Abstract
Background
Carbapenemase-producing Enterobacteriaceae (CPE), including KPC-producing Klebsiella pneumoniae (KPC-Kpn), are an increasing threat to patient safety.
Objectives
To use WGS to investigate the extent and complexity of carbapenemase gene dissemination in a controlled KPC outbreak.
Materials and methods
Enterobacteriaceae with reduced ertapenem susceptibility recovered from rectal screening swabs/clinical samples, during a 3 month KPC outbreak (2013–14), were investigated for carbapenemase production, antimicrobial susceptibility, variable-number-tandem-repeat profile and WGS [short-read (Illumina), long-read (MinION)]. Short-read sequences were used for MLST and plasmid/Tn4401 fingerprinting, and long-read sequence assemblies for plasmid identification. Phylogenetic analysis used IQTree followed by ClonalFrameML, and outbreak transmission dynamics were inferred using SCOTTI.
Results
Twenty patients harboured KPC-positive isolates (6 infected, 14 colonized), and 23 distinct KPC-producing Enterobacteriaceae were identified. Four distinct KPC plasmids were characterized but of 20 KPC-Kpn (from six STs), 17 isolates shared a single pKpQIL-D2 KPC plasmid. All isolates had an identical transposon (Tn4401a), except one KPC-Kpn (ST661) with a single nucleotide variant. A sporadic case of KPC-Kpn (ST491) with Tn4401a-carrying pKpQIL-D2 plasmid was identified 10 months before the outbreak. This plasmid was later seen in two other species and other KPC-Kpn (ST14,ST661) including clonal spread of KPC-Kpn (ST661) from a symptomatic case to nine ward contacts.
Conclusions
WGS of outbreak KPC isolates demonstrated blaKPC dissemination via horizontal transposition (Tn4401a), plasmid spread (pKpQIL-D2) and clonal spread (K. pneumoniae ST661). Despite rapid outbreak control, considerable dissemination of blaKPC still occurred among K. pneumoniae and other Enterobacteriaceae, emphasizing its high transmission potential and the need for enhanced control efforts.
Introduction
Carbapenemase-producing Enterobacteriaceae (CPE) are an increasing threat to patient safety and the function of healthcare institutions in Europe and worldwide.1 Genes encoding carbapenemases may be integrated in the bacterial chromosome but are most often located on mobile elements, such as plasmids or transposons, which are transferable between bacterial strains, species and genera.1,2 Thus, clinical outbreaks are often complex, comprising varying degrees of clonal, plasmid or transposon-mediated gene spread. Klebsiella pneumoniae carbapenemases (KPC), encoded by blaKPC alleles, have disseminated globally, primarily due to the clonal spread of KPC-producing K. pneumoniae (KPC-Kpn) strains, such as sequence type (ST) 258.1,3,4 However, in the last decade, dispersal of the transposable Tn4401 element harbouring blaKPC5–7 amongst diverse plasmid structures, bacterial clones and species has increasingly been observed.2,8
KPC-Kpn have spread rapidly and extensively in some countries. In Italy, widespread nosocomial dissemination followed the introduction of KPC in 2008, leading to one-third of invasive K. pneumoniae isolates being carbapenem resistant in 2014.9 In the USA, KPC has spread to most states. In both countries, the outbreaks were predominantly due to transmission of KPC-Kpn ST258, and other clonal group 258 STs, such as ST512.1,10
KPC was first recognized in the UK in 2003.11 Since then, the incidence of KPC-producing Enterobacteriaceae (KPC-E) has risen,12 with 664 isolates referred to PHE’s Antimicrobial Resistance and Healthcare Associated Infections (AMRHAI) Reference Unit in 2015.13 Twenty-three blaKPC variants have been described, but most UK isolates carry either blaKPC-2, blaKPC-3 or occasionally blaKPC-4.14 More than 95% of UK KPC-E originate from northwest England where transmission has principally occurred horizontally via variants of an epidemic plasmid (pKpQIL).12,14 When seen outside northwest England, previous UK blaKPC isolates have been typically associated with K. pneumoniae clonal group 258.12,14,15
Whole-genome sequencing (WGS) is a highly discriminatory typing technique used to investigate pathogen transmission in nosocomial outbreaks. Genetic relatedness amongst strains is determined by nucleotide-level variation.16–18 However, analysing plasmid structures using short-read sequencing remains challenging,19 limiting the complete elucidation of blaKPC transmission networks. Recently, data from long-read platforms, particularly PacBio’s single molecule real-time (SMRT) sequencers, have enabled plasmid-level analyses, in some instances leading to an improved understanding of horizontal blaKPC spread.8,20 Oxford Nanopore Technology’s (ONT) long-read MinION sequencers represent a potentially quicker and more cost-effective alternative.
In 2013, Leeds Teaching Hospitals NHS Trust (LTH), UK, experienced a short-lived poly-species KPC-E outbreak on a single hospital ward. We investigated the molecular epidemiology of this temporo-spatially limited outbreak using WGS and detailed hospital admissions data to investigate the likely transmission mechanisms. The characterization of blaKPC plasmid structures was facilitated by MinION sequencing data, marking the first application of MinION long-read sequencing technology to tracking blaKPC plasmid diversity in a clinical outbreak. We also attempted to elucidate the infection prevention and control interventions likely to have brought the outbreak under control.
Materials and methods
Setting
LTH is a large tertiary referral centre (>2000 beds), including a 32-bed supra-regional liver unit with eight single rooms, and six 4-bed bays. Patients are regularly referred from outside the region (including northwest England) for liver transplantation. The LTH liver unit experienced only rare, sporadic cases of infection/colonization with KPC-E before 2013 (one case >18 months prior to outbreak), with no previous evidence of patient-to-patient CPE transmission. Admission CPE screening using rectal swabs was commenced for all LTH liver unit admissions in 2011 following the introduction of UK CPE guidance.21
KPC-E outbreak and case definitions
An outbreak was declared in October 2013 following reference laboratory confirmation of three LTH liver unit patients carrying KPC-Kpn with the same variable-number-tandem-repeat (VNTR)22,23 profile (6,4,2,0,2,2,2,3,1). The initial outbreak definition included patients with KPC-Kpn with this VNTR type, as it was rare in the UK (19/922 national isolates; J. F. Turton AMRHAI unpublished data, 2012–13). However, this definition was almost immediately broadened to include all KPC-E once plasmid dispersal was suspected.
Clinical cases were defined as patients with KPC-E isolated from a clinical sample who had signs and symptoms consistent with infection. Colonized cases were defined as patients who had KPC-E identified on rectal screening without clinical evidence of infection. Details on ward movements, sampling and clinical outcomes were collected retrospectively from electronic healthcare records and clinical notes.
Infection prevention and control interventions
A multifaceted infection prevention and control strategy was immediately implemented when the outbreak was detected and continued until the outbreak was declared over (January 2014). The strategy targeted five main areas: hand hygiene, cohorting and isolation, screening, environmental cleaning, and education (Table S1, available as Supplementary data at JAC Online). Interventions were regularly reviewed in multidisciplinary outbreak team meetings. The outbreak was limited to patients on a single ward and interventions were targeted to this location.
CPE screening including microbiology methods
At the outbreak onset, enhanced rectal screening was introduced, i.e. for all patients on admission and discharge from the liver unit (including to/from other wards), and weekly following admission. Rectal CPE screening swabs were cultured using ESBL chromogenic agar with a 10 μg ertapenem disc (Oxoid, Basingstoke, UK) since KPC producers are typically resistant to cephalosporins. Species identification (using MALDI-TOF) was performed for all Enterobacteriaceae from clinical samples and screening swabs with reduced ertapenem susceptibility (zone diameter <26 mm, by EUCAST disc testing24); isolates were phenotypically investigated for carbapenem resistance using the Modified Hodge test, Rosco discs (KPC/Metallo-B-Lactamase Confirm kit, Rosco, Denmark) and by determining ertapenem MICs (Etest, bioMérieux, UK). All potential CPE-producing isolates were sent to the AMRHAI Reference Unit for PCR investigation for blaOXA-48, blaIMP, blaNDM, blaVIM and blaKPC.25–28 Plasmids were typed using a specifically designed pKpQIL-like plasmid PCR29 targeting six signature sites.
DNA extraction and sequencing
DNA was extracted from a single colony after subculture of frozen KPC-E isolates using a commercial kit (Quickgene, Fujifilm) as per the manufacturer’s instructions, with an additional mechanical lysis step following chemical lysis (FastPrep, MP Biomedicals). WGS used Illumina Hiseq 2500 sequencing technology, with a sequencing depth of ∼100 × per sample. We selected five isolates for MinION sequencing: one random dominant strain KPC-Kpn-ST661 isolate and four with blaKPC plasmid structures that were thought to be distinct (short-read WGS-based analysis, three patients). Genomic DNA was isolated using the Qiagen Genomic-tip 100/G kit (Qiagen, Germany) following the manufacturer’s recommendations. Isolates were prepared for 2D sequencing using the SQK-NSK007 sequencing kit with no DNA shearing and modifications to minimize DNA fragmentation and preserve plasmid structures (see Supplementary data: Methods).
Sequence processing and analysis
Species identification was performed on short-read Illumina data using Kraken30 with a reduced database comprising human, bacterial and viral genomes. Reads were then mapped to species-specific reference sequences and base calling was performed as previously described.31 Resistance mechanisms were identified using resistType (https://github.com/hangphan/resistType), a method combining both assembly/BLASTn and mapping of sequencing reads to a panel of known reference mechanisms, including both chromosomal and acquired mechanisms.
Consensus FASTA sequences from the pipeline were used to reconstruct phylogenetic trees for each species with IQTree,32 using a GTR + G model and a maximum parsimony starting tree. The phylogeny was corrected for recombination using ClonalFrameML with default parameters.33 Short-read sequences were assembled using SPAdes34 (version 3.6) and the assemblies used for in silico multilocus sequence-typing (MLST), plasmid Inc typing and Tn4401 typing by BLASTn. For plasmid typing, all publicly available complete plasmids were downloaded from NCBI (query term: plasmids AND Enterobacteriaceae AND complete sequence), deduplicated, and plasmids carrying blaKPC alleles were extracted. Additional fully closed plasmid sequences were obtained from a global KPC study.35 These plasmid sequences were then used as references to identify similar blaKPC-carrying plasmid structures in the outbreak using BLASTn.
MinION sequencing data were processed by poretools36 to extract 2D reads. Multiple approaches to generate assemblies were applied using both MinION long-read and Illumina short-read data, namely hybridSPAdes,34 npScarf37 and Canu38 (Supplementary data: Methods). A plasmid was defined as circularized if it had >100 bp overlapping ends with 100% sequence identity for hybridSPAdes/npScarf assemblies, and >1 kbp overlapping ends at >99% sequence identity for Canu assemblies.
We applied SCOTTI,39 a structured coalescent-based tool for reconstructing transmission within outbreaks, to the dominant outbreak KPC-Kpn ST661 isolates, combining epidemiological and chromosomal genomic data. We masked the recombinant regions detected by ClonalFrameML, and used the resulting genome alignment as input to SCOTTI, together with the first date where a KPC-E isolate was detected in a patient, and the start and end date of each patient’s infection risk period (Supplementary data: Methods). We used a π prior distribution for the mutation rate (mean 2 × 10–6)31 and a uniform prior distribution between 14 and 16 for the number of hosts (also allowing possible non-sampled hosts).
All short-read and long-read WGS data are available in the NCBI repository under project number PRJNA353334.
Results
Twenty identified patients (6 cases, 14 colonized; Table 1) were included in the outbreak analysis; 17 were detected in LTH during the outbreak period, 2 cases were identified after transfer from LTH to other UK hospitals and 1 patient was identified (by the AMRHAI Reference Unit) as a potential index case due to it sharing a KPC-Kpn-associated pKpQIL-D2 variant plasmid29 (December 2012; case 1). The first KPC-E of each species for each patient was analysed, giving rise to 23 distinct KPC-E isolates (all blaKPC-positive) including: 20 K. pneumoniae (6 distinct STs; 15 ST661 and 5 STs with a single isolate), 1 Citrobacter freundii, 1 Klebsiella oxytoca and 1 Enterobacter cloacae. Two patients (cases 6 and 12) had two distinct KPC-E and one patient (case 2) had two K. pneumoniae isolates (with differing antimicrobial susceptibility profiles) analysed.
Table 1.
Summary of cases and isolates involved in the LTH KPC outbreak
| Case | First KPC positive sample | Sample type | Clinical status | Isolate species | VNTR type | Sequence type | Plasmid type | Replicon type | Tn4401 type | Clinical outcome |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 13/12/12 | rectal screen | colonized | K. pneumoniae | 8,5,4,2,1,1,3,2,1 | Kpne-ST-491 | pKpQIL-D2 | IncFIB(K),IncFIB(pKPHS1), IncHI2,IncHI2A,IncR | Tn4401a | recovered |
| 2 | 10/07/13 | rectal screen | colonized | K. pneumoniae | 6,4,-,0,-,2,2,3,1 | Kpne-ST-661 | pKpQIL-D2 | IncFIB(K),IncFII,IncFII,IncR | Tn4401a | recovered |
| 2 | 10/07/13 | rectal screen | colonized | K. pneumoniae | 6,4,-,0,-,2,2,3,1 | Kpne-ST-661 | pKpQIL-D2 | Col(MG828),Col156,IncFIB(K), IncFII,IncFII,IncR | Tn4401a | recovered |
| 3 | 13/07/13 | rectal screen | colonized | K. pneumoniae | 6,4,-,0,-,2,2,3,1 | Kpne-ST-661 | pKpQIL-D2 | IncFIB(K),IncR | Tn4401a | recovered |
| 4 | 03/10/13 | intra-operative swab | case | K. pneumoniae | 6,4,2,0,-,2,2,3,1 | Kpne-ST-661 | pKpQIL-D2 | IncFIB(K),IncR | Tn4401a | died <30 days |
| 5 | 09/10/13 | rectal screen | colonized | K. pneumoniae | 6,4,-,0,-,2,2,3,1 | Kpne-ST-661 | pKpQIL-D2 | IncFIB(K),IncR | Tn4401a | recovered |
| 6 | 13/10/13 | sputum | colonized | E. cloacae | NA | Eclo-ST-144 | pKpQIL-D2 | IncFIB(K),IncR | Tn4401a | recovered |
| 6 | 13/10/13 | sputum | colonized | K. pneumoniae | NA | Kpne-ST-661 | pKpQIL-D2 | IncFIB(pECLA),IncR | Tn4401a | – |
| 7 | 17/10/13 | sputum | colonized | K. pneumoniae | 6,4,2,0,2,2,2,3,1 | Kpne-ST-661 | pKpQIL-D2 | IncFIB(K),IncR | Tn4401a | recovered |
| 8 | 20/10/13 | mid-stream urine | case | K. pneumoniae | 6,4,2,0,2,2,2,3,1 | Kpne-ST-661 | pKpQIL-D2 | IncFIB(K),IncR | Tn4401a | died <30 days |
| 9 | 25/10/13 | rectal screen | colonized | K. pneumoniae | 6,4,2,0,2,2,2,3,1 | Kpne-ST-661 | pKpQIL-D2 | IncFIB(K),IncR | Tn4401a [986G>T] | recovered |
| 10 | 25/10/13 | rectal screen | colonized | K. oxytoca | NA | Koxy-NF | pKpQIL-D2 | IncFIB(K),IncR | Tn4401a | recovered |
| 11 | 01/11/13 | rectal screen | colonized | K. pneumoniae | 6,4,2,0,2,2,2,3,1 | Kpne-ST-661 | pKpQIL-D2 | Col(IMGS31),IncN,IncR | Tn4401a | recovered |
| 12 | 04/11/13 | rectal screen | colonized | C. freundii | NA | Cfre-NF | distinct | IncFIA(HI1),IncFIB(K), IncFII(Yp),IncN,IncR | Tn4401a | recovered |
| 12 | 04/11/13 | rectal screen | colonized | K. pneumoniae | NA | Kpne-ST-250 | distinct | IncFIB(K),IncR | Tn4401a | – |
| 13 | 08/11/13 | rectal screen | colonized | K. pneumoniae | distinct | Kpne-ST-14 | pKpQIL-D2 | IncFIB(K),IncR | Tn4401a | recovered |
| 14 | 19/11/13 | rectal screen | colonized | K. pneumoniae | 6,4,2,0,2,2,2,3,1 | Kpne-ST-661 | pKpQIL-D2 | IncFIB(K),IncR | Tn4401a | recovered |
| 15 | 25/11/13 | blood culture | case | K. pneumoniae | 6,4,2,0,2,2,2,3,1 | Kpne-ST-661 | pKpQIL-D2 | IncFIB(K),IncR | Tn4401a | died <30 days |
| 16 | 19/12/13 | rectal screen | colonized | K. pneumoniae | distinct | Kpne-ST-1820 | distinct | IncFIB(K) | Tn4401a | recovered |
| 17 | 07/01/14 | rectal screen | colonized | K. pneumoniae | 6,4,2,0,2,2,2,3,1 | Kpne-ST-661 | pKpQIL-D2 | IncFIB(K),IncFII(K),IncR | Tn4401a | recovered |
| 18 | 08/01/14 | rectal screen | colonized | K. pneumoniae | distinct | Kpne-ST-321 | pKpQIL-D2 like | IncFIB(K),IncFIB(pKPHS1),IncR | Tn4401a | recovered |
| 19 | 16/01/14 | rectal screen | colonized | K. pneumoniae | 6,4,2,0,2,2,2,3,1 | Kpne-ST-661 | pKpQIL-D2 | IncFIB(K),IncR | Tn4401a | recovered |
| 20 | 20/01/14 | mid-stream urine | case | K. pneumoniae | 6,4,2,0,2,2,2,3,1 | Kpne-ST-661 | pKpQIL-D2 | IncFIB(K),IncR | Tn4401a | recovered |
VNTR, variable-number-tandem repeat; NA, not available.
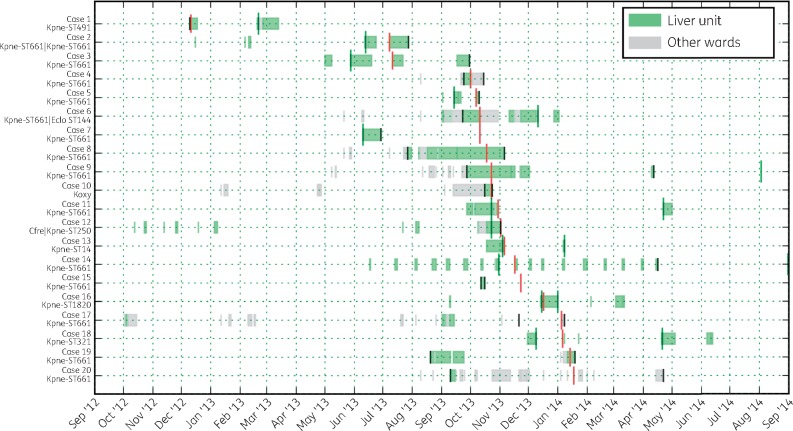
The median patient age was 56 years (range 32–76), and 12/20 (60%) patients were male. 8/20 (40%) patients were liver transplant recipients. Most (19/20) cases (cases 2–20) spent time on the LTH liver unit between July 2013 and January 2014 and were presumed nosocomial acquisitions (Figure 1). blaKPC was detected >48 h post-admission for 9 cases. For the remaining 10 patients (excluding the index case), typically with short-lived but regular admissions to the unit, there was a median 25 days (range 10–125, IQR 16–73) between the most recent admission to the unit and first blaKPC detection. Six patients experienced clinical infection associated with a KPC-producing isolate July 2013–January 2014; three had KPC-E bacteraemia, and one each had urinary tract, lower respiratory tract or surgical wound infections. The 30 day all-cause mortality among KPC-affected patients was 3/20 (15%).
Figure 1.
Patient admissions to LTH liver unit and other LTH wards during the outbreak period. Red vertical marks are the time of first KPC detection in the patients, black vertical marks denote the starts and ends of the infection risk periods defined for SCOTTI, green vertical marks denote the closest KPC-negative screening time points (where applicable) before or after the first KPC-positive screening result. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
The outbreak was deemed successfully controlled due to the lack of any new KPC-E colonization episodes/cases in LTH over the subsequent 21 months. The KPC-positive patients were not formally followed up after discharge as many were resident outside of Leeds. Of the 17 surviving discharged KPC-positive patients, 3/6 opportunistically screened ≥3 months post-diagnosis remained KPC-positive and 2/3 screened ≥6 months post-diagnosis remained positive. One patient remained KPC-positive 511 days post-diagnosis. One patient had three negative rectal swabs over a 6 month period, suggestive of blaKPC clearance;21 the remainder had insufficient local screens to confirm/refute loss of carriage.
WGS demonstrates multiple modes of transmission
Sequence data were available for all (n = 23) isolates (Figure 2). Twenty-two of 23 isolates had an identical transposon (Tn4401a,5 as in pIT-01C03, GenBank accession: HG969995.1), and one had just one single nucleotide variant (SNV) resulting in transposon type Tn4401a[986G > T]. This mutation results in a stop codon at position 261 in the resolvase/phage integrase protein facilitating site-specific recombination which could affect Tn4401 mobility.
Figure 2.
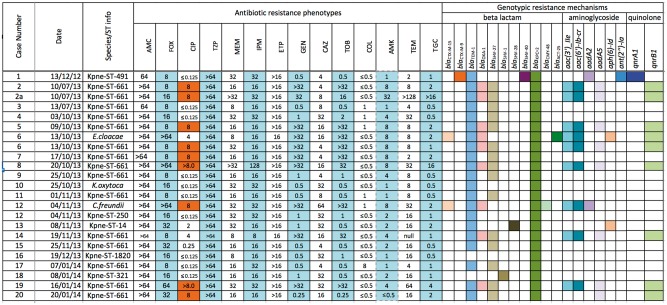
Phenotypic sensitivity and antimicrobial resistance gene prediction results of 23 strains from LTH KPC outbreak (MICs: mg/L measured by agar dilution at AMRHAI). AMC, co-amoxiclav; FOX, cefoxitin; CIP, ciprofloxacin; TZP, piperacillin/tazobactam; MEM, meropenem; IPM, imipenem; ETP, ertapenem; GEN, gentamicin; CAZ, ceftazidime; TOB, tobramycin; COL, colistin; AMK, amikacin; TEM, temocillin; TGC, tigecycline. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Variable phenotypic susceptibilities and antimicrobial resistance genotypes were noted amongst study isolates (Figure 2). All isolates carried blaKPC-2; no other carbapenem resistance mechanisms were demonstrated. Many also carried the ESBL gene blaSHV-27 (n = 15) and the β-lactamase genes blaTEM-1 (n = 21) and ΔblaOXA-9 (n = 21). Other broad or extended-spectrum β-lactamase genes observed included blaOXA-1 (n = 8), blaCTX-M-1 (n = 2) blaCTX-M-9 (n = 1) and blaCMY-48 (n = 1). Some isolates also carried aminoglycoside resistance genes. The ciprofloxacin-resistant phenotype in 12 isolates (>0.5 mg/L) was inconsistently explained by relevant genotypes. High-level ciprofloxacin resistance (≥8.0 mg/L) was seen in nine isolates of which seven carried qnrB1. For the remainder, chromosomal gyrA mutations (S83Y/F) and uncharacterized changes in efflux pump activity might have contributed to resistance. Colistin resistance (8 mg/L) was observed in one isolate (case 17). WGS data of this isolate revealed the presence of neither mcr-1 nor mcr-2 but N42D amino acid change was observed in the chromosomal gene mgrB whose alterations (e.g. gene disruption by insertion sequences, missense or nonsense point mutations, and small deletions) can confer colistin resistance.40 The N42D mutation has not, however, to our knowledge been previously described as causative.
The WGS-based plasmid typing results mostly agreed with PCR-based typing (19/20 PCR-positive cases). Nineteen isolates yielded assemblies with contigs highly similar to plasmid pKpQIL-D229 (>95% sequence length match/>99% sequence identity). pKpQIL-D2 is an IncR/IncFII blaKPC-2 plasmid, differing from pKpQIL in the partitioning and replication regions, without the pKpQIL signature replicon IncFIB(pQIL), and also harbouring blaTEM-1 and ΔblaOXA-9. pKPC18_LTH from case 18 (Kpne-ST321) was identified as pKpQIL-D2 by PCR but did not meet our predefined WGS-based plasmid typing thresholds for a match. However, on more detailed review, this isolate’s assembly included contigs matching 91% of pKpQIL-D2 at 99% sequence identity, and would be consistent with it being classified as a pKpQIL-D2-like plasmid affected by large indels (see long read results below). The remaining three isolates had distinct plasmid structures with significant matches to the NCBI plasmid database (threshold: sequence length match >75%; sequence identity >95%). Most isolates carried plasmid replicon types IncR (22/23) and IncFIB (22/23).
blaKPC/Tn4401 plasmid characterization from MinION long-read sequencing
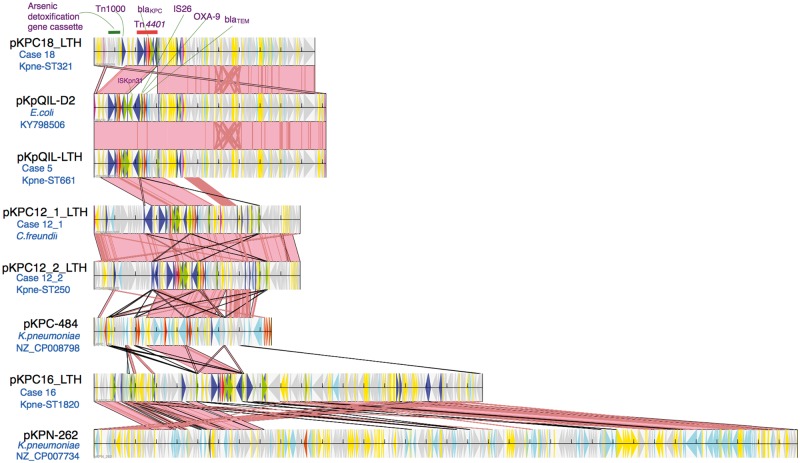
MinION sequencing coverage for the five selected isolates ranged from 26× to 43× (Table S2), enough to facilitate complete assembly of the blaKPC/Tn4401-carrying plasmids in 4/5 isolates, either by hybridSPAdes (n = 1) or Canu (n = 3, Table S3). The closed structure of the blaKPC/Tn4401-carrying plasmid of the KPC-Kpn ST661 isolates, pKpQIL-LTH (117 kb), was 99% identical to the IncR/IncFII pKpQIL-D2 (Figure 3). The blaKPC/Tn4401-carrying plasmids from C. freundii (pKPC12_1_LTH, 99.3 kb) and KPC-Kpn ST250 (pKPC12_2_LTH, 99.3 kb) from case 12 showed nearly identical plasmid structure (>99% sequence length match and sequence identity; Figure 3). This was a novel blaKPC-harbouring IncN/IncR plasmid, most closely related to pKPC-484 (GenBank accession: CP008798.1, 85.5 kb, Maryland, USA; 56% sequence length match, 99% sequence identity). The distinct structure of this plasmid compared with the dominant outbreak pKpQIL-D2 plasmid (43% sequence length match including regions flanking Tn4401, 99% sequence identity) implicates either independent import of this plasmid from an unknown source, or a novel local transfer event of blaKPC/Tn4401 into a new plasmid backbone, likely via recombination. The presence of these near-identical plasmids in two different species suggests cross-species transmission of this novel plasmid structure within this patient.
Figure 3.
Outbreak KPC plasmids and contigs derived from long-read sequencing of isolates and their alignment to the most closely genetically matched complete plasmid sequences from NCBI. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
The blaKPC/Tn4401 plasmid pKPC18_LTH from case 18 (KPC-Kpn ST321, Canu assembly, 106 kb) was an IncR/IncFII pKpQIL-D2-like plasmid (91% sequence length match, 99% sequence identity; Figure 3). It differed from pKpQIL-D2 by the deletion of the ΔblaOXA-9–blaTEM-1 gene cassette and the insertion of an arsenic detoxification gene cassette into the pKpQIL-D2 plasmid backbone. This patient from northwest England was admitted to LTH in December 2013, and was first found to be colonized with a KPC-positive isolate in January 2014. Therefore, this patient might have carried this plasmid prior to admission to LTH or acquired it locally from an unidentified source with large indel events potentially driven by selection pressures.
In the remaining case with a distinct plasmid (KPC-Kpn ST1820, case 16), manual inspection of the hybridSPAdes assembly implicated that there was an assembly error, incorrectly merging the blaKPC/Tn4401 plasmid contig with the chromosomal contig (Figure S1). After error correction, the revised blaKPC/Tn4401 contig pKPC16_LTH (187 kb) was 83% similar to pKPN-262 (Figure 3) harbouring the pKpQIL-like IncFII replicon and IncFIB(K) as opposed to IncFIB(pQil) generally found in pKpQIL-like plasmids. The patient harbouring pKPC16_LTH was admitted for liver transplant in December 2013 from Manchester and had negative screening results for blaKPC on admission. This could imply either a horizontal transfer of the blaKPC/Tn4401 element from pKpQIL-D2 outbreak plasmid into another plasmid backbone or an independent acquisition from a local unknown source.
Transmission analysis
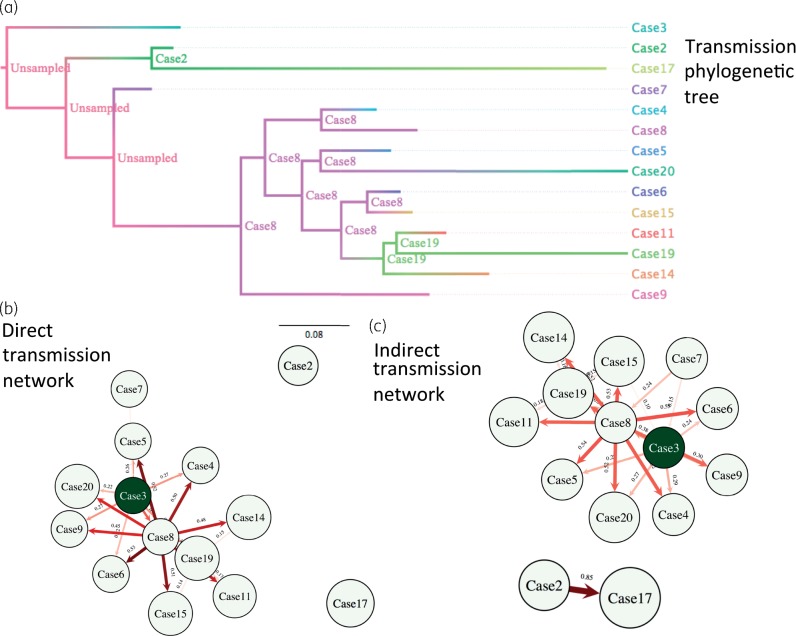
WGS-based chromosomal sequences and detailed epidemiological data were used to estimate probable blaKPC transmission routes, with the aim of differentiating between ward-based dispersal and importation from high-risk areas outside, but associated with, Leeds. A potential index case of KPC-Kpn (ST491) with the pKpQIL-D2 Tn4401a-carrying plasmid was identified 10 months before the outbreak. This plasmid was later seen in other KPC-Kpn (ST14, ST661) and two other species, consistent with unrecognized plasmid dispersal, and subsequently a strain–plasmid (K. pneumoniae ST661-pKpQIL-LTH, Figure S2) outbreak nearly a year later. The transmission network and transmission phylogenetic tree (Figure 4) suggested that a single case (case 8) was potentially central to dissemination of ST661-pKpQIL-LTH following probable acquisition from case 3. This symptomatic case was in a shared bay for a prolonged period prior to KPC detection (Figure 1), potentially transmitting to nine cases (with >0.4 probability), of which six were within 2–4 SNPs (cases 4–7, 11 and 15). Patient-to-patient transmission was not demonstrated between other cases in the outbreak, for which KPC acquisition was more likely to relate to unidentified sources, such as unscreened patients, staff or environmental reservoirs. Cases 2 and 17 (7 SNPs apart) plausibly represented an indirect transmission link, probably through multiple transmission chains. Although case 17 was >10 SNPs different from other clonal strains, case 2 was 4–11 SNPs apart from the remaining cases. Due to large differences in hospital stays and times from colonization to sampling, coalescent times within hosts vary, giving rise to a broad range of SNP distances, and therefore absolute SNP distance thresholds are not informative and often bias to discriminate direct transmission links.
Figure 4.
Transmission analysis results inferred by SCOTTI for the dominant outbreak strain KPC-Kpne ST661 using consensus chromosomal sequences and epidemiological data. The ‘maximum clade credibility’ of the posterior coalescent tree (scale in years) was inferred by SCOTTI, taking into account epidemiological data and transmission processes. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Discussion
This is the first report of the detailed molecular epidemiology of a successfully controlled KPC-E outbreak in the UK. This study also marks the first application of MinION long-read sequencing to investigate blaKPC plasmid diversity in a clinical outbreak. This nosocomial outbreak, involving 23 isolates from just 20 patients, demonstrated blaKPC dissemination via multiple routes, including clonal spread of a bacterial strain in conjunction with a pKpQIL-D2 plasmid (K. pneumoniae ST661, 15 cases), horizontal transfer of the outbreak pKpQIL-D2 plasmid to other STs/species (5 cases) most likely following its introduction via an index case 10 months previously, and either horizontal transposition (Tn4401a transmission from the outbreak pKpQIL-D2 plasmid to other distinct plasmids) or external import of blaKPC/Tn4401 plasmids. This small outbreak included striking genetic diversity (six K. pneumoniae lineages, cross-species plasmid transmission within patients, four distinct plasmid types) and highlights the relative impact of symptomatic patients, non-K. pneumoniae KPC-E, environmental reservoirs and unscreened individuals in blaKPC dissemination (despite high-density contact-screening). The initial outbreak definition capturing only KPC-Kpn with the same VNTR type failed to demonstrate the extent of blaKPC transmission, and reaffirms the need for consideration of multispecies involvement early in putative outbreaks. Patient-to-patient KPC-E transmission outside the liver unit could not be excluded but was thought unlikely as no new cases/colonized patients were detected outside the liver unit during the outbreak, or subsequently.
This outbreak demonstrates the difficulty in identifying transmission events between CPE cases, even with the aid of high-intensity screening, short-read WGS data and long-read plasmid sequencing. We demonstrated a high colonized : infected patient ratio (3:1) compared with other outbreaks,41 possibly reflecting more intensive screening or a lower rate of clinical infection in colonized individuals. Our analysis demonstrates probable dissemination of ST661-pKpQIL-D2 from a central source to nine cases, and indirect transmission for the remainder, supporting the existence of alternative unscreened reservoirs. Long-read MinION sequencing proved to be beneficial for high-resolution plasmid tracking, but still suffered from sequencing and bioinformatics limitations, resulting in an erroneous result in one assembly. Improvements in sequencing protocols to enrich for longer reads, better bioinformatics tools (assemblers) independent of highly skilled bioinformatics support, and cost reductions are required to implement routine WGS-based plasmid tracking. Additionally, although same-day, direct from sample, turnaround time as demonstrated for Mycobacterium tuberculosis diagnosis42 has not yet been achieved for Enterobacteriaceae species, current developments in culture-based laboratory protocols allow sample preparation for long-read sequencing to be completed in ∼8 h (6 h DNA extraction; 10 min or 1.5 h options for sequence library preparation; ONT). Subsequently, 80% of sequencing data can be obtained within the first 10 h of a 48 h sequencing run,42 and bioinformatics analysis can be performed in 4–5 h. This allows potential turnaround times of <24 h from culture positivity to initial outbreak analysis.
Nosocomial CPE outbreaks may evolve into regional endemicity if transmission chains between patients/other reservoirs cannot be interrupted. Patient transfer from high-prevalence KPC-E centres in northwest England to the Leeds supra-regional liver unit were frequent during this outbreak, representing multiple opportunities for reintroductions. A recent study suggests the most probable source of CPE is from readmission of patients from the same hospital/region rather than transfer between regions.15 Despite detailed genetic analysis it was difficult to differentiate clearly between importation of resistance vectors and patient-to-patient KPC-E transmission in this outbreak. However, the rare plasmid-type, dissemination of a rare lineage (ST661) and the fact that the outbreak was successfully controlled suggests local transmission was the dominant source of acquisition. Given that all patients on the unit were screened following detection of the outbreak, undetected human reservoirs were a less likely source of KPC acquisition than environmental persistence. Hydrogen peroxide vapour treatment of the unit on two occasions, along with strict patient cohorting and screening, may have contributed significantly to halting the outbreak.
This outbreak resulted in changes to clinical care: transplant assessments were transferred to the outpatient setting and included CPE screening, and strict algorithms for admission screening/isolation on the liver unit, discharge CPE screening and carbapenem-sparing antibiotic prescribing strategies were implemented. It was not possible to evaluate the relative impact of each of these interventions on reducing CPE transmission.
There were several limitations of this study, mostly relating to the potential to have underestimated cases/colonization episodes. Phenotypic tests were used to identify potential CPE producers during the outbreak; since then, more sensitive and specific PCR-based CPE detection has been implemented.43,44 There was a lack of high-intensity screening of asymptomatic patients prior to outbreak detection (July–October 2013). However, routine CPE admission screening was in place during this period. Repeat KPC-E isolates from individual patients were not available. Analysis of the evolution of KPC-E strains within individuals over time may have provided further information about the genetic mechanisms associated with blaKPC dispersal. Finally, there are few publically available UK KPC-carrying whole-genome and plasmid sequences, limiting our ability to contextualize the Leeds strains.
We emphasize that there is considerable potential for rapid dissemination of carbapenemase genes among Klebsiella spp. and between Enterobacteriaceae, despite the implementation of extensive infection control interventions. Tracking transmission networks is challenging, even with detailed epidemiological and WGS data, due to the mobility and evolution of mobile genetic elements. Publicly available national CPE sequence data will enable local institutions to contextualize outbreak strains and identify the role of importation versus patient-to-patient transmission in response to increases in CPE incidence and outbreaks.
Supplementary Material
Acknowledgments
Funding
The research was funded by the National Institute for Health Research Health Protection Research Unit (NIHR HPRU) in Healthcare Associated Infections and Antimicrobial Resistance at Oxford University in partnership with Public Health England (PHE) [HPRU-2012–10041]. J. M. is an NIHR academic clinical fellow. N. S. is a PHE/University of Oxford clinical lecturer. D. W. E. is an NIHR clinical lecturer. D. W. C. and T. E. A. P. are NIHR senior investigators.
Transparency declarations
None to declare.
Disclaimer
The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR, the Department of Health or PHE.
Supplementary data
Methods, Tables S1–3 and Figures S1 and S2 appear as Supplementary data at JAC Online.
References
- 1. Munoz-Price LS, Poirel L, Bonomo RA. et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis 2013; 13: 785–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mathers AJ, Cox HL, Kitchel B. et al. Molecular dissection of an outbreak of carbapenem-resistant enterobacteriaceae reveals intergenus KPC carbapenemase transmission through a promiscuous plasmid. MBio 2011; 2: e00204–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Andrade LN, Curiao T, Ferreira JC. et al. Dissemination of blaKPC-2 by the spread of Klebsiella pneumoniae clonal complex 258 clones (ST258, ST11, ST437) and plasmids (IncFII, IncN, IncL/M) among Enterobacteriaceae species in Brazil. Antimicrob Agents Chemother 2011; 55: 3579–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cuzon G, Naas T, Truong H. et al. Worldwide diversity of Klebsiella pneumoniae that produce β-lactamase blaKPC-2 gene. Emerg Infect Dis 2010; 16: 1349–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cuzon G, Naas T, Nordmann P. Functional characterization of Tn4401, a Tn3-based transposon involved in blaKPC gene mobilization. Antimicrob Agents Chemother 2011; 55: 5370–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Naas T, Cuzon G, Villegas MV. et al. Genetic structures at the origin of acquisition of the β-lactamase blaKPC gene. Antimicrob Agents Chemother 2008; 52: 1257–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kanamori H, Parobek CM, Juliano JJ. et al. A prolonged outbreak of KPC-3-producing Enterobacter cloacae and Klebsiella pneumoniae driven by multiple mechanisms of resistance transmission at a large academic burn center. Antimicrob Agents Chemother 2017; 61: pii: e01516-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Conlan S, Thomas PJ, Deming C. et al. Single-molecule sequencing to track plasmid diversity of hospital-associated carbapenemase-producing Enterobacteriaceae. Sci Transl Med 2014; 6: 254ra126.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Control ECfDPa. Annual Report of the European Antimicrobial Resistance Surveillance Network (EARS-net) 2014. http://ecdc.europa.eu/en/publications/_layouts/forms/Publication_DispForm.aspx?List=4f55ad51-4aed-4d32-b960-af70113dbb90&ID=1400.
- 10. Lin MY, Lyles-Banks RD, Lolans K. et al. The importance of long-term acute care hospitals in the regional epidemiology of Klebsiella pneumoniae carbapenemase-producing Enterobacteriaceae. Clin Infect Dis 2013; 57: 1246–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Palepou M-FI, Woodford N, Hope R. et al. Novel class A carbapenemase, KPC-4, in an Enterobacter isolate from Scotland. In: Fifteenth European Congress of Clinical Microbiology and Infectious Diseases, Copenhagen Abstract 1134_01_20. Clin Microbiol Infect 2005; 11: Suppl 2: S106. [Google Scholar]
- 12. Public Health England. English Surveillance Programme for Antimicrobial Utilisation and Resistance (ESPAUR) 2010-2014 2015. https://www.gov.uk/government/uploads/system/uploads/attachment_data/file/477962/ESPAUR_Report_2015.pdf.
- 13. Public Health England. Carbapenemase-Producing Enterobacteriaceae: Laboratory Confirmed Cases, 2003 to 2015 https://www.gov.uk/government/publications/carbapenemase-producing-enterobacteriaceae-laboratory-confirmed-cases/carbapenemase-producing-enterobacteriaceae-laboratory-confirmed-cases-2003-to-2013.
- 14. Findlay J, Hopkins KL, Doumith M. et al. KPC enzymes in the UK: an analysis of the first 160 cases outside the North-West region. J Antimicrob Chemother 2016; 71: 1199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Donker T, Henderson KL, Hopkins KL. et al. The relative importance of large problems far away versus small problems closer to home: insights into limiting the spread of antimicrobial resistance in England. BMC Med 2017; 15: 86.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Eyre DW, Golubchik T, Gordon NC. et al. A pilot study of rapid benchtop sequencing of Staphylococcus aureus and Clostridium difficile for outbreak detection and surveillance. BMJ Open 2012; 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Walker TM, Clp CL, Harrell RH. et al. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect Dis 2013; 13: 137–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stoesser N, Giess A, Batty EM. et al. Genome sequencing of an extended series of NDM-producing Klebsiella pneumoniae isolates from neonatal infections in a Nepali hospital characterizes the extent of community- versus hospital-associated transmission in an endemic setting. Antimicrob Agents Chemother 2014; 58: 7347–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Arredondo-Alonso S, Willems RJ, Schurch AC On the (im)possibility to reconstruct plasmids from whole genome short-read sequencing data. Bioarvx2016; doi:10.1101/086744. [DOI] [PMC free article] [PubMed]
- 20. Conlan S, Park M, Deming C. et al. Plasmid dynamics in KPC positive Klebsiella pneumoniae during long-term patient colonization. MBio 2016; 7: e00742–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Public Health England. Carbapenemase-Producing Enterobacteriaceae: Early Detection, Management and Control Toolkit for Acute Trusts 2013. https://www.gov.uk/government/uploads/system/uploads/attachment_data/file/329227/Acute_trust_toolkit_for_the_early_detection.pdf.
- 22. Turton JF, Perry C, Elgohari S. et al. PCR characterization and typing of Klebsiella pneumoniae using capsular type-specific, variable number tandem repeat and virulence gene targets. J Med Microbiol 2010; 59: 541–7. [DOI] [PubMed] [Google Scholar]
- 23. Al-Agamy MH, Shibl AM, Elkhizzi NA. et al. Persistence of Klebsiella pneumoniae clones with OXA-48 or NDM carbapenemases causing bacteraemias in a Riyadh hospital. Diagn Microbiol Infect Dis 2013; 76: 214–6. [DOI] [PubMed] [Google Scholar]
- 24. European Committee on Antimicrobial Susceptibility Testing (EUCAST). 2017. Breakpoint Tables for Interpretation of MICs and Zone Diameters http://www.eucast.org/clinical_breakpoints/.
- 25. Yigit H, Queenan AM, Anderson GJ. et al. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother 2001; 45: 1151–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Poirel L, Heritier C, Tolun V. et al. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother 2004; 48: 15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mushtaq S, Irfan S, Sarma JB. et al. Phylogenetic diversity of Escherichia coli strains producing NDM-type carbapenemases. J Antimicrob Chemother 2011; 66: 2002–5. [DOI] [PubMed] [Google Scholar]
- 28. Ellington MJ, Kistler J, Livermore DM. et al. Multiplex PCR for rapid detection of genes encoding acquired metallo-β-lactamases. J Antimicrob Chemother 2007; 59: 321–2. [DOI] [PubMed] [Google Scholar]
- 29. Doumith M, Findlay J, Hirani H. et al. Major role of pKpQIL-like plasmids in the early dissemination of KPC-type carbapenemases in the UK. J Antimicrob Chemother 2017; 72: 2241–8. [DOI] [PubMed] [Google Scholar]
- 30. Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol 2014; 15: R46.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stoesser N, Sheppard AE, Pankhurst L. et al. Evolutionary history of the global emergence of the Escherichia coli epidemic clone ST131. MBio 2016; 7: e02162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nguyen LT, Schmidt HA, von Haeseler A. et al. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 2015; 32: 268–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Didelot X, Wilson DJ. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput Biol 2015; 11: e1004041.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bankevich A, Nurk S, Antipov D. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012; 19: 455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stoesser N, Peirano G, Anson LW. et al. Genomic epidemiology of global Klebsiella pneumoniae carbapenemase (KPC)-producing Escherichia coli. Sci Rep 2017; 7: 5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Loman NJ, Quinlan AR. Poretools: a toolkit for analyzing nanopore sequence data. Bioinformatics 2014; 30: 3399–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cao MD, Nguyen SH, Ganesamoorthy D. et al. Scaffolding and completing genome assemblies in real-time with nanopore sequencing. Nat Commun 2017; 8: 14515.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Berlin K, Koren S, Chin CS. et al. Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat Biotechnol 2015; 33: 623–30. [DOI] [PubMed] [Google Scholar]
- 39. De Maio N, Wu CH, Wilson DJ. SCOTTI: efficient reconstruction of transmission within outbreaks with the structured coalescent. PLoS Comput Biol 2016; 12: e1005130.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cannatelli A, Giani T, D’Andrea MM. et al. MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob Agents Chemother 2014; 58: 5696–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wendt C, Schutt S, Dalpke AH. et al. First outbreak of Klebsiella pneumoniae carbapenemase (KPC)-producing K. pneumoniae in Germany. Eur J Clin Microbiol Infect Dis 2010; 29: 563–70. [DOI] [PubMed] [Google Scholar]
- 42. Votintseva AA, Bradley P, Pankhurst L. et al. Same-day diagnostic and surveillance data for tuberculosis via whole-genome sequencing of direct respiratory samples. J Clin Microbiol 2017; 55: 1285–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ellington MJ, Findlay J, Hopkins KL. et al. Multicentre evaluation of a real-time PCR assay to detect genes encoding clinically relevant carbapenemases in cultured bacteria. Int J Antimicrob Agents 2016; 47: 151–4. [DOI] [PubMed] [Google Scholar]
- 44. Hammoudi D, Moubareck CA, Sarkis DK. How to detect carbapenemase producers? A literature review of phenotypic and molecular methods. J Microbiol Methods 2014; 107: 106–18. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.