Abstract
Objectives: Gram-negative bacteria harbouring the mcr-1 plasmid are resistant to the ‘last-line’ polymyxins and have been reported worldwide. Our objective was to define the impact of increasing the initial polymyxin B dose intensity against an mcr-1-harbouring strain to delineate the impact of plasmid-mediated polymyxin resistance on the dynamics of bacterial killing and resistance.
Methods: A hollow fibre infection model (HFIM) was used to simulate polymyxin B regimens against an mcr-1-harbouring Escherichia coli (MIC 8 mg/L) over 10 days. Four escalating polymyxin B ‘front-loading’ regimens (3.33, 6.66, 13.3 or 26.6 mg/kg for one dose followed by 1.43 mg/kg every 12 h starting 12 h later) simulating human pharmacokinetics were utilized in the HFIM. A mechanism-based, mathematical model was developed using S-ADAPT to characterize bacterial killing.
Results: The 3.33 mg/kg ‘front-loading’ regimen resulted in regrowth mirroring the growth control. The 6.66, 13.3 and 26.6 mg/kg ‘front-loading’ regimens resulted in maximal bacterial reductions of 1.91, 3.79 and 6.14 log10 cfu/mL, respectively. Irrespective of the early polymyxin B exposure (24 h AUC), population analysis profiles showed similar growth of polymyxin B-resistant subpopulations. The HFIM data were well described by the mechanism-based model integrating three subpopulations (susceptible, intermediate and resistant). Compared with the susceptible subpopulation of mcr-1-harbouring E. coli, the resistant subpopulation had an approximately 10-fold lower rate of killing due to polymyxin B treatment.
Conclusions: Manipulating initial dose intensity of polymyxin B was not able to overcome plasmid-mediated resistance due to mcr-1 in E. coli. This reinforces the need to develop new combinatorial strategies to combat these highly resistant Gram-negative bacteria.
Introduction
The polymyxins [polymyxin B and polymyxin E (colistin)] are critically important as the last line of defence against carbapenem-resistant Enterobacteriaceae.1 The recent worldwide incursion of Gram-negative bacteria harbouring the plasmid-mediated polymyxin resistance gene, mcr-1, is a cause for significant public health concern. Disconcertingly, the mcr-1-encoding plasmids display a proclivity for horizontal transfer among Escherichia coli, Klebsiella pneumoniae and Salmonella enterica, making an already challenging treatment landscape more desperate.2 Since its initial description in November 2015, polymyxin resistance in E. coli through plasmid-mediated mcr-1 has been reported throughout the world and may lead to the rise of pan-drug resistance.3,4 A more recent study characterized bacterial strains co-harbouring New Delhi metallo-β-lactamases, oxacillinases, K.pneumoniae carbapenemases and Verona integron-encoded metallo-β-lactamases together with MCR-1, creating an increased urgency to develop new therapeutic options to combat this new frontier of Gram-negative resistance.5–8
There is a paucity of data regarding the activity of polymyxins against mcr-1-harbouring organisms. Since many of the mcr-1 strains display polymyxin MICs only a single dilution higher than the EUCAST breakpoint of 2 mg/L (i.e. 4 mg/L), a strategy to combat plasmid-mediated polymyxin resistance may be to increase the polymyxin dose intensity very early in therapy. Therefore, we sought to define the pharmacodynamic impact of increasing initial polymyxin B intensity against an mcr-1-harbouring E. coli using a hollow fibre infection model (HFIM) and to develop a mechanism-based mathematical model. Based on results from previous studies, a ‘front-loading’ structure for polymyxin administration produces exposures conducive to maximal bacterial killing, while minimizing the toxicity.9,10 Thus, in the face of mcr-1, ‘front-loading’ polymyxin B was evaluated to improve the likelihood of bactericidal activity to combat plasmid-mediated polymyxin resistance.
Methods
Bacterial isolates, antibiotics and media
The gene coding for MCR-1 (GenBank accession number KP347127) containing Met2Val and Tyr179Met amino acid substitutions was ligated into the low-copy pGDP1 plasmid, a derivative of pBR322 (New England BioLabs) containing a constitutive β-lactamase Pbla promoter. The construct was confirmed by sequencing (central MOBIX facility, McMaster University) and transformed into E. coli BW25113 (WT strain and parent strain of the Keio collection). E. coli strain BW25113-mcr1a grown in CAMHB was utilized for all experiments. MICs were determined in duplicate for each strain via broth microdilution according to the CLSI.11
Static time–kill studies
Static time–kill studies were conducted in CAMHB for both the mcr-1-harbouring strain and the isogenic WT. The studies were conducted at 37°C over 24 h with samples taken at 0, 1, 2, 4, 8 and 24 h.12 We utilized a starting inoculum of ∼106 cfu/mL and exposed each strain to 0, 0.125, 0.25, 0.5, 1, 2, 4, 8, 16, 32 and 64 mg/L polymyxin B. Bactericidal activity was defined as a ≥99.9% reduction in bacterial counts.
HFIM
An HFIM was utilized to profile the total population in response to escalating polymyxin B regimens, as previously described.12 Similar to the time–kill studies, a starting inoculum of ∼106 cfu/mL was utilized. Population analysis profiles (PAPs) were performed to track resistant subpopulations over 240 h using samples obtained during the HFIM experiments by plating samples onto Mueller–Hinton agar (MHA) plates containing 0.5, 2, 8, 16 or 32 mg/L polymyxin B followed by incubation for 48 h at 37°C. Polymyxin B (Sigma–Aldrich, St Louis, MO, USA; lot number WXBB4470V) stock solutions were created every 48 h throughout the entire experiment. To validate the polymyxin B concentrations in the HFIM, samples were obtained over 48 h and then analysed using an LC single-quadrupole MS method, as described previously; the assay had good reproducibility (coefficient of variation ≤10%) and accuracy (observed concentrations were within 10% of target concentrations).13
Regimens were simulated for polymyxin B (t1/2 = 8 h, fraction unbound = 42%) based on a population pharmacokinetic study in 24 adult critically ill patients who received physician-selected, intravenous polymyxin B dosage regimens ranging from 0.45 to 3.38 mg/kg/day.14 Four regimens were simulated in the HFIM. The baseline ‘front-loading’ regimen (3.33 mg/kg for one dose followed by 1.43 mg/kg every 12 h starting 12 h later) was derived from a clinically achievable polymyxin B exposure from the population pharmacokinetic study where the highest fAUC0–24 was 53.5 mg·h/L. The initial (0 h) polymyxin B dose of the baseline ‘front-loading’ regimen was scaled ×2, ×4 and ×8 to produce a dose–response relationship that could be mathematically modelled. This approach was implemented to evaluate the initial dose intensity in relation to bacterial killing and properly inform the mathematical model. All polymyxin B regimens were designed to follow a ‘front-loading’ structure that introduced the largest exposure of polymyxin B within the first 24 h to maximize bacterial killing as follows:12
Regimen I (baseline regimen): polymyxin B ‘front-loading’ (3.33 mg/kg for 1 dose followed by 1.43 mg/kg every 12 h starting 12 h later, with an fAUC0–24 of 48.2 mg·h/L across the first day and an fAUC of 35.9 mg·h/L).
Regimen II (2 × baseline regimen): polymyxin B ‘front-loading’ (6.66 mg/kg for one dose followed by 1.43 mg/kg every 12 h starting 12 h later, with an fAUC0–24 of 84.9 mg·h/L across the first day and an fAUC of 35.9 mg·h/L).
Regimen III (4 × baseline regimen): polymyxin B ‘front-loading’ (13.3 mg/kg for one dose followed by 1.43 mg/kg every 12 h starting 12 h later, with an fAUC0–24 of 158 mg·h/L across the first day and an fAUC of 35.9 mg·h/L).
Regimen IV (8 × baseline regimen): polymyxin B ‘front-loading’ (26.6 mg/kg for one dose followed by 1.43 mg/kg every 12 h starting 12 h later, with an fAUC0–24 of 304 mg·h/L across the first day and an fAUC of 35.9 mg·h/L).
Regimens II, III and IV were proof-of-concept, supra-therapeutic doses designed to test the hypothesis that increased initial polymyxin B exposure would result in increased killing and suppression of resistance.15
Mechanism-based mathematical model
The data were modelled in S-ADAPT, utilizing S-ADAPT_TRAN for pre- and post-processing.16,17 Hollow fibre and time–kill data for the mcr-1-harbouring strain were modelled simultaneously in order to provide a more accurate portrayal of model parameters across study design. In addition, in order to provide a baseline comparison in parameter values, the WT strain time-kill data were modelled using the same structural model. The model structure consisted of susceptible, intermediately resistant and fully resistant subpopulations, each capable of independent proliferation, as previously described (Figure S1, available as Supplementary data at JAC Online).18 Bacterial replication was controlled using a logistic growth function that was determined by the total population and the estimated maximum population of bacteria present. The model was modified from the original model published by Bulitta et al.18 to permit different maximum bacterial population sizes between the static time–kills and HFIM, which are typically seen between these experimental designs. A second-order killing function was implemented, driven by the effective polymyxin B concentration as determined by displacement of magnesium and calcium cations at surface binding receptors. The displacement of these ions was modelled to be required for polymyxin to insert itself into and destabilize the membrane, inducing bacterial cell death.
Results
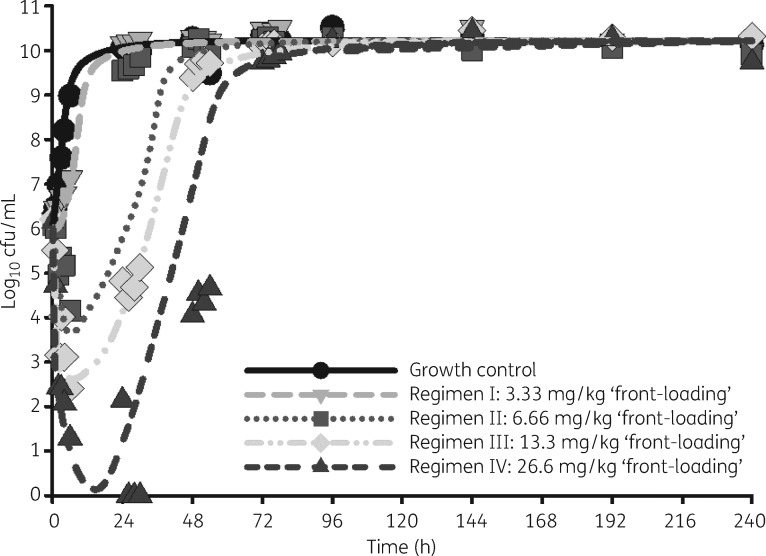
Observed polymyxin B concentrations in the HFIM from the 48 h pharmacokinetic validation study were found to be in good agreement with expected concentrations (R2 > 0.95) (Figure S2). The WT E. coli BW25113 (parent strain from the Keio collection) had a polymyxin B MIC <0.25 mg/L, whereas the isogenic E. coli BW25113-mcr1a strain had an MIC of 8 mg/L for both polymyxin B and colistin, but was expectedly susceptible to all other clinically relevant antibiotics. In the HFIM, as a response to treatment with polymyxin B, the baseline regimen I appeared to have a similar shape to the growth control but with a slight delay in growth rate. Regimens II, III and IV delayed regrowth by 24, 48 and 72 h, respectively. With regard to net reduction in bacterial counts, regimens II, III and IV resulted in maximal reductions of 1.91, 3.79 and 6.14 log10 cfu/mL, respectively. Only regimen IV caused undetectable bacterial counts, which were seen between 26 and 30 h (Figure 1).
Figure 1.
Observed (symbols) and model-fitted (lines) viable total counts for the growth control and each of the four treatment arms in the HFIM: regimen I, polymyxin B ‘front-loading’ (3.33 mg/kg for one dose followed by 1.43 mg/kg every 12 h starting 12 h later); regimen II, polymyxin B ‘front-loading’ (6.66 mg/kg for one dose followed by 1.43 mg/kg every 12 h starting 12 h later); regimen III, polymyxin B ‘front-loading’ (13.3 mg/kg for one dose followed by 1.43 mg/kg every 12 h starting 12 h later); and regimen IV, polymyxin B ‘front-loading’ (26.6 mg/kg for one dose followed by 1.43 mg/kg every 12 h starting 12 h later).
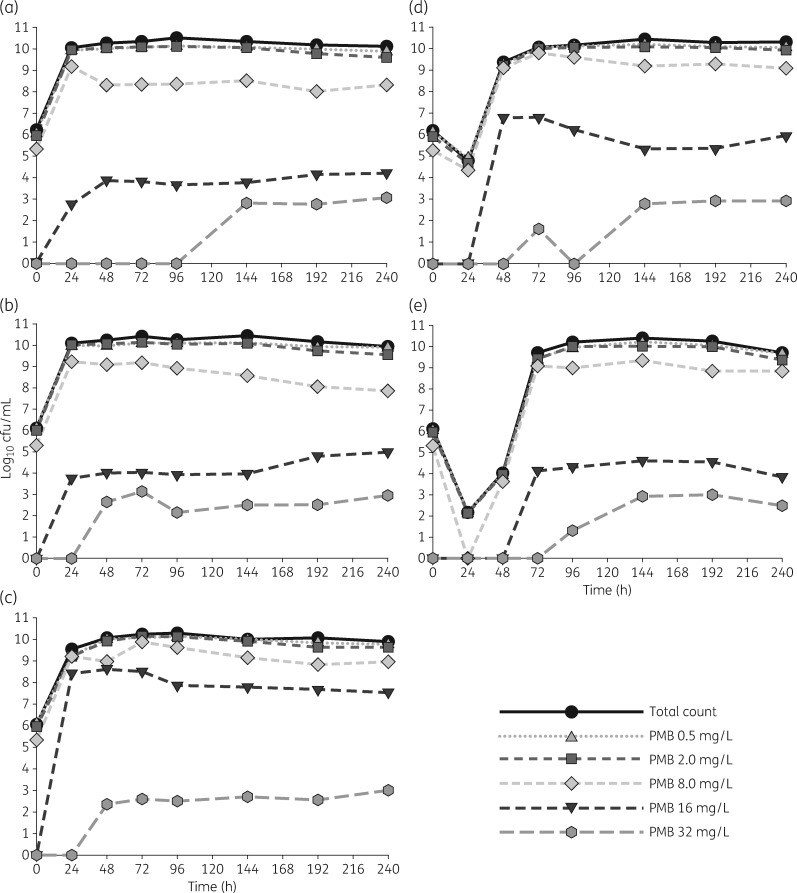
Baseline PAPs from the start of the HFIM (i.e. PAPs taken at 0 h) showed that the bacteria growing on MHA containing a polymyxin B concentration of 2 mg/L constituted 61.5 ± 11.7% of the total counts, meaning the majority of the bacteria were at or above the EUCAST breakpoint of 2 mg/L before the experiment started (Figure 2). Beyond this, subpopulations capable of growing on 8 mg/L polymyxin B were within approximately 2 log10 cfu/mL of total counts throughout the duration of the experiment and for each arm. Of additional interest, all arms, including the growth control, eventually developed resistant subpopulations capable of growing on polymyxin B PAPs of 32 mg/L ≥24 h after regrowth had already occurred.
Figure 2.
Real-time PAPs of resistance changes over 240 h for the growth control (a) and each of the four regimens: regimen I, polymyxin B ‘front-loading’ (3.33 mg/kg for one dose followed by 1.43 mg/kg every 12 h starting 12 h later) (b); regimen II, polymyxin B ‘front-loading’ (6.66 mg/kg for one dose followed by 1.43 mg/kg every 12 h starting 12 h later) (c); regimen III, polymyxin B ‘front-loading’ (13.3 mg/kg for one dose followed by 1.43 mg/kg every 12 h starting 12 h later) (d); and regimen IV, polymyxin B ‘front-loading’ (26.6 mg/kg for one dose followed by 1.43 mg/kg every 12 h starting 12 h later) (e). The total count for each 24 h sample is overlaid using a black continuous line with circles to provide a point of reference for each of the PAP concentrations. PMB, polymyxin B.
The differential pharmacodynamic activity of polymyxin B against the three bacterial subpopulations is well described by the variation in the second-order kill constants. The model determined the second-order kill constants of polymyxin B to be 0.373, 0.0455 and 0.0323 L/(mg·h) for the susceptible, intermediate and resistant subpopulations of the mcr-1-harbouring strain, respectively (Table 1). The near 10-fold difference that exists between susceptible and resistant subpopulations underscores the potential for heterogeneity in resistance conferred by mcr-1-harbouring bacteria, which may have stimulated colony formation on 32 mg/L polymyxin B PAP plates. This compares with the second-order kill constants of the polymyxin-susceptible WT, which were determined to be 1380, 61 and 1.1 L/(mg·h) for the susceptible, intermediate and resistant subpopulations, respectively. At a sufficiently high bacterial density, the killing effect of polymyxin could be completely inhibited (maximum inhibition of killing, >0.99); whereas high bacterial densities also could significantly inhibit the maximal growth rate (maximum inhibition of growth rate, >0.96). Graphs of the final fits of the time–kill data can be found in Figure S3.
Table 1.
Parameter estimates
|
E. coli BW25113-mcr1a |
E. coli BW25113a |
|||||
|---|---|---|---|---|---|---|
| Parameter | Symbol | Units | mean value | SE% | mean value | SE% |
| Polymyxin B parameters | ||||||
| shape parameter | Hilleff | – | 1.65 | 15.5 | 1.65b | – |
| fraction of cations displaced for 50% effective polymyxin B concentration | EC50 | – | 0.748 | 29.2 | 0.748b | – |
| cation dissociation | KD,CAc | μM | 200 | – | 200 | – |
| polymyxin B dissociation | KD,PMBc | μM | 0.0003 | – | 0.0003 | – |
| divalent cation concentration | CCAc | μM | 1.14 | – | 1.14 | – |
| Signal molecules | ||||||
| max growth inhibition | IMAX,Growth | – | 0.964 | 13.1 | 0.897 | 11.1 |
| max killing inhibition | IMAX,Kill | – | 0.998 | 5.90 | 0.998 | 7.14 |
| signal molecules for 50% inhibition of growth | Log(IC50,growth) | – | 8.88 | 1.41 | 8.61 | 1.78 |
| signal molecules for 50% inhibition of killing | Log(IC50,kill) | – | 6.76 | 3.53 | 8.06 | 2.17 |
| signal molecule decay rate constant | koutc | 1/h | 1 | – | 1 | – |
| initial signal molecules relative to inoculum | Log(Sigi) | – | −3.55 | 8.52 | −3.44 | 5.37 |
| Bacterial parameters | ||||||
| initial inoculum | Log(Inoc) | log cfu/mL | 6.01 | 0.504 | 6.01 | 0.0689 |
| fraction of inoculum with intermediate resistance | Log(MFI) | – | −2.91 | 5.24 | −4.49 | 1.70 |
| fraction of inoculum with high resistance | Log(MFR) | – | −6.53 | 2.83 | −7.18 | 0.111 |
| cell division rate constant | k21c | 1/h | 50 | – | 50 | – |
| half-life of growth at low bacterial density | ||||||
| susceptible subpopulation | MGTS | min | 39.8 | 2.36 | 45.4 | 8.85 |
| intermediate subpopulation | MGTI | min | 131 | 6.95 | 48.2 | 6.27 |
| resistant subpopulation | MGTR | min | 87.9 | 18.8 | 97.3 | 4.7 |
| max population, HFIM | Log(cfumax),HFIM | log cfu/mL | 10.2 | 0.456 | – | – |
| max population, time–kill | Log(cfumax),TK | log cfu/mL | 8.72 | 0.432 | 8.89 | 1.36 |
| polymyxin-driven kill constants | ||||||
| susceptible subpopulation | k2,S | L/(mg·h) | 0.373 | 13.7 | 1380 | 13.3 |
| intermediate subpopulation | k2,I | L/(mg·h) | 0.0455 | 25.3 | 61.0 | 12.7 |
| resistant subpopulation | k2,R | L/(mg·h) | 0.0323 | 13.7 | 1.1 | 34.8 |
| Residual errord | σ | log cfu/mL | 0.257 | 4.82 | 0.379 | 9.4 |
WT.
Fixed to values obtained from mcr-1 fitting to improve model stability of the fit. Because the strains are isogenic, the cation dissociation process was assumed to be similar.
Fixed parameter.
Additive residual error model implemented.
Discussion
Polymyxin resistance that is chromosomally mediated (e.g. mgrB and pmrAB) typically results from a number of transcriptomic and metabolomic changes in the regulatory pathways that alter the functional form of LPS.15,19 The most relevant of these changes produces a modified outer membrane surface charge of the bacteria, reducing polymyxin binding. In E. coli, polymyxin resistance can largely be attributed to changes in the pharmacological target itself through the addition of 4-amino-4-deoxy-l-arabinose (L-ara4N) or phosphoethanolamine (PEA) to the lipid A portion of LPS.20 It has also been shown that alterations in transporter structure or expression can result in reduced polymyxin susceptibility, specifically among the resistance–nodulation–cell division (RND) family.20 The mcr-1 gene codes for a PEA transferase, which works to reduce the overall binding affinity for the polymyxins through LPS modifications with PEA.4 Although alterations to LPS by chromosomally encoded pathways and MCR-1 can have the same functional consequences, there may be differences in gene expression between the two pathways that alter the pharmacodynamics of polymyxin therapy alone or in combination.
Since the first reported incidence of bacteria containing mcr-1, a number of polymyxin-resistant isolates containing the gene have been reported worldwide.6 The majority of clinical surveillance studies have reported mcr-1 strains with polymyxin MICs of 4 or 8 mg/L, near the EUCAST breakpoint of 2 mg/L.21–23 This provided the major impetus for the current study, which was to define the pharmacodynamic impact of increasing initial dose intensity of polymyxin B against mcr-1-harbouring E. coli. Our data indicate that although the polymyxin MIC may appear to be modestly elevated above the breakpoint, there are resistant subpopulations capable of overcoming much higher polymyxin B exposures than would be predicted by the nominal MIC.
Given the variable mechanisms by which LPS is modified to induce polymyxin resistance, there was a need to explore how mcr-1 plasmid-mediated resistance would alter the dynamics of killing and resistance in response to polymyxin exposure.4,19 Our findings suggest that, irrespective of the initial dose intensity, polymyxin B monotherapies will not be sufficient to eradicate mcr-1 E. coli. Interestingly, at the highest polymyxin B PAP concentration of 32 mg/L, there was an unexpected late growth of resistant subpopulations in both treatment and growth control arms, which arose after the total count reached its maximum. However, it does not appear that polymyxin B exposure will significantly alter polymyxin-resistant subpopulation growth patterns. With regard to the 32 mg/L PAPs, there is a more rapid time to bacterial growth following exposure to regimen A and regimen B compared with the unexposed growth control, which may make exposure to these polymyxin B regimens clinically significant. The subpopulations that grew on lower-concentration PAPs may have an impact on high-inoculum infections, where there will be a greater absolute number of resistant organisms. That is, the changes in resistance due to therapy may end up being different temporally between low-inoculum and high-inoculum infections. By comparison, resistance via chromosomal mutations in response to polymyxin therapy has typically been observed with much larger proliferations of polymyxin resistance, with the potential for post-exposure polymyxin B or colistin MICs to reach 128 mg/L.24 As discussed by Liu et al.,4 there is a significant risk of spread of mcr-1 to other Enterobacteriaceae. The stability of polymyxin resistance in mcr-1-harbouring E. coli, despite intensive drug exposure, suggests that polymyxin B may be used in combinations without concern for resistance amplification. With the increased reporting of mcr-1-harbouring bacteria that are also capable of producing carbapenemases, development of optimized combination therapies will soon be imperative. For bacteria containing mcr-1 and that have polymyxin MICs near the breakpoint, polymyxin B may have enough activity to produce a synergistic interaction if given strategically in combination to potentiate a clinically relevant response by another agent.24 In particular, the recent resurgence in the development of novel polymyxins as adjuvants may hold significant promise even in the face of plasmid-mediated polymyxin-resistant strains.25,26
This study is limited by the fact that a single MCR-1 strain was explored in the HFIM and there was no comparison with other polymyxin resistance mechanisms. Additionally, this study lacks confounding mechanisms of resistance to other classes of antibiotics, specifically carbapenemases, which would ordinarily indicate the use of polymyxin antibiotics. Future studies should better describe mcr-1-medated resistance in co-existence with carbapenemases, such as the metallo-β-lactamases (e.g. New Delhi metallo-β-lactamase, Verona integron-encoded metallo-β-lactamase or imipenemase). Despite this, the information gained is helpful in understanding the unique behaviours of a strain resistant to polymyxins via mcr-1. Additionally, we acknowledge that the high-intensity polymyxin B regimens utilized in the HFIM have not been studied in humans and are not necessarily indicative of an in vivo system. But, by using a proof-of-concept approach against an mcr-1-harbouring strain that is otherwise susceptible, we were able to study the specific pharmacodynamic impact of this new mechanism of resistance at an unprecedented resolution.
Overall, this study found that the dynamics of resistance in an E. coli strain harbouring mcr-1 may not be as pronounced as previously reported for chromosomally mediated polymyxin resistance. Further, polymyxin B monotherapy, even at high initial doses, is unlikely to provide sustained bacterial killing. Our findings support future studies that should be focused on the development of novel combinations, especially in the face of future, truly pan-resistant infections.
Supplementary Material
Acknowledgments
Funding
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number R01AI111990. N. M. S. is supported, in part, by the American Foundation for Pharmaceutical Education.
Transparency declarations
None to declare.
Disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Supplementary data
Supplementary data, including Figures S1 to S3, are available at JAC Online.
References
- 1. Collignon PC, Conly JM, Andremont A. et al. World Health Organization ranking of antimicrobials according to their importance in human medicine: a critical step for developing risk management strategies to control antimicrobial resistance from food animal production. Clin Infect Dis 2016; 63: 1087–93. [DOI] [PubMed] [Google Scholar]
- 2. Zeng KJ, Doi Y, Patil S. et al. Emergence of the plasmid-mediated mcr-1 gene in colistin-resistant Enterobacter aerogenes and Enterobacter cloacae. Antimicrob Agents Chemother 2016; 60: 3862–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McGann P, Snesrud E, Maybank R. et al. Escherichia coli harboring mcr-1 and blaCTX-M on a novel IncF plasmid: first report of mcr-1 in the United States. Antimicrob Agents Chemother 2016; 60: 4420–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu Y-Y, Wang Y, Walsh TR. et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 2016; 16: 161–8. [DOI] [PubMed] [Google Scholar]
- 5. Mediavilla JR, Patrawalla A, Chen L. et al. Colistin- and carbapenem-resistant Escherichia coli harboring mcr-1 and blaNDM-5, causing a complicated urinary tract infection in a patient from the United States. mBio 2016; 7: e01191-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Falgenhauer L, Waezsada SE, Yao Y. et al. Colistin resistance gene mcr-1 in extended-spectrum β-lactamase-producing and carbapenemase-producing Gram-negative bacteria in Germany. Lancet Infect Dis 2016; 16: 282–3. [DOI] [PubMed] [Google Scholar]
- 7. Poirel L, Kieffer N, Liassine N. et al. Plasmid-mediated carbapenem and colistin resistance in a clinical isolate of Escherichia coli. Lancet Infect Dis 2016; 16: 281. [DOI] [PubMed] [Google Scholar]
- 8. Mulvey MR, Mataseje LF, Robertson J. et al. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 2016; 16: 289–90. [DOI] [PubMed] [Google Scholar]
- 9. Tsuji BT, Landersdorfer CB, Lenhard JR. et al. Paradoxical effect of polymyxin B: high drug exposure amplifies resistance in Acinetobacter baumannii. Antimicrob Agents Chemother 2016; 60: 3913–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rao GG, Ly NS, Haas CE. et al. New dosing strategies for an old antibiotic: pharmacodynamics of front-loaded regimens of colistin at simulated pharmacokinetics in patients with kidney or liver disease. Antimicrob Agents Chemother 2014; 58: 1381–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-fifth Informational Supplement M100-S25. CLSI, Wayne, PA, USA, 2015. [Google Scholar]
- 12. Lenhard JR, Bulitta JB, Connell TD. et al. High-intensity meropenem combinations with polymyxin B: new strategies to overcome carbapenem resistance in Acinetobacter baumannii. J Antimicrob Chemother 2016; 72: 153–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cheah S-E, Bulitta JB, Li J. et al. Development and validation of a liquid chromatography-mass spectrometry assay for polymyxin B in bacterial growth media. J Pharm Biomed Anal 2014; 92: 177–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sandri AM, Landersdorfer CB, Jacob J. et al. Population pharmacokinetics of intravenous polymyxin B in critically ill patients: implications for selection of dosage regimens. Clin Infect Dis 2013; 57: 524–31. [DOI] [PubMed] [Google Scholar]
- 15. Cheah S-E, Johnson MD, Zhu Y. et al. Polymyxin resistance in Acinetobacter baumannii: genetic mutations and transcriptomic changes in response to clinically relevant dosage regimens. Sci Rep 2016; 6: 26233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bulitta JB, Bingolbali A, Shin BS. et al. Development of a new pre- and post-processing tool (SADAPT-TRAN) for nonlinear mixed-effects modeling in S-ADAPT. AAPS J 2011; 13: 201–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bauer RJ. S-ADAPT/MCPEM User's Guide Berkeley, CA, USA: Biomedical Simulation Resource, 2011. [Google Scholar]
- 18. Bulitta JB, Yang JC, Yohonn L. et al. Attenuation of colistin bactericidal activity by high inoculum of Pseudomonas aeruginosa characterized by a new mechanism-based population pharmacodynamic model. Antimicrob Agents Chemother 2010; 54: 2051–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mahamad Maifiah MH, Cheah SE, Johnson MD. et al. Global metabolic analyses identify key differences in metabolite levels between polymyxin-susceptible and polymyxin-resistant Acinetobacter baumannii. Sci Rep 2016; 6: 22287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Baron S, Hadjadj L, Rolain JM. et al. Molecular mechanisms of polymyxin resistance: knowns and unknowns. Int J Antimicrob Agents 2016; 48: 583–91. [DOI] [PubMed] [Google Scholar]
- 21. Castanheira M, Griffin MA, Deshpande LM. et al. Detection of mcr-1 among Escherichia coli clinical isolates collected worldwide as part of the SENTRY Antimicrobial Surveillance Program in 2014 and 2015. Antimicrob Agents Chemother 2016; 60: 5623–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Doumith M, Godbole G, Ashton P. et al. Detection of the plasmid-mediated mcr-1 gene conferring colistin resistance in human and food isolates of Salmonella enterica and Escherichia coli in England and Wales. J Antimicrob Chemother 2016; 71: 2300–5. [DOI] [PubMed] [Google Scholar]
- 23. Coetzee J, Corcoran C, Prentice E. et al. Emergence of plasmid-mediated colistin resistance (MCR-1) among Escherichia coli isolated from South African patients. S Afr Med J 2016; 106: 35–6. [DOI] [PubMed] [Google Scholar]
- 24. Bergen PJ, Bulman ZP, Landersdorfer CB. et al. Optimizing polymyxin combinations against resistant Gram-negative bacteria. Infect Dis Ther 2015; 4: 391–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wright GD. Antibiotic adjuvants: rescuing antibiotics from resistance. Trends Microbiol 2016; 24: 862–71. [DOI] [PubMed] [Google Scholar]
- 26. Hackel M, Lister T, Parr TR Jr. et al. In vitro activity of Spr741 against recent clinical isolates of Escherichia coli and Klebsiella pneumoniae In: Abstracts of ASM Microbe 2016, Boston, MA, USA. Abstract Number 500. American Society for Microbiology, Washington, DC, USA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.