Abstract
Objectives: MRSA is a leading cause of hospital-associated infection. Acquired resistance is encoded by the mecA gene or its homologue mecC, but little is known about the evolutionary dynamics involved in gain and loss of resistance. The objective of this study was to obtain an expanded understanding of Staphylococcus aureus methicillin resistance microevolution in vivo, by focusing on a single lineage.
Methods: We compared the whole-genome sequences of 231 isolates from a single epidemic lineage [clonal complex 30 (CC30) and spa-type t018] of S. aureus that caused an epidemic in the UK.
Results: We show that resistance to methicillin in this single lineage was gained on at least two separate occasions, one of which led to a clonal expansion around 1995 presumably caused by a selective advantage. Resistance was, however, subsequently lost in vivo by nine strains isolated between 2008 and 2012. We describe the genetic mechanisms involved in this loss of resistance and the imperfect relationship between genotypic and phenotypic resistance.
Conclusions: The recent re-emergence of methicillin susceptibility in this epidemic lineage suggests a significant fitness cost of resistance and reduced selective advantage following the introduction in the mid-2000s of MRSA hospital control measures throughout the UK.
Introduction
Staphylococcus aureus is a commensal bacterium frequently colonizing the nose and skin, but also a potential pathogen, causing diseases ranging from mild skin infections to septicaemia. Worldwide, S. aureus is a leading cause of hospital-associated infections, exacerbated by strains resistant to commonly used antibiotics. MRSA is resistant to most β-lactam antibiotics, including penicillins and cephalosporins.1 MRSA genomes are typically distinguishable from MSSA by the presence of the mecA gene or its homologue mecC. In the UK, healthcare-associated MRSA came to the fore in the 1990s mostly in the form of the two epidemic clones EMRSA-15 and EMRSA-16, the presence of which declined after 2005.2 Genome sequence analysis to detect mecA enables the resistance phenotype to be predicted with high, although imperfect, accuracy.3,4 The mecA gene is part of the SCCmec cassette that can be inserted into the staphylococcal chromosome and inherited vertically, or transferred between lineages via horizontal gene transfer.5 Most MRSA lineages evolved from MSSA ancestors after gaining SCCmec, providing a selective advantage, which likely contributed to their worldwide spread. However, little is known about the fitness cost of resistance and the dynamics of SCCmec acquisition or about the re-emergence of genomic and phenotypic susceptibility. Additionally, there are reports of phenotypic resistance in the absence of mecA and conversely of phenotypic susceptibility in the presence of apparently functional mecA,3 although the underlying mechanisms are poorly understood.
In order to shed new light on these important issues, we compared whole-genome sequences of 231 isolates (197 MRSA, 34 MSSA) sampled from across England between 1997 and 2013. All isolates belonged to the clinically important clonal complex 30 (CC30) and to spa-type t018. This collection includes the healthcare-associated MRSA clone known as EMRSA-16 (ST36-SCCmecII).
Materials and methods
Isolates
We selected 231 isolates (see Table S1, available as Supplementary data at JAC Online) obtained from clinical specimens (one isolate per patient), which all belonged to both spa-type t018 (as determined using spa-typing) and CC30 (as determined based on genome sequences). Forty-eight isolates were from carriage screening swabs and 183 from diagnostic samples, including 167 from blood cultures. Isolates originated from Brighton (131), Oxford (47), London (19), elsewhere in southern England (21) and the Midlands and northern England (13). Thirty-nine isolates were obtained from material archived at the PHE reference laboratory, Colindale. Ten isolates had been collected by the UK Clinical Infection Research Group. STs represented were: ST36 (213), ST30 (15), ST34 (2) and ST38 (1). The methicillin susceptibility of the isolates was assessed phenotypically on primary testing as part of routine diagnostic laboratory procedures. Methicillin susceptibility was subsequently reassessed by disc diffusion (cefoxitin) and Etest (oxacillin). Isolates were stored, cultured, identified and sequenced as described elsewhere.3,6
Bioinformatics methods
The sequenced reads were assembled both de novo and by reference-based mapping against MRSA2527 using a previously described bioinformatics pipeline.8 STs were determined in silico based on the de novo assemblies. The phylogeny was built using PhyML,9 corrected for the effect of recombination using ClonalFrameML10 and dated using previously described methodology.11 The dating process relied on the sampling date of each sample and on a mutation rate that was assumed to be 8.4 mutations per year per genome.12–14
Ethics statement
Isolate storage and data collection was approved in Brighton by the Brighton and Sussex University Hospitals (BSUH) NHS Trust Research and Development office as a service evaluation, involving anonymized data from patient records and not requiring formal ethical review. Isolates were collected for epidemiological studies covered by Statutory Instrument Regulations 2002 No. 1438, section (iii) ‘Communicable disease and other risks to public health (Health Service Control of Patient Information)’ of Section 60 of the Health and Social Care Act and therefore did not require research ethics committee approval.
Results
Phylogenetic distribution of resistance
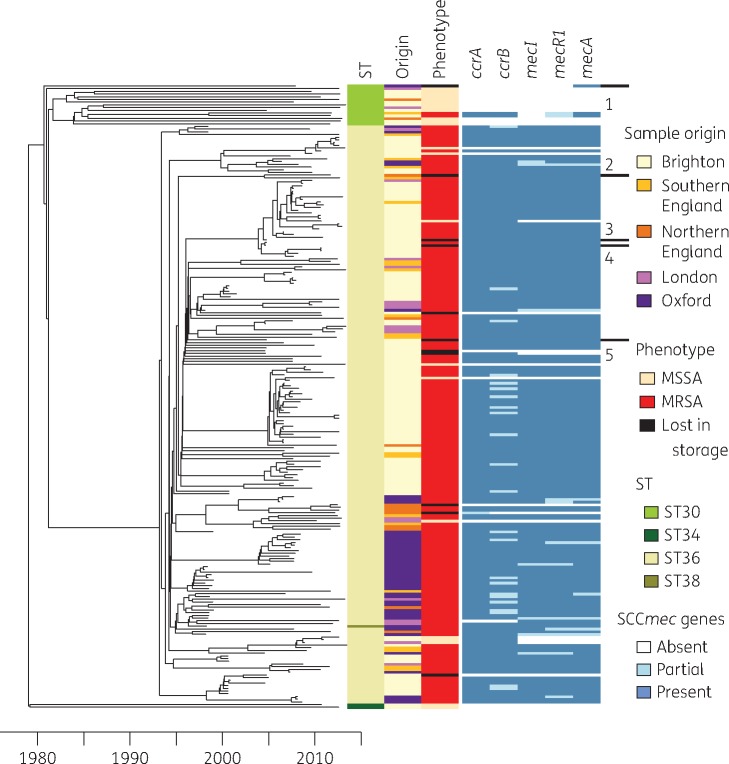
A dated phylogeny was constructed using the genome sequences of all isolates (Figure 1). As expected, the samples cluster in accordance with MLST ST as determined in silico. The most recent common ancestor for the entire lineage dates to 1978, with divergence thereafter of branches leading to ST30, ST34 and ST36. Most isolates belong to ST36 (EMRSA-16) whose most recent common ancestor was dated to 1993. This is more recent than a previous estimate of 1975,14 but our result is in good agreement with the timing of the first observations in the UK of ST36.2 The unique ST38 sequence nests within the ST36 clade, indicating its direct derivation from ST36. Both available ST34 isolates were MSSA. Most ST30 isolates were methicillin susceptible although two were methicillin resistant following the acquisition of SCCmecIV. Most ST36 isolates were methicillin resistant, with many branches diverging close to the most recent common ancestor, suggesting rapid clonal expansion associated with a fitness advantage conferred by the loss of susceptibility. Resistance acquisition by ST30 and ST36 cannot be dated more accurately than between 1980 and 1995 as both events occurred on long branches. Surprisingly, within the predominantly resistant ST36 lineage were 19 MSSA isolates. Ten of these could be explained by loss of resistance during storage,15 because the isolates had been found to be resistant in susceptibility tests performed directly after isolation. In contrast, the remaining nine isolates had been identified as MSSA at the time of primary culture (Figure 1).
Figure 1.
Dated phylogenetic tree showing the relationship between all 231 S. aureus genomes. The panel on the right shows a number of properties of the genomes, namely (from left to right) the MLST ST, geographical location of origin (origin), phenotypic resistance status (MRSA, MSSA or loss of resistance during storage) and presence/absence of five genes typically present in SCCmec type II (ccrA, ccrB, mecI, mecR1 and mecA). The five genomes for which phenotypic and genotypic resistance data were discrepant are labelled 1–5. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Discrepancies between resistance phenotype and genotype
The mecA gene, encoding resistance to β-lactam antibiotics,16 is located in the SCCmec cassette, which represents a hotspot of recombination.8 Many different alleles of SCCmec have been described, differing in the number and type of genes present.5 In our dataset we found two different SCCmec types, each paired to a different ST: ST36 harbours a type II cassette, with mecR1, mecI, ccrA, ccrB and mecA;1 and ST30 harbours a type IV cassette, lacking the mecI gene and having a partial mecR1. In general we found concordance between resistance phenotype and genotype (Table 1). SCCmec does not have to be complete to be functional.17 We found partial SCCmec in seven isolates. In one case only the ccrA/B genes were missing and the strain was phenotypically resistant; in all other cases only the ccrA/B genes were present and the strain was phenotypically susceptible.
Table 1.
Summary of genotypic and phenotypic methicillin resistance status for all 231 isolates described in this study
| Genotype | Phenotype |
||
|---|---|---|---|
| resistant | susceptible | ||
| ST36 | SCCmec (type II) | 191 | 5 |
| partial SCCmec (type II) | 1 | 5 | |
| no SCCmec | 0 | 9 | |
| ST30 | SCCmec (type IV) | 2 | 0 |
| partial SCCmec (type IV) | 0 | 1 | |
| no SCCmec | 0 | 14 | |
| ST34 | SCCmec | 0 | 0 |
| partial SCCmec | 0 | 0 | |
| no SCCmec | 0 | 2 | |
| ST38 | SCCmec (type II) | 1 | 0 |
| partial SCCmec | 0 | 0 | |
| no SCCmec | 0 | 0 | |
Five isolates were methicillin susceptible despite the presence of the mecA gene, and all of them had lost methicillin resistance during storage (labelled 1–5 in Figure 1). An ST30 isolate (labelled 1 in Figure 1) had the gene, but lacked the rest of the operon, which might explain its susceptibility. The entire operon was present in the other four discrepant isolates. One isolate (labelled 2 in Figure 1) shows deletion of a single base-pair in mecA, resulting in a frameshift, premature stop codon and gene inactivation. In the remaining three discrepant isolates (labelled 3–5 in Figure 1) the mecA gene is identical to the functional mecA genes present in resistant isolates. Other SCCmec genes in these discrepant isolates do not exhibit any particular differences from resistant isolates in the collection. Analysis of genes previously described as interacting with SCCmec (blaZ, blaI, blaR1, femA and femB) yielded no conclusive result. Similarly, analysis of polymorphic sites known to be associated with resistance yielded no significant result.
Re-emergence of susceptibility
The distribution of susceptible isolates within the timed tree shows that the ancestral resistant phenotype was lost in vivo in nine strains isolated in Brighton (n = 7) and London (n = 2) between 2008 and 2012. Three of these formed a genetic cluster whilst the others were genetically distant, with their nearest neighbours being MRSA. Methicillin susceptibility therefore re-emerged independently on at least seven separate occasions within the ancestrally resistant ST36, and this was confirmed using ancestral state reconstruction. Within ST36, the dates of the nine MSSA isolates were significantly more recent than the dates of the MRSA isolates (P < 0.01, Kolmogorov–Smirnov test), suggesting that re-emergence of susceptibility was linked with MRSA-specific control measures introduced in the UK in the mid-2000s.2 Interestingly, we found several different molecular mechanisms that led to the loss of the resistant phenotype in vivo or in storage. The most frequent genetic background for the susceptible phenotype (9 genomes out of the total 19) was loss of the entire SCCmec cassette. In six of the susceptible samples we were able to detect only part of the cassette, but no resistance-associated mec genes (mecA, mecI or mecR1). In one genome (labelled 2 in Figure 1) the entire SCCmec cassette was present, but the susceptibility can be explained by a deletion causing a frameshift and loss of function in the mecA gene. Finally, there remain three cases (labelled 3–5 in Figure 1) for which we were unable to find a genetic explanation for the phenotypic loss of resistance, as described above.
Discussion
By comparing 197 MRSA and 34 MSSA genomes, representing a single epidemic lineage (CC30) of S. aureus, we were able to show that ST36 (corresponding to EMRSA-16) gained SCCmec before the mid-1990s and subsequently underwent clonal expansion (Figure 1). Loss of methicillin resistance during the storage retrieval process is well documented15 and we found 10 examples of this in our study. More surprisingly, we also recorded many examples of loss of methicillin resistance in vivo affecting multiple sublineages within ST36 and occurring after 2008, at a time when MRSA control measures were being implemented in UK hospitals. These observations suggest that methicillin resistance originally provided a selective advantage to ST36 compared with other members of CC30, including the putative methicillin-susceptible ST36 ancestor, which does not feature in our dataset. However, resistance may impart a fitness cost,18 which has apparently not been overcome by compensatory mutations. When the fitness cost exceeds the selective advantage of resistance, susceptible strains are expected to re-emerge. Recent initiatives to limit β-lactam usage, including restricted prescribing of cephalosporins, may partly explain our observations.19 Further work will be needed to determine to what extent our observation is unique to the ST36 lineage we studied, or whether similar dynamics of resistance loss occur for all MRSA, which would for example explain why Swedish MSSA outbreak isolates contained remnants of SCCmec.20
We have demonstrated multiple disparate mechanisms to explain reversion from MRSA to MSSA and our detection of a cluster of three susceptible genetically related isolates suggests that such strains are transmissible and have the potential to spread. As we have shown, the prediction of phenotypic resistance from genomic sequence data has yet to be perfected, although increasing interest in this subject suggests that it will improve rapidly.3,4 More accurate resistance prediction, combined with reductions in sequencing costs and turnaround times may allow more targeted use of antibiotics and facilitate antibiotic stewardship. Our findings represent an encouraging observation for MRSA control efforts and more generally for the control of antibiotic-resistant pathogens.
Funding
This work was funded by the UK National Institute for Health Research (NIHR) Health Protection Research Units (HPRU) in Modelling Methodology at Imperial College London (grant HPRU-2012-10080) and in Healthcare-Associated Infections and Antimicrobial Resistance at the University of Oxford (HPRU-2012-10041).
Transparency declarations
None to declare.
Supplementary data
Table S1 is available as Supplementary data at JAC Online.
Supplementary Material
References
- 1. Chambers HF, Deleo FR.. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol 2009; 7: 629–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wyllie D, Paul J, Crook DW.. Waves of trouble: MRSA strain dynamics and assessment of the impact of infection control. J Antimicrob Chemother 2011; 66: 2685–8. [DOI] [PubMed] [Google Scholar]
- 3. Gordon NC, Price JR, Cole K. et al. Prediction of Staphylococcus aureus antimicrobial resistance by whole-genome sequencing. J Clin Microbiol 2014; 52: 1182–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bradley P, Gordon NC, Walker TM. et al. Rapid antibiotic resistance predictions from genome sequence data for S. aureus and M. tuberculosis. Nat Comm 2015; 6: 18564.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ito T, Hiramatsu K, Oliveira DC. et al. Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob Agents Chemother 2009; 53: 4961–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Miller RM, Price JR, Batty EM. et al. Healthcare-associated outbreak of meticillin-resistant Staphylococcus aureus bacteraemia: role of a cryptic variant of an epidemic clone. J Hosp Infect 2014; 86: 83–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Holden MTG, Feil EJ, Lindsay JA. et al. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc Natl Acad Sci USA 2004; 101: 9786–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Everitt RG, Didelot X, Batty EM. et al. Mobile elements drive recombination hotspots in the core genome of Staphylococcus aureus. Nat Commun 2014; 5: 3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guindon S, Dufayard J-F, Lefort V. et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 2010; 59: 307–21. [DOI] [PubMed] [Google Scholar]
- 10. Didelot X, Wilson DJ.. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput Biol 2015; 11: e1004041.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Didelot X, Eyre DW, Cule M. et al. Microevolutionary analysis of Clostridium difficile genomes to investigate transmission. Genome Biol 2012; 13: R118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harris SRR, Feil EJ, Holden MT. et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science 2010; 327: 469–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Young BC, Golubchik T, Batty EM. et al. Evolutionary dynamics of Staphylococcus aureus during progression from carriage to disease. Proc Natl Acad Sci USA 2012; 109: 4550–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McAdam PR, Templeton KE, Edwards GF. et al. Molecular tracing of the emergence, adaptation, and transmission of hospital-associated methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci USA 2012; 109: 9107–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. van Griethuysen A, van Loo I, van Belkum A. et al. Loss of the mecA gene during storage of methicillin-resistant Staphylococcus aureus strains. J Clin Microbiol 2005; 43: 1361–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chambers HF. Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clin Microbiol Rev 1997; 10: 781–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Suzuki E, Kuwahara-Arai K, Richardson JF. et al. Distribution of mec regulator genes in methicillin-resistant Staphylococcus clinical strains. Antimicrob Agents Chemother 1993; 37: 1219–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Didelot X, Walker AS, Peto TE. et al. Within-host evolution of bacterial pathogens. Nat Rev Microbiol 2016; 14: 150–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Llewelyn MJ, Hand K, Hopkins S. et al. Antibiotic policies in acute English NHS trusts: implementation of ‘Start Smart-Then Focus’ and relationship with Clostridium difficile infection rates. J Antimicrob Chemother 2014; 70: 1230–5. [DOI] [PubMed] [Google Scholar]
- 20. Lindqvist M, Isaksson B, Grub C. et al. Detection and characterisation of SCCmec remnants in multiresistant methicillin-susceptible Staphylococcus aureus causing a clonal outbreak in a Swedish county. Eur J Clin Microbiol Infect Dis 2012; 31: 141–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.