillin-resistant Streptococcus pneumoniae.7,8 Lantibiotics with similar antibacterial properties have been obtained after incorporation of Br into NAI-107, leading to NAI-108,9 or after semi-synthetic conversion, leading to NAI-857DA and related compounds.10 NAI-857DA and NAI-107/108 share a similar peptide scaffold and identical topology of thioether rings, with the first two N-terminal rings shared with nisin (Figure 1). A rapid bactericidal activity9–11 and a prolonged half-life in plasma12 are consistent with the efficacy of NAI-107 in different experimental models of infection in rodents13 and insects.14 The ratio between the area under the concentration–time curve and the MIC has been proposed as the pharmacodynamic index predictive of efficacy in the mouse.12
Figure 1.
Structures of NAI-107 (top, where X = Cl), NAI-108 (top, where X = Br) and NAI-857DA (bottom). Note that NAI-107 and NAI-108 are obtained directly from fermentation, whereas NAI-857DA is obtained by converting the natural lantibiotic NAI-857 into its diamide.8,10
The scope of this study was to characterize the in vitro properties of NAI-107 with respect to the propensity to select for resistance, its activity against biofilms and non-dividing cells of S. aureus, and its activity against Gram-negative pathogens, alone or in combination with polymyxin B. Where appropriate, the properties of NAI-108 and NAI-857DA were also investigated.
Materials and methods
Materials and methods are available as Supplementary data at JAC Online.
Results
Activity against fastidious Gram-negative pathogens
Most antibiotics targeting lipid II are large molecules that cannot cross the outer membrane barrier present in Gram-negative bacteria.15 However, in contrast to most clinically used drugs targeting Gram-positive pathogens, NAI-107 extends its antibacterial spectrum to some fastidious Gram-negative bacteria (Table 1), including Neisseria meningitidis (MIC range 0.06–1 mg/L), Moraxella catarrhalis (MIC range 0.25–1 mg/L) and Haemophilus influenzae (MIC range 8–16 mg/L). Although only a few strains were tested in comparison, NAI-107 was considerably more active than nisin (Table 1), suggesting that the activity against these bacterial species is not a general property of lantibiotics. No activity was seen against individual strains of Salmonella Enteritidis, Enterobacter cloacae and Proteus mirabilis (data not shown).
Table 1.
MIC (mg/L) or MIC ranges (mg/L) of NAI-107 for selected Gram-negative pathogens
| Species | Number of tested strains | NAI-107 | Vancomycin | Teicoplanin | Linezolid | Nisin |
|---|---|---|---|---|---|---|
| H. influenzaea | 18 | 8–16 | 64 to >128 | 64–128 | 8–32 | >128d |
| M. catarrhalisb | 8 | 0.25–1 | 32–64 | 4–32 | 4–8 | 1–4d |
| N. meningitidisc | 4 | 0.06–1 | 32 to >128 | 16–128 | 8–32 | 8e |
Tested strains include ATCC strains 49247, 9334, 19418 and 9006, and 14 clinical isolates collected in Italy and the UK.
Tested strains include ATCC 8176 and seven clinical isolates collected in the USA and the UK.
Tested strains include ATCC 13804, 13090, 13102 and 13113.
Only two strains tested with this antibiotic.
Only one strain tested with this antibiotic.
Interestingly, NAI-107 was highly active against Neisseria gonorrhoeae, including isolates with intermediate or high resistance to penicillin. Against 18 tested strains, the observed MICs ranged from 0.015 to 2 mg/L (Table 2), with no apparent correlation to a strain's susceptibility to penicillin. Similar results were observed with NAI-108, which, as previously observed with Gram-positive pathogens,9 was usually twice as active as NAI-107 (Table 2). In contrast, NAI-857DA was substantially less active than the other two lantibiotics (Table 2).
Table 2.
MICs (mg/L) of NAI-107, NAI-108, NAI-857DA and penicillin for N. gonorrhoeae
| Strain codea | NAI-107 | NAI-108 | NAI-857DA | Penicillin |
|---|---|---|---|---|
| ATCC 49226 | 0.25 | 0.125 | 2 | 1 |
| L1596 | 0.125 | 0.125 | 4 | ≤0.015 |
| L1599 | 0.015 | 0.015 | 0.25 | 0.25 |
| L1601 | 0.5 | 0.25 | 8 | >32 |
| L1602 | 0.25 | 0.125 | 8 | >32 |
| L1603 | 1 | 1 | 16 | >32 |
| L1604 | 0.5 | 0.25 | 4 | >32 |
| L1605 | 0.25 | 0.125 | 8 | >32 |
| ND755 | 1 | 1 | 8 | 2 |
| ND756 | 0.25 | 0.125 | 2 | 0.5 |
| ND757 | 1 | 0.5 | 16 | 4 |
| ND758 | 1 | 0.5 | 8 | 4 |
| ND759 | 0.25 | 0.125 | 2 | 0.25 |
| ND760 | 1 | 0.5 | 8 | 4 |
| ND761 | 1 | 0.5 | 16 | 4 |
| ND762 | 0.5 | 0.25 | 16 | 2 |
| ND763 | 1 | 0.5 | 8 | 2 |
| ND764 | 2 | 1 | 8 | 2 |
Strains with an L or ND prefix are clinical isolates collected in Italy or the USA.
NAI-107 was essentially inactive against three strains each of the Gram-negative pathogens Escherichia coli, K. pneumoniae and P. aeruginosa, with no measurable MICs at the highest concentration tested (Table 3). Some activity could, however, be observed against A. baumannii, with one strain showing an MIC of 32 mg/L (Table 4). In about half of 12 independent A. baumannii isolates, NAI-108 was slightly more active than NAI-107, with 16 mg/L as the lowest observed MIC (Table 4). It should be noted that none of the analysed strains was resistant to polymyxin (Table 4; see also below).
Table 3.
In vitro synergism with polymyxin B
| Species | Strain code | Polymyxin |
NAI-107 |
FIC index | Polymyxin (mg/L) at 2 mg/L NAI-107a | |||
|---|---|---|---|---|---|---|---|---|
| MIC (mg/L) | FIC | MIC (mg/L) | concentration of NAI-107 in the most synergistic combination (mg/L) | FIC | ||||
| A. baumannii | L373b | 0.5 | 0.03 | 128 | 8 | 0.06 | 0.09 | 0.03 |
| L3030c | 0.125 | 0.06 | 128 | 4 | 0.03 | 0.09 | 0.016 | |
| L2859b | 0.25 | 0.125 | 32 | 0.5 | 0.016 | 0.14 | 0.06 | |
| E. coli | L47d | 0.125 | 0.03 | >256 | 4 | ≤0.008 | 0.04 | 0.008 |
| ATCC 25922 | 0.125 | 0.03 | >256 | 8 | ≤0.016 | 0.05 | 0.004 | |
| ND480b | 0.25 | 0.06 | >256 | 2 | ≤0.004 | 0.07 | 0.016 | |
| K. pneumoniae | L3392b | 0.06 | 0.25 | >256 | 16 | ≤0.03 | 0.28 | 0.03 |
| ND484e | 0.25 | 0.06 | >64 | 4 | ≤0.03 | 0.09 | 0.03 | |
| ND443b | 0.25 | 0.06 | >256 | 2 | ≤0.004 | 0.07 | 0.016 | |
| P. aeruginosa | ATCC 10145 | 0.125 | 0.125 | >256 | 4 | ≤0.008 | 0.13 | 0.03 |
| ATCC 25668 | 0.125 | 0.125 | >256 | 32 | ≤0.06 | 0.19 | 0.06 | |
| ATCC 27853 | 0.25 | 0.25 | >256 | 8 | ≤0.016 | 0.27 | 0.125 | |
The table reports the concentrations of polymyxin (as fractions of the MIC) and those of NAI-107 giving the lowest calculated FIC index for each strain.
Lowest polymyxin concentration required for growth inhibition in the presence of 2 mg/L NAI-107.
Clinical isolate collected in Italy.
Clinical isolate collected in the UK.
Strain from historical Lepetit collection.
Clinical isolate collected in the USA.
Table 4.
Antibacterial activities against A. baumannii
| Strain codea | MIC (mg/L) |
|||
|---|---|---|---|---|
| NAI-107 | NAI-108 | polymyxin | colistin | |
| L256 | 128 | 128 | 1 | 1 |
| L2831 | 128 | 32 | ≤0.06 | 1 |
| L2859 | 32 | 16 | ≤0.06 | 0.125 |
| L2860 | 128 | 128 | 0.125 | 0.5 |
| L364 | >128 | >128 | ≤0.06 | 0.5 |
| L373 | 128 | 64 | 0.25 | 1 |
| L3030 | 128 | 128 | 0.25 | 0.25 |
| ATCC 17904 | 128 | 128 | ≤0.06 | ≤0.06 |
| L756 | 128 | 128 | ≤0.06 | ≤0.06 |
| ND021808 | 128 | 128 | ≤0.06 | ≤0.06 |
| ND045309 | 64 | 32 | ≤0.06 | ≤0.06 |
| ND048710 | 128 | 64 | ≤0.06 | 0.25 |
| ND049010 | 128 | 128 | 0.125 | 0.25 |
| WT 19606 | >128 | NT | NT | 0.5 |
| AB167 (lpxC)b | 2–4 | NT | NT | >128 |
| AB176 (lpxD)b | 2–4 | NT | NT | >128 |
| CR17 (pmrA)b | >128 | NT | NT | 64 |
NT, not tested.
Strains with an L or ND prefix are clinical isolates collected in Italy.
Strains described by García-Quintanilla et al.19
The acquisition of polymyxin resistance in Gram-negative species involves LPS modifications16–18 and vancomycin is active against some LPS-deficient A. baumannii mutants.19 We thus tested the LPS-deficient A. baumannii mutants AB167R and AB176R (defective in LPS formation) and CR17 (which adds phosphoethanolamine to LPS), as well as the polymyxin-susceptible WT ATCC 19606.20 The LPS-deficient strains proved susceptible to NAI-107 (Table 4), with MICs of 2–4 mg/L, while the WT and the pmrA mutant were not susceptible (MIC >128 mg/L). Therefore, the activity of NAI-107 against Gram-negative species should be achievable via permeabilization of the outer membranes.
Synergism with polymyxin
Antibiotics (e.g. vancomycin, daptomycin) that are not effective against Gram-negative bacteria can become active if the permeability barrier provided by the outer membrane is weakened by sub-inhibitory concentrations of polymyxin or colistin.21–24 These drugs have become the last options to treat infections by MDR Gram-negative pathogens, but they also have significant toxicity. If polymyxin or colistin concentrations could be substantially reduced in effective combinations with NAI-107, the toxic effect would be less severe.
We thus tested combinations of NAI-107 and polymyxin against three independent isolates each of the target pathogens A. baumannii, E. coli, K. pneumoniae and P. aeruginosa. Against A. baumannii, the fractional inhibitory concentrations (FICs) of polymyxin and NAI-107 were 0.03–0.125 and 0.016–0.06, respectively, with FIC indexes ranging from 0.09 to 0.14 (Table 3). Similarly, polymyxin and NAI-107 formed synergistic combinations against all the tested strains of the other examined species: the lowest FICs of polymyxin against E. coli, K. pneumoniae and P. aeruginosa were 0.03–0.06, 0.06–0.25 and 0.125–0.25, respectively, with growth inhibition requiring NAI-107 concentrations of 2–8, 2–16 and 4–32 mg/L, respectively (Table 3). Since NAI-107 was inactive against these strains, FIC indexes were essentially determined by the polymyxin FIC and ranged from 0.28 (for K. pneumoniae L3392) to 0.04 (for E. coli L47).
We also evaluated the lowest polymyxin concentration required to inhibit the growth of the analysed strains in the presence of 2 mg/L NAI-107, a concentration that can be readily maintained after systemic administration in rodents.12,13 With the exception of P. aeruginosa ATCC 27853, for which the polymyxin concentration was 0.125 mg/L (i.e. 0.5 × MIC), in all other cases ≤0.06 mg/L polymyxin was sufficient to inhibit growth in the presence of 2 mg/L NAI-107 (Table 3).
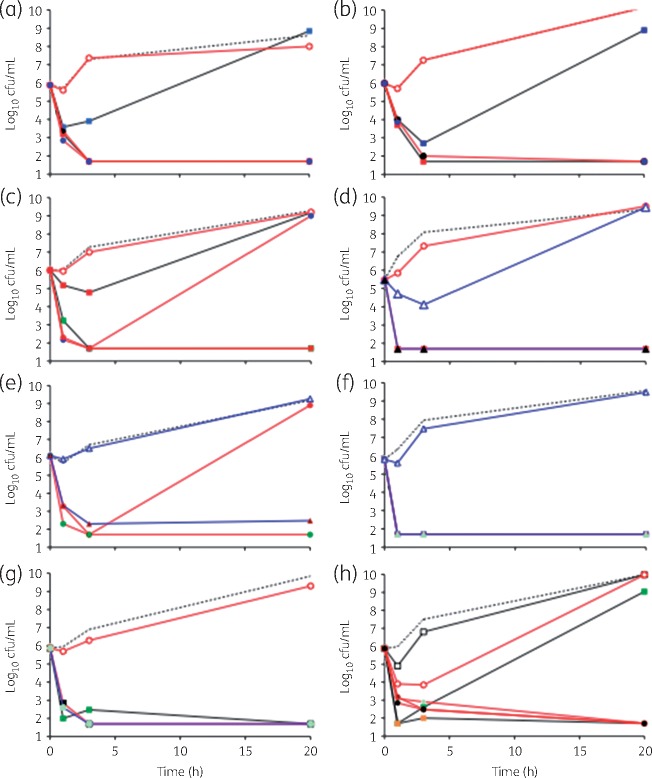
Polymyxin is known to rapidly kill susceptible Gram-negative bacteria,25 as does NAI-107 for Gram-positives.11 We thus tested different growth-inhibiting combinations of polymyxin and NAI-107 for their ability to reduce the viable bacterial counts in dynamic chequerboards. We did observe a decrease in viable counts in combinations containing NAI-107 and sub-inhibitory concentrations of polymyxin but not in cultures treated with polymyxin alone. At 0.031 mg/L polymyxin (i.e. 0.25 × MIC), the viable count for A. baumannii L3030 was undistinguishable from that of the untreated control (Figure 2a). However, addition of as little as ≤0.5 mg/L NAI-107 was sufficient to decrease viable counts by at least four orders of magnitude (i.e. below the detection limit of 50 cfu/mL) for up to 20 h (Figure 2a and Table S1). Similarly, in the presence of 0.016 mg/L polymyxin, NAI-107 caused a concentration-dependent decrease in viable counts, with 2 mg/L NAI-107 being sufficient to decrease the viable counts below the detection limit, whereas 1 mg/L NAI-107, despite an initial decrease in viable counts, was unable to prevent full growth at 20 h (Figure 2a). In bactericidal combinations viable counts decreased in a time-dependent manner, with 1 h of incubation sufficient to decrease cfu/mL by 2.5–3.5 log, whereas a 3 h incubation was required to reduce cfu/mL below the detection limit (Figure 2a).
Figure 2.
Killing of A. baumannii L3030 by NAI-107 (a), NAI-108 (b), NAI-857DA (c) or vancomycin (d), each in combination with polymyxin, and killing by NAI-107 in combination with polymyxin of A. baumannii L373 (e), E. coli ATCC 25922 (f), K. pneumoniae ND484 (g) or P. aeruginosa ATCC 10145 (h). Untreated controls are represented by broken lines. Polymyxin concentrations are represented as follows: 0.016 mg/L, squares and black lines; 0.031 mg/L, circles and red lines; and 0.062 mg/L, triangles and blue lines. Cultures containing polymyxin only are represented by open symbols, while filled symbols indicate the presence of a combination antibiotic (NAI-107, NAI-108, NAI-857DA or vancomycin) at the following concentrations: 0.125 mg/L, light green; 0.25 mg/L, purple; 0.5 mg/L, black; 1 mg/L, blue; 2 mg/L, red; 4 mg/L, green; and 8 mg/L, orange. The limit of detection was 50 cfu/mL.
Next, we used A. baumannii L3030 to test the effects of the lantibiotics NAI-108 and NAI-857DA, and of vancomycin. As seen for NAI-107, NAI-108 showed time- and concentration-dependent killing, with 2 and ≤0.5 mg/L NAI-108 being sufficient to decrease viable counts below the detection limit in the presence of 0.016 and 0.031 mg/L polymyxin, respectively (Figure 2b). In contrast, at least 2 mg/L NAI-857DA (Figure 2c) or 2 mg/L vancomycin (Figure 2d) was necessary for reducing viable counts of A. baumannii L3030 below the detection limit in the presence of 0.031 mg/L polymyxin. Lowering the concentration of polymyxin to 0.016 mg/L required 4 and 64 mg/L NAI-857DA (Figure 2c) and of vancomycin (Table 5), respectively, for observing a complete bactericidal effect for the entire duration of the experiment. Only when polymyxin concentrations were raised to 0.062 mg/L (i.e. 0.5 × MIC) did we observe a bactericidal effect at vancomycin concentrations of ≤0.5 mg/L (Figure 2d). It should be noted that 0.062 mg/L polymyxin alone did have a transient effect on cell viability, with a small decrease in viable cells up to 3 h, followed by full growth by 20 h (Figure 2d). Overall, these results are consistent with the general trend observed with the lantibiotics shown in Figure 1, with the hydrophobic lantibiotics NAI-107 and NAI-108 showing comparable activities, whereas the hydrophilic NAI-857DA was slightly less active than the other two compounds.8 Vancomycin was less potent than NAI-107 in synergistic combinations with polymyxin. Table 5 describes a summary of the results observed with growth and viable counts in chequerboard experiments. Usually, in the presence of a given polymyxin FIC, the lowest concentration of antibiotic able to reduce viable counts below the detection limit at 3 h coincided with the concentration that was able to prevent growth (Table 5).
Table 5.
Effect of polymyxin-containing combinations on growth and viability of A. baumannii L3030
| Polymyxin (mg/L) | NAI-107 (mg/L) |
NAI-108 (mg/L) |
NAI-857DA (mg/L) |
Vancomycin (mg/L) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| growth inhibition | killing ata |
growth inhibition | killing ata |
growth inhibition | killing ata |
growth inhibition | killing ata |
|||||||||
| 1 h | 3 h | 20 h | 1 h | 3 h | 20 h | 1 h | 3 h | 20 h | 1 h | 3 h | 20 h | |||||
| 0.062 | ≤0.5 | NT | NT | NT | ≤0.25 | NT | NT | NT | ≤0.13 | NT | NT | ≤0.13 | 0.5 | 0.5 | 0.5 | 0.5 |
| 0.031 | ≤0.5 | 4 | ≤0.5 | ≤0.5 | ≤0.25 | 8 | 2 | 0.5 | 0.5 | >2 | 1 | 2 | 2 | ≤4 | ≤4 | ≤4 |
| 0.016 | 2 | 8 | 2 | 2 | 0.5 | 8 | 2 | 2 | 4 | >8 | 4 | 4 | 64 | NT | NT | NT |
| 0.008 | 8 | 16 | 4 | 8 | 4 | 16 | 4 | 8 | 8 | >8 | 8 | >8 | 128 | NT | NT | NT |
| 0.004 | 4 | 16 | 4 | 4 | 16 | 32 | 16 | 32 | 32 | NT | NT | NT | 128 | NT | NT | NT |
| 0.002 | 8 | 16 | 16 | 16 | 32 | NT | NT | NT | 64 | NT | NT | NT | 128 | NT | NT | NT |
NT, not tested.
At the indicated polymyxin concentration, the table reports for each compound the lowest concentration able to inhibit growth and the lowest concentration necessary to reduce the number of viable cells below the detection limit (50 cfu/mL) in replicated microtitre plate experiments.
Lowest concentration required to reduce the number of viable counts below the detection limit after 1, 3 or 20 h. Note that the symbol ≤ is used only when cfu/mL was not determined from cultures that were fully inhibited at concentrations lower than indicated. cfu/mL at time zero ranged from 3 × 105 (vancomycin experiment) to 1 × 106 (NAI-857DA experiment).
We next expanded the evaluation of NAI-107 using selected strains from Table 3. As seen with A. baumannii L3030, within 1–3 h NAI-107 was able to significantly reduce viable counts (below or close to the detection limit) of A. baumannii L373 (Figure 2e), E. coli ATCC 25922 (Figure 2f), E. coli L47 (data not shown), K. pneumoniae ND484 (Figure 2g) and P. aeruginosa ATCC 10145 (Figure 2h), most of the time at the same concentrations as those sufficient to cause growth inhibition (Table S2). With P. aeruginosa ATCC 10145, 0.5 × MIC polymyxin caused a transient decrease in viable counts, followed by regrowth. The addition of ≥0.125 mg/L NAI-107 prevented regrowth (Figure 2h).
Activity against S. aureus biofilms
NAI-107 and related lantibiotics are active against most Gram-positive pathogens, including MRSA and GISA.7,9,10 In order to evaluate activity against biofilms, four S. aureus strains were selected for their ability to form biofilms (Table S1). Against these strains, NAI-107, NAI-108 and NAI-857DA were able to inhibit growth of planktonic cells detaching from the biofilms, with minimal biofilm-inhibiting concentrations (MBICs) comparable to MICs (Table 6). The minimal biofilm-eradicating concentration (MBEC) values were usually 16–32 mg/L, with the exception of NAI-857DA, which showed an MBEC of 2 mg/L against a single strain. Rifampicin and amoxicillin, used as positive and negative controls, respectively, behaved as expected (Table 6).
Table 6.
Activities against S. aureus biofilms (values in mg/L)
| Microorganism | Parameter | NAI-107 | NAI-108 | NAI-857DA | Rifampicin | Amoxicillin |
|---|---|---|---|---|---|---|
| S. aureus L3988 | MIC | 1 | NT | 1 | 0.002 | 16 |
| MBIC | 2 | NT | 2 | 0.007 | >128 | |
| MBEC | 16 | NT | 2 | 0.03 | >128 | |
| S. aureus L3797 | MIC | 4 | 8 | 8 | 0.002 | 128 |
| MBIC | 8 | 16 | 32 | 0.002 | >128 | |
| MBEC | 16 | 16 | 32 | 0.03 | >128 | |
| S. aureus USA300 | MIC | 0.5 | 1 | 1 | 0.002 | 64 |
| MBIC | 0.125 | 0.5 | 1 | 0.002 | >128 | |
| MBEC | 16 | 32 | 32 | 0.03 | >128 | |
| S. aureus L1400 | MIC | 1 | 2 | 2 | 0.002 | 128 |
| MBIC | 1 | 2 | 2 | 0.002 | 128 | |
| MBEC | 32 | 16 | 16 | 0.06 | >128 |
See the Materials and methods section for details.
NAI-107 effectively kills non-dividing S. aureus cells
Daptomycin and oritavancin were shown to kill S. aureus cells in stationary phase, whereas vancomycin was inactive.26,27 Therefore, we compared the effect of our lantibiotics on exponentially growing and non-dividing cells of S. aureus, using vancomycin as control. After determining the MICs of the compounds (Table 7), time–kill experiments were initially performed on the community-acquired MRSA strain USA300 and the glycopeptide-intermediate S. aureus (GISA) strain Mu50.28
Table 7.
MICs for the S. aureus strains used in time–kill experiments
| MIC (mg/L) |
||||
|---|---|---|---|---|
| vancomycin | NAI-107 | NAI-108 | NAI-857DA | |
| USA300 | 2 | 0.25 | 1–2* | 1 |
| Mu3 | 4*–8 | 4 | 8*–16 | 4*–8 |
| Mu50 | 1 | 2 | 8 | 4 |
MICs were determined for three individual clones. When more than one value was found, the MIC values used for further study are those found in two of three determinations and are designated by an asterisk.
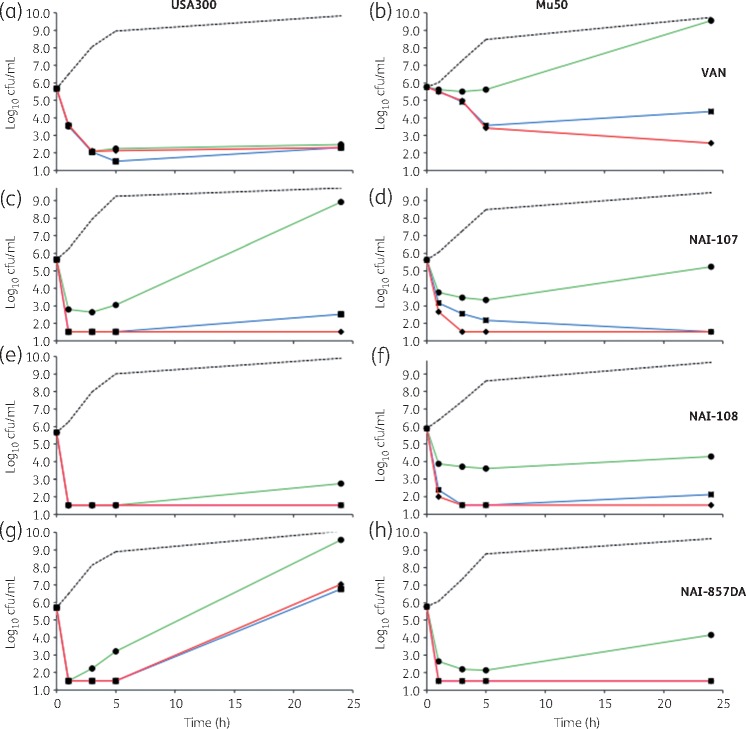
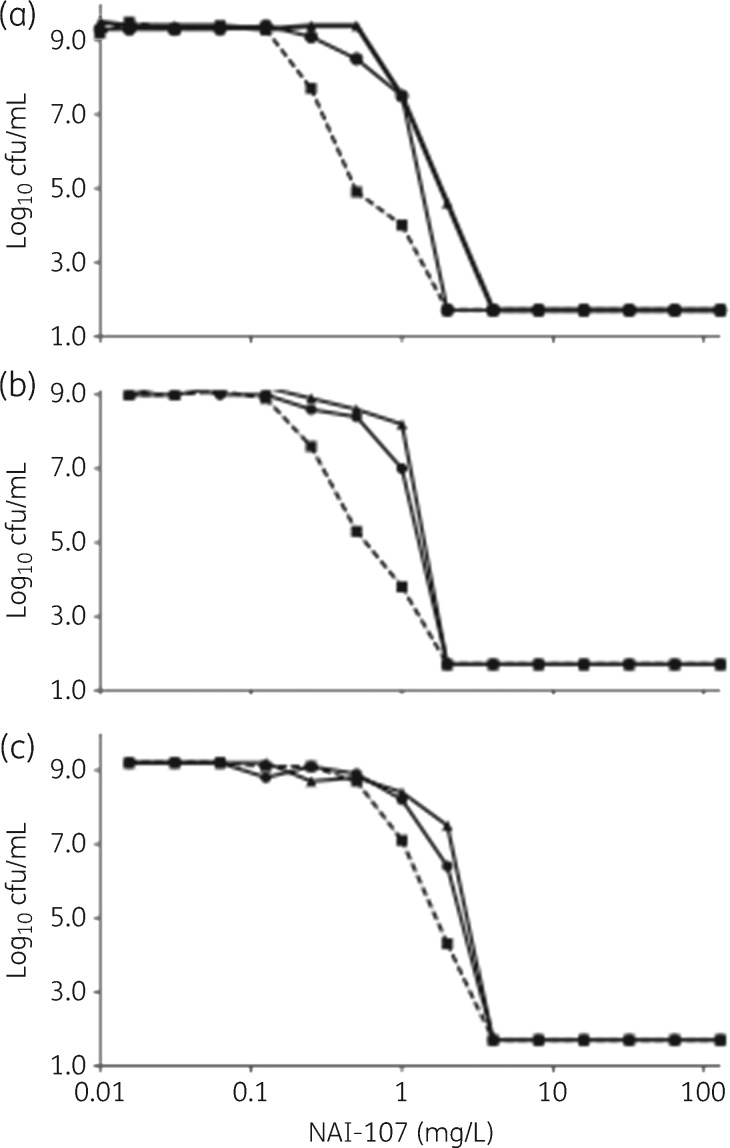
Vancomycin demonstrated killing of exponentially growing USA300 by >99.9% within 3 h at 1 × MIC (2 mg/L), with no increase in viable counts seen up to 24 h (Figure 3a). Treatment of USA300 with the three lantibiotics resulted in reduction in bacterial titres of at least 99% within 1 h at all concentrations tested. However, viable counts at 24 h were comparable to untreated controls at 1 × MIC NAI-107 (0.25 mg/L), but not at higher concentrations (Figure 3c), and in the presence of all concentrations of NAI-857DA (1–5 mg/L; Figure 3g). This phenomenon was not observed with NAI-108 at all concentrations tested (2–10 mg/L; Figure 3e).
Figure 3.
Killing of exponentially growing S. aureus USA300 (a, c, e and g) and Mu50 (b, d, f and h) by vancomycin (VAN; a and b), NAI-107 (c and d), NAI-108 (e and f) or NAI-857DA (g and h), as indicated. Compounds were added at 1 × MIC (green lines and circles), 3 × MIC (blue lines and squares) or 5 × MIC (red lines and diamonds). Broken lines denote untreated controls. Data represent the average of three independent treatments. Standard deviations are omitted for clarity. The limit of detection was 33 cfu/mL. Note that one of three USA300 cultures treated with 5 × MIC NAI-857DA showed no growth and these data were not considered in calculating the average cfu/mL in (h).
Against Mu50, vancomycin showed an effect pronouncedly different from that seen against USA300 (Figure 3b). At 1 × MIC (4 mg/L), there was no change in the number of viable cells for the first 5 h, and by 24 h the strain had reached the same density as the untreated control. Increasing vancomycin concentration to 3 × or 5 × MIC resulted in slow killing up to 5 h, but viable counts could still be observed at 24 h (Figure 3b). In contrast, the response of USA300 and Mu50 to the lantibiotics was similar: at 3 × or 5 × MIC, NAI-107, NAI-108 and NAI-857DA reduced viable counts of Mu50 (Figure 3d, f and h).
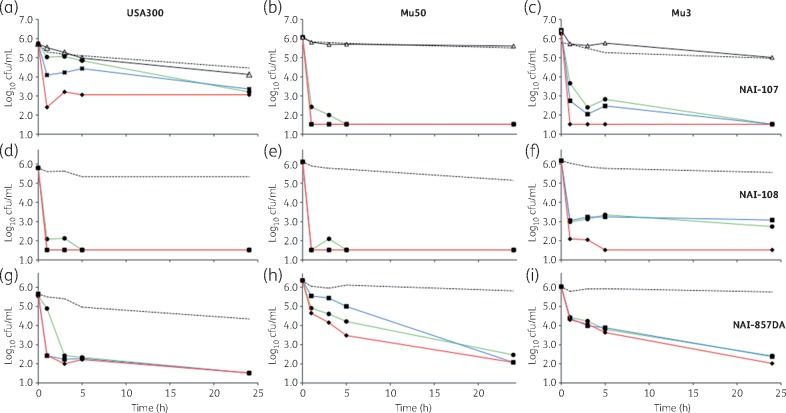
We next examined the activity of NAI-107, NAI-108 and NAI-857DA against non-dividing cells resuspended in PBS at ∼5 × 105 cfu/mL. In these experiments, we also included the GISA strain Mu3.28 NAI-107, NAI-108 and NAI-857DA were tested at 3 ×, 5 × and 10 × MIC, whereas vancomycin, which was expected to have little activity, was used as a control at 10 × MIC only. Against USA300, NAI-107 showed time- and concentration-dependent killing of non-dividing cells, with cfu/mL decreased by 99% within 5 h at 5 × MIC and within 1 h at 10 × MIC (Figure 4a). Against Mu50 (Figure 4b) and Mu3 (Figure 4c), NAI-107 treatment resulted in ≥99% reduction of viable cells within 1–5 h, with the effect lasting the remainder of the experiment. Similar results were observed when USA300 was resuspended in spent medium rather than in PBS (data not shown).
Figure 4.
Killing of non-dividing cells of S. aureus strain USA300 (a, d and g), Mu50 (b, e and h) and Mu5 (c, f and i) by NAI-107 (a–c), NAI-108 (d–f) or NAI-857DA (g–i), as indicated. Antibiotics were added at 3 × MIC (green lines and circles), 5 × MIC (blue lines and squares) or 10 × MIC (red lines and diamonds). Broken lines denote untreated controls. Vancomycin was added at 10 × MIC and is shown only for (a), (b) and (c) (black lines and open triangles). Overnight cultures were diluted to ∼5 × 105 in pre-heated PBS (37 °C). Experiments were performed in triplicate. Standard deviations are omitted for clarity. The limit of detection was 33 cfu/mL. Note that one of three Mu50 cultures treated with NAI-857DA showed growth equivalent to controls and these data were not considered in calculating the average cfu/mL in (h).
When exposed to NAI-108, viable cells of USA300 (Figure 4d) and Mu50 (Figure 4e) dropped below the detection limit within 1 h at all concentrations tested. Only with Mu3 did we observe concentration-dependent killing, with 10 × MIC NAI-108 required to reduce viable counts below 0.1% of the starting titre (Figure 4f). When exposed to NAI-857DA, USA300 cells were killed within 3 h at all tested concentrations (Figure 4g), Mu50 cells were resistant to killing (Figure 4h) and strain Mu3 appeared to have an intermediate behaviour, with time-dependent and concentration-independent killing (Figure 4i).
Altogether, the above results indicate that dividing and non-dividing cells of the MRSA strain USA300 are rapidly killed by NAI-107 and by NAI-108 at equivalent multiples of the MIC, whereas this strain is able to eventually escape killing by NAI-857DA in growth medium but not in PBS. Under non-dividing conditions, the GISA strains Mu3 and Mu50 are rapidly killed by NAI-107 and by NAI-108, but only partially and slowly by NAI-857DA.
NAI-107 effectively kills non-dividing A. baumannii cells
Inspired by the above results, we exposed non-dividing cells of A. baumannii L3030 in PBS to combinations of NAI-107 and polymyxin. Under these conditions, the untreated strain was stable (just a few duplications observed during the incubation period) and killed by ≥0.5 mg/L polymyxin (Figure 5). Notwithstanding an MIC of 128 mg/L, NAI-107 alone was able to kill the strain within 3 h at ≥16 mg/L, with no detectable cells observed after 20 h. A similar killing effect by 16 mg/L NAI-107 was observed against non-dividing cells of A. baumannii L2859 (data not shown). It should be noted that the activities against dividing and non-dividing cells were measured under similar conditions, with the main difference represented by the presence of growth medium versus PBS. Furthermore, NAI-107 in PBS led to a decrease in cfu/mL after 3 h (Figure 5), at the same time that an increase in viable counts was observed in growth medium (Figure 2a). None of the other Gram-negatives tested (E. coli ATCC 25922, K. pneumoniae L3392 and P. aeruginosa ATCC 27853) was killed by NAI-107 under non-dividing conditions (data not shown).
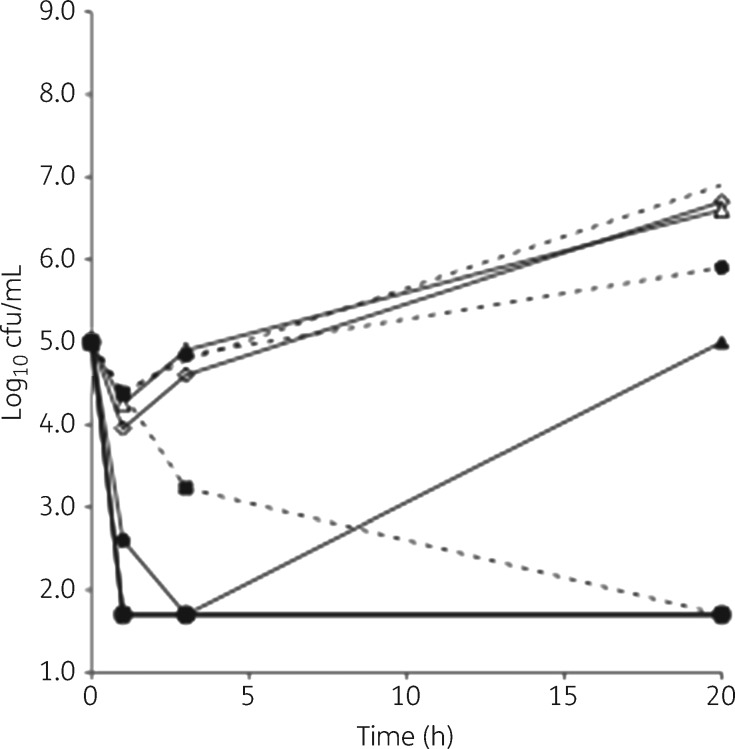
Figure 5.
Killing of non-dividing A. baumannii L3030 by NAI-107 alone or in combination with polymyxin. Untreated controls are represented by broken lines with no symbols. Continuous lines and open symbols denote cultures treated with polymyxin alone at 1.0 (circles), 0.125 (diamonds) or 0.031 (triangles) mg/L. Broken lines and filled symbols denote cultures treated with NAI-107 alone at 16 (squares) or 8 (diamonds) mg/L. Continuous lines and filled symbols denote cultures treated with the following combinations: circles, 0.125 mg/L polymyxin and 2 mg/L NAI-107; triangles, 0.031 mg/L polymyxin and 4 mg/L NAI-107; and squares, 0.016 mg/L polymyxin and 4 mg/L NAI-107. Note that cells were resistant to DMSO (the solvent used to dissolve NAI-107) up to 3%.
Consistent with the data observed with growing cells, combinations of NAI-107 and polymyxin were highly effective in killing non-dividing A. baumannii L3030: at polymyxin concentrations of 0.125 or 0.06 mg/L, ≤2 mg/L NAI-107 was sufficient to reduce the number of viable counts below the detection limit (Figure 5). When 4 mg/L NAI-107 was used, killing was observed at 0.016 mg/L polymyxin, the lowest concentration tested (Figure 5).
Lack of in vitro resistance in Gram-positives
In contrast to molecules targeting cellular proteins, antibiotics binding to lipid II do not select resistant mutants by direct plating.15,29 Consistently, no spontaneous resistance mutants of two GISA strains, two MRSA strains and one E. faecalis VanA strain were observed at 10 × MIC NAI-107, indicating a frequency lower than 10−10 (Table S3).
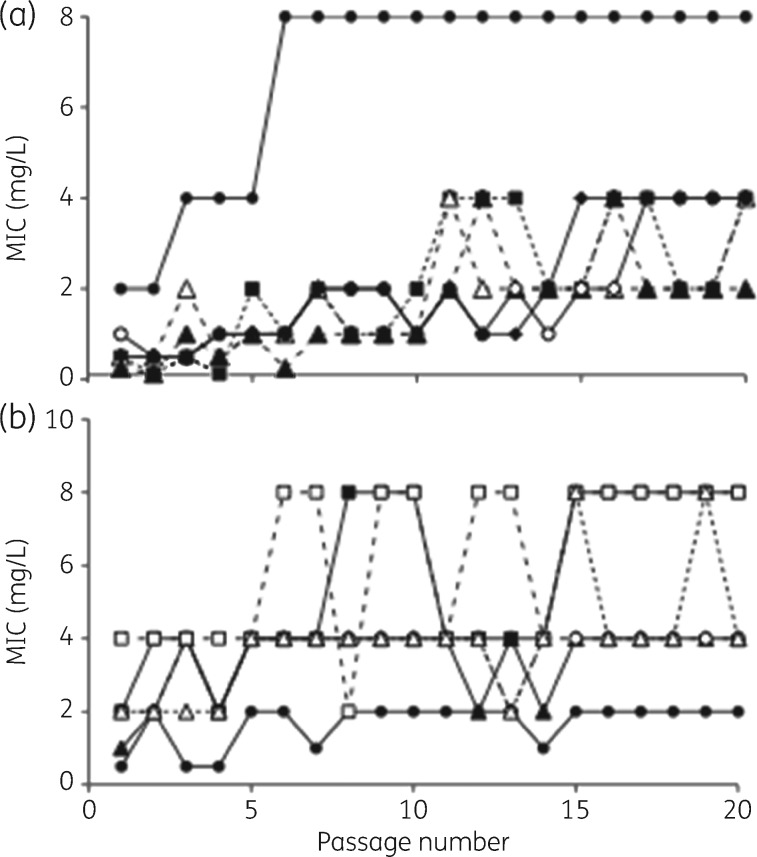
Using 12 strains (1 MSSA, 2 MRSA, 3 GISA, 3 E. faecalis VanA and 3 E. faecium VanA), 20 serial subcultures in the presence of subinhibitory concentrations yielded only modest increases in the NAI-107 MIC, which appeared to stabilize at 2- or 4-fold the initial value, with isolated spikes at 8-fold (Figure 6). Population profile analyses of cultures emerging after 1 passage, 10 passages or 20 passages indicated that the procedure had probably enriched for a subpopulation of cells already present in the initial culture, with no significant differences seen after 10 or 20 passages (Figure 7).
Figure 6.
Variation in NAI-107 MICs after serial passages of cultures growing at 0.5 × MIC. (a) S. aureus strains: squares and broken line, MSSA strain L819; triangles and broken line, MRSA strains L1400 (open symbols) and L4064 (filled symbols); and continuous line, GISA strains L3797 (filled circles), L3798 (open circles) and L4062 (filled diamonds). (b) Enterococcus spp. VanA: filled symbols and broken line, E. faecalis strains A533 (circles), J1 (squares) and A8 (triangles); and open symbols and continuous line, E. faecium strains A6349 (circles), B518 (squares) and D561 (triangles).
Figure 7.
Population analysis profiles of cultures of strains MSSA L819 (a), GISA L4064 (b) and MRSA L4061 (c) emerging after 1 passage (squares, broken line), 10 passages (circles, continuous line) or 20 passages (triangles, continuous line). The limit of detection in these experiments was 50 cfu/mL.
Discussion
Gram-negative bacteria can be sensitized to different antibiotics by sub-inhibitory concentrations of polymyxin or colistin.30 We could find only a few reports addressing the ability of polymyxin-based combinations to kill the target pathogens: these studies were performed at 1 mg/L polymyxin and at a single, high concentration (20 mg/L) of vancomycin,21 teicoplanin31 or telavancin.22 In this study, we show that, in addition its strong synergism, essentially the same combinations of NAI-107 and polymyxin that inhibit growth also reduce viable counts below the detection limit of the Gram-negative pathogens.
Few studies have investigated antibacterial activity against non-dividing S. aureus cells. Mascio et al.27 demonstrated that daptomycin kills MRSA in stationary phase, with a mechanism that does not require energy or protein synthesis. In their elegant studies, Müller et al.32 demonstrated that daptomycin binds to high-fluidity regions in the membrane, thus preventing the function of proteins that specifically localize to those regions. Combining the two observations, we surmise that high-fluidity regions are present in non-dividing cells. Unlike vancomycin, oritavancin kills non-dividing S. aureus cells, and septum staining of stationary phase cells is affected by oritavancin and not by vancomycin.26
NAI-107 forms 1:1 or 2:1 complexes with bactoprenol–pyrophosphate-coupled precursors of the bacterial cell wall, such as lipid II. In whole cells, NAI-107 does not form nisin-like pores in the bacterial membrane, but binding of NAI-107 to lipid II is followed by a slow membrane depolarization.11 Current evidence indicates that NAI-107, like nisin, binds to the pyrophosphate moiety of lipid II,11 a site distinct from the binding site of glycopeptides. Indeed, vancomycin does not kill non-dividing S. aureus,26 as confirmed here, consistent with the fact that the d-Ala-d-Ala moiety (i.e. vancomycin’s target) is not present in mature peptidoglycan.33
Altogether, the observed killing by NAI-107 of MRSA and GISA cells under non-dividing conditions is consistent with the hypothesis that killing requires interaction with the membrane. In the case of NAI-107, we believe this interaction occurs after an initial docking on lipid II or other bactoprenol–pyrophosphate-based intermediates. Indeed, it has been recently reported that peptidoglycan recycling is important for survival in the stationary phase of Gram-positive bacteria, including S. aureus.34 Although not demonstrated by these authors, such recycling might involve pyrophosphate-based carriers similar to lipid II, and would explain NAI-107's activity against non-dividing cells. A similar explanation might apply also to the reported killing of non-dividing cells by oritavancin.26
We were surprised to observe that NAI-107 can also kill non-dividing cells of two A. baumannii strains at sub-MIC values. This phenomenon might be a general feature of this species, as it was not observed with E. coli, K. pneumoniae and P. aeruginosa. Thus, it is tempting to speculate that the permeability of the A. baumannii outer membrane might change as the cells go from a non-dividing to a dividing state, allowing NAI-107 to reach its target under the former conditions. Among the four pathogenic species in Table 3, A. baumannii is the only species for which NAI-107 showed occasionally a measured MIC. In this respect, it has been reported that A. baumannii is generally more permeable to large antibiotics such as novobiocin and erythromycin, possibly connected with the ability of Acinetobacter spp. to use long-chain fatty acids as growth substrates.35
A growing body of evidence indicates the importance of killing persisters and non-dividing cells in an infection setting.36 Overall, the results of the current work, along with previously published results on the efficacy of NAI-107 in experimental models of infection,12,13 indicate that this lantibiotic represents a promising option in addressing the serious threat of antibiotic resistance in Gram-positive pathogens, with rapid killing of actively dividing and non-dividing MRSA and GISA cells. In addition, the activity of NAI-107 against N. gonorrhoeae, its ability to kill non-dividing A. baumannii and its strong synergism with polymyxin might provide additional therapeutic options for treating infections by A. baumannii, E. coli, K. pneumoniae and P. aeruginosa. From the limited comparisons reported here, it appears that NAI-108 shares the same properties as NAI-107.
Supplementary Material
Acknowledgements
We are grateful to Alessandra Polissi for valuable advice and to research technicians Michaela Anna Lederer and Michelle Halling Sørensen for media preparation and help with cfu determinations at the University of Copenhagen.
Funding
This work received funding from the European Union (contracts 634588 for H2020 NoMorFilm and 245066 for FP7-KBBE-2009–3 LAPTOP), by the Danish Council For Independent Research (DFF) grant # 11-106387, by the Danish National Research Foundation (DNFR 120) - Centre for Bacterial Stress Response and Persistence at the University of Copenhagen, and by the Juchum Foundation. T. T. T. was also supported by Læge Sofus Carl Emil Friis og hustru Olga Doris Friis' Legat and Kirsten og Freddy Johansens Fond.
Transparency declarations
E. G., S. M., M. S. and S. D. are employees of NAICONS Srl. C. B., M. S. and S. D. are employees of KtedoGen Srl. S. M., M. S., D. J. and S. D. own shares of NAICONS Srl, which may be financially affected by the conclusions of the present article. T. T. T. and A. L.-O.: none to declare.
Supplementary data
Materials and methods and Tables S1 to S3 are available as Supplementary data at JAC Online.
References
- 1. Brown E, Wright G.. Antibacterial drug discovery in the resistance era. Nature 2016; 529: 336–43. [DOI] [PubMed] [Google Scholar]
- 2. Marston H, Dixon D, Knisely J. et al. Antimicrobial resistance. JAMA 2016; 316: 1193–204. [DOI] [PubMed] [Google Scholar]
- 3. Butler MS, Blaskovich MA, Cooper MA.. Antibiotics in the clinical pipeline in 2013. J Antibiot 2013; 66: 571–91. [DOI] [PubMed] [Google Scholar]
- 4. Rice LB. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J Infect Dis 2009; 197: 1079–81. [DOI] [PubMed] [Google Scholar]
- 5. Liu C, Graber CJ, Karr M. et al. A population-based study of the incidence and molecular epidemiology of methicillin-resistant Staphylococcus aureus disease in San Francisco, 2004–2005. Clin Infect Dis 2008; 46: 1637–46. [DOI] [PubMed] [Google Scholar]
- 6. Arnison PG, Bibb MJ, Bierbaum G. et al. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep 2013; 30: 108–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jabes D, Brunati C, Guglierame P. et al. In vitro antibacterial profile of the new lantibiotic NAI-107. In: Abstracts of the 49th Interscience Conference of Antimicrobial Agents and Chemotherapy, San Francisco, USA, CA, 12–15 September 2009. Abstract F1-1502. American Society for Microbiology, Washington, DC, USA.
- 8. Maffioli SI, Cruz JC, Monciardini P. et al. Advancing cell wall inhibitors towards clinical applications. J Ind Microbiol Biotechnol 2016; 43: 177–84. [DOI] [PubMed] [Google Scholar]
- 9. Cruz JC, Iorio M, Monciardini P. et al. Brominated variant of the lantibiotic NAI-107 with enhanced antibacterial potency. J Nat Prod 2015; 78: 2642–7. [DOI] [PubMed] [Google Scholar]
- 10. Maffioli SI, Monciardini P, Catacchio B. et al. Family of class I lantibiotics from actinomycetes and improvement of their antibacterial activities. ACS Chem Biol 2015; 10: 1034–42. [DOI] [PubMed] [Google Scholar]
- 11. Münch D, Müller A, Schneider T. et al. The lantibiotic NAI-107 binds to bactoprenol-bound cell wall precursors and impairs membrane functions. J Biol Chem 2014; 289: 12063–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lepak AJ, Marchillo K, Craig WA. et al. In vivo pharmacokinetics and pharmacodynamics of the lantibiotic NAI-107 in a neutropenic murine thigh infection model. Antimicrob Agents Chemother 2015; 59: 1258–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jabes D, Brunati C, Candiani GP. et al. Efficacy of the new lantibiotic NAI-107 in experimental infections induced by multidrug-resistant Gram-positive pathogens. Antimicrob Agents Chemother 2011; 55: 1671–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thomsen TT, Mojsoska B, Cruz JC. et al. The lantibiotic NAI-107 efficiently rescues Drosophila melanogaster from infection with methicillin-resistant Staphylococcus aureus USA300. Antimicrob Agents Chemother 2016; 60: 5427–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schneider T, Sahl HG.. An oldie but a goodie—cell wall biosynthesis as antibiotic target pathway. Int J Med Microbiol 2010; 300: 161–9. [DOI] [PubMed] [Google Scholar]
- 16. Lee H, Hsu FF, Turk J. et al. The PmrA-regulated pmrC gene mediates phosphoethanolamine modification of lipid A and polymyxin resistance in Salmonella enterica. J Bacteriol 2004; 186: 4124–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Moffatt JH, Harper M, Harrison P. et al. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob Agents Chemother 2010; 54: 4971–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jochumsen N, Marvig RL, Damkiær S. et al. The evolution of antimicrobial peptide resistance in Pseudomonas aeruginosa is shaped by strong epistatic interactions. Nat Commun 2016; 7: 13002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. García-Quintanilla M, Carretero-Ledesma M, Moreno-Martínez P. et al. Lipopolysaccharide loss produces partial colistin dependence and collateral susceptibility to azithromycin, rifampicin and vancomycin in Acinetobacter baumannii. Int J Antimicrob Agents 2015; 46: 696–702. [DOI] [PubMed] [Google Scholar]
- 20. García-Quintanilla M, Pulido MR, Moreno-Martínez P. et al. Activity of host antimicrobials against multidrug-resistant Acinetobacter baumannii acquiring colistin resistance through loss of lipopolysaccharide. Antimicrob Agents Chemother 2014; 58: 2972–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gordon NC, Png K, Wareham DW.. Potent synergy and sustained activity of a vancomycin-colistin combination versus multidrug-resistant strains of Acinetobacter baumannii. Antimicrob Agents Chemother 2010; 54: 5316–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hornsey M, Longshaw M, Phee L. et al. In vitro activity of telavancin in combination with colistin versus Gram negative bacterial pathogens. Antimicrob Agents Chemother 2012; 56: 3080–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Galani I, Orlandou K, Moraitou H. et al. Colistin/daptomycin: an unconventional antimicrobial combination synergistic in vitro against multidrug-resistant Acinetobacter baumannii. Int J Antimicrob Agents 2014; 43: 370–4. [DOI] [PubMed] [Google Scholar]
- 24. Bergen PJ, Bulman ZP, Landersdorfer CB. et al. Optimizing polymyxin combinations against resistant Gram-negative bacteria. Infect Dis Ther 2015; 4: 391–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vaara M. Novel derivatives of polymyxins. J Antimicrob Chemother 2013; 68: 1213–9. [DOI] [PubMed] [Google Scholar]
- 26. Belley A, Neesham-Grenon E, McKay G. et al. Oritavancin kills stationary-phase and biofilm Staphylococcus aureus cells in vitro. Antimicrob Agents Chemother 2009; 53: 918–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mascio CT, Alder JD, Silverman JA.. Bactericidal action of daptomycin against stationary-phase and nondividing Staphylococcus aureus cells. Antimicrob Agents Chemother 2007; 51: 4255–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hiramatsu K, Aritaka N, Hanaki H. et al. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet 1997; 350: 1670–3. [DOI] [PubMed] [Google Scholar]
- 29. Ling LL, Schneider T, Peoples AJ. et al. A new antibiotic kills pathogens without detectable resistance. Nature 2015; 517: 455–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lenhard JR, Nation RL, Tsuji BT.. Synergistic combinations of polymyxins. Int J Antimicrob Agents 2016; 48: 607–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wareham DW, Gordon NC, Hornsey M.. In vitro activity of teicoplanin combined with colistin versus multidrug-resistant strains of Acinetobacter baumannii. J Antimicrob Chemother 2011; 66: 1047–51. [DOI] [PubMed] [Google Scholar]
- 32. Müller A, Wenzel M, Strahl H. et al. Daptomycin inhibits cell envelope synthesis by interfering with fluid membrane microdomains. Proc Natl Acad Sci USA 2016; 113: E7077–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vollmer W, Blanot D, de Pedro MA.. Peptidoglycan structure and architecture. FEMS Microbiol Rev 2008; 32: 149–67. [DOI] [PubMed] [Google Scholar]
- 34. Borisova M, Gaupp R, Duckworth A. et al. Peptidoglycan recycling in Gram-positive bacteria is crucial for survival in stationary phase. mBio 2016; 7: e00923-16.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zgurskaya HI, Löpez CA, Gnanakaran S.. Permeability barrier of Gram-negative cell envelopes and approaches to bypass it. ACS Infect Dis 2015; 1: 512–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Harms A, Maisonneuve E, Gerdes K.. Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 2016; 354: aaf4268.. [DOI] [PubMed] [Google Scholar]
- 37. Maffioli SI, Iorio M, Sosio M. et al. Characterization of the congeners in the lantibiotic NAI-107 complex. J Nat Prod 2014; 77: 79–84. [DOI] [PubMed] [Google Scholar]
- 38. Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically—Seventh Edition: Approved Standard M7-A7 CLSI, Wayne, PA, USA, 2006.
- 39. Clinical and Laboratory Standards Institute. Methods for Antimicrobial Susceptibility Testing of Anaerobic Bacteria—Sixth Edition: Approved Standard M11-A6 CLSI, Wayne, PA, USA, 2006.
- 40. Christensen GD, Simpson WA, Younger JJ. et al. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J Clin Microbiol 1985; 22: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stefanovic S, Vukovic D, Hola V. et al. Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS 2007; 115: 891–9. [DOI] [PubMed] [Google Scholar]
- 42. Pillai SK, Moellering RC, Eliopolus GM.. Antimicrobial combinations In: Lorian V, ed. Antibiotics in Laboratory Medicine, 5th edn.Philadelphia, PA: Lippincott Williams and Wilkins, 2005; 365–440. [Google Scholar]
- 43. Wiegand I, Hilpert K, Hancock RE.. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 2008; 3: 163–75. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.