Abstract
Background
fosA3 is the most commonly reported plasmid-mediated fosfomycin resistance gene among Enterobacteriaceae.
Objectives
To identify the origin of fosA3.
Methods
The chromosome of Kluyvera georgiana clinical strain YDC799 was fully sequenced with single-molecule real-time sequencing. Comparative genetic analysis was performed for K. georgiana YDC799, K. georgiana type strain ATCC 51603 and representative fosA3-carrying plasmids. fosA genes were cloned in Escherichia coli to confirm function.
Results
K. georgiana YDC799 harboured fosA (designated fosAKG) and blaCTX-M-8 on the chromosome. The genetic environments surrounding fosA3 and bounded by IS26 were nearly identical with the corresponding regions of K. georgiana YDC799 and ATCC 51603. The amino acid sequence of FosAKG from YDC799 and K. georgiana ATCC 51603 shared 99% and 94% identity with FosA3, respectively. Cloned FosAKG conferred fosfomycin resistance with an MIC of >1024 mg/L for E. coli.
Conclusions
The plasmid-mediated fosA3 gene was likely mobilized from the chromosome of K. georgiana by an IS26-mediated event.
Introduction
Fosfomycin exhibits a broad-spectrum activity against Gram-positive and -negative bacteria by covalent modification of the N-acetylglucosamine enolpyruvyl transferase (MurA) active site.1,2 FosA enzymes are Mn2+ and K+-dependent glutathione S-transferases that inactivate fosfomycin in some Gram-negative bacteria rendering them resistant to this agent.3 The fosA gene can be encoded on plasmids or chromosomes. The most commonly reported plasmid-mediated fosA is fosA3, which is widely distributed among ESBL-producing Escherichia coli in East Asia.4,5 However, the origin of fosA3 is unknown. Based on the sequence similarity of FosA3 with FosA encoded on the draft genome of Kluyvera georgiana ATCC 51603, we hypothesized that this species may serve as the reservoir of fosA3.
Methods
Bacterial strains
The following strains were used in this study: E. coli YD472, which carries fosA3;6K. georgiana YDC799, which was identified in an autopsy culture at the University of Pittsburgh Medical Center in 2017; and K. georgiana ATCC 51603, which was purchased from ATCC.
Susceptibility testing
Fosfomycin MICs were determined by the agar dilution method using Mueller–Hinton agar supplemented with and without 25 mg/L glucose-6-phosphate according to CLSI guidelines.7E. coli ATCC 25922 was used as the quality control strain. Inhibition of FosA activity by sodium phosphonoformate, an inhibitor of FosA,8,9 was examined using the disc diffusion method.10
PCR and cloning of fosA
PCR was performed in E. coli YD472, K. georgiana YDC799 and K. georgiana ATCC 51603 using the following set of primers: FosA3_BamHI_F, GCGGATCCATGCTGCAGGGATTGAATC and FosA3_HindIII_R, GCAAGCTTAAGCTGAACTAACCCGTCA. To confirm the role of the promoter region in the expression of FosA, we used the following forward primer to generate PCR products of fosA with their promoter region: FosA3pr_BamHI_F, GCGGATCCATTTTATCGGGCGTATGAA. The PCR products were digested with restriction enzymes BamHI and HindIII, and ligated with cloning vector pBC-SK (Thermo Scientific, Waltham, MA, USA), which were then introduced into E. coli TOP10 (Thermo Scientific). Transformants were identified by growth on lysogeny broth agar plates containing 30 mg/L chloramphenicol and 20 mg/L fosfomycin, and the sequences were confirmed by Sanger sequencing.
WGS
K. georgiana YDC799 was subjected to WGS using single-molecule real-time (SMRT) sequencing performed on an RS II instrument (Pacific Biosciences, Menlo Park, CA, USA) at the Yale Center for Genome Analysis.11 A total of 56948 sub-reads with an average read length of 10258 bp were obtained. De novo assembly of the reads using the hierarchical genome assembly process (HGAP 3.0) available in the SMRT Analysis v2.3 software generated one contig with 94× coverage. This contig was then circularized and polished using the resequencing protocol in SMRT Analysis with two passes to reach the final consensus accuracy of 100%. The sequence was annotated through the Rapid Annotations using Subsystem Technology (RAST) Server.12 The annotated sequence of YDC799 was compared with that of K. georgiana ATCC 51603 (LXEU01000090.1) as well as those of representative fosA3-carrying plasmids harboured by E. coli YD472 (KR078259.1), E. coli 5CRE51 (CP021177.1) and E. coli 21TF (JQ343849.1) by using NCBI BLAST analysis. E. coli 5CRE51 was identified in Taiwan and harbours fosA3 and blaNDM-9 on a single plasmid. E. coli 21TF was identified in Korea and harbours an IncN-type plasmid carrying fosA3 and blaCTX-M-14.13 Antimicrobial resistance genes in YDC799 were identified by ResFinder.14
Results
Antimicrobial susceptibility
E. coli YD472 carrying fosA3 exhibited a high fosfomycin MIC of >1024 mg/L. K. georgiana YDC799 and ATCC 51603 had fosfomycin MICs of 32 and 0.5 mg/L in the presence of glucose-6-phosphate, respectively (Table 1).7 However, E. coli transformants harbouring fosA3 and fosAKG (fosA from both K. georgiana YDC799 and ATCC 51603), either with or without native promoters, were all highly resistant to fosfomycin with an MIC of >1024 mg/L. The expansion of the growth inhibition zone around the fosfomycin discs for K. georgiana YDC799 and E. coli transformants carrying fosAKG upon the addition of sodium phosphonoformate corroborated FosA activity (Table 1).8,9
Table 1.
Characteristics of the E. coli and K. georgiana strains used in this study
| Strains | Characteristics | MIC with G6P (mg/L) | MIC without G6P (mg/L) | Zone diameter (mm) | Zone diameter with PPF (mm) |
|---|---|---|---|---|---|
| E. coli YD472 | contains fosA3 on plasmid | >1024 | >1024 | 6 | 6 |
| K. georgiana YDC799 | contains fosAKG on the chromosome | 32 | >1024 | 18 | 24 |
| K. georgiana ATCC 51603 | contains fosAKG on the chromosome | 0.5 | 128 | 32 | 34 |
| E. coli TOP10 (pFosA3) | carries fosA3 from YD472 on pBC-SK | >1024 | >1024 | 6 | 18 |
| E. coli TOP10 (pFosAKG-YDC799) | carries fosAKG from YDC799 on pBC-SK | >1024 | >1024 | 6 | 18 |
| E. coli TOP10 (pFosAKG-ATCC 51603) | carries fosAKG from ATCC 51603 on pBC-SK | >1024 | >1024 | 6 | 22 |
| E. coli TOP10 (pBC-SK) | vector control | 1 | 128 | 40 | 40 |
| E. coli TOP10 | 1 | 64 | 38 | 38 |
MICs were determined by the agar dilution method with and without 25 mg/L glucose-6-phosphate (G6P). For disc testing, 1 mg of sodium phosphonoformate (PPF) was added to fosfomycin discs.
Sequence of K. georgiana FosA (FosAKG)
We ran a BLASTn search of fosA3 against the assembled genome sequences of K. georgiana YDC799 (this study; CP022114.1) and K. georgiana ATCC 51603 (NZ_LXEU00000000.1). As a result, we identified sequences with high similarity to fosA3 in K. georgiana YDC799 and K. georgiana ATCC 51603, which were named fosAKG-YDC799 and fosAKG-ATCC 51603, respectively. The deduced amino acid sequences of FosA from K. georgiana YDC799 (FosAKG-YDC799) and K. georgiana ATCC 51603 (FosAKG-ATCC 51603) shared 99% (136/138 amino acids) and 94% (130/138 amino acids) identity with FosA3, respectively (Figure 1). The next closest FosA sequence available in GenBank was that of Kluyvera ascorbata WCH1410 with an identity of 93% (129/138 amino acids). Therefore, FosA3 is most closely related to FosAKG from K. georgiana among known chromosomally encoded FosA. All active site residues were conserved (H7, T9, W46, C48, S50, H67, K93, N95, S97, E98, G99, S101, Y103, E113 and R122) across the FosA3 and FosAKG proteins (Figure 1).
Figure 1.
Amino acid alignment of FosA proteins in E. coli YD472, K. georgiana YDC799 and K. georgiana ATCC 51603. FosAKG-ATCC 51603, FosA from K. georgiana ATCC 51603; FosAKG-YDC799, FosA from K. georgiana YDC799.
Location and genetic environments of fosAKG in K. georgiana YDC799 and ATCC 51603
Since the only publicly available K. georgiana genome sequence (strain ATCC 51603) consists of 163 contigs, we performed SMRT sequencing of K. georgiana YDC799 to determine the location and genetic environment of fosAKG. SMRT sequencing with a single cell resulted in a closed chromosome after de novo assembly. There was no evidence of extra-chromosomal genetic elements, including plasmids. The YDC799 chromosome was ∼5.0 Mbp in size with a GC content of 55.0% and encoded 7557 genes, including hypothetical ones.
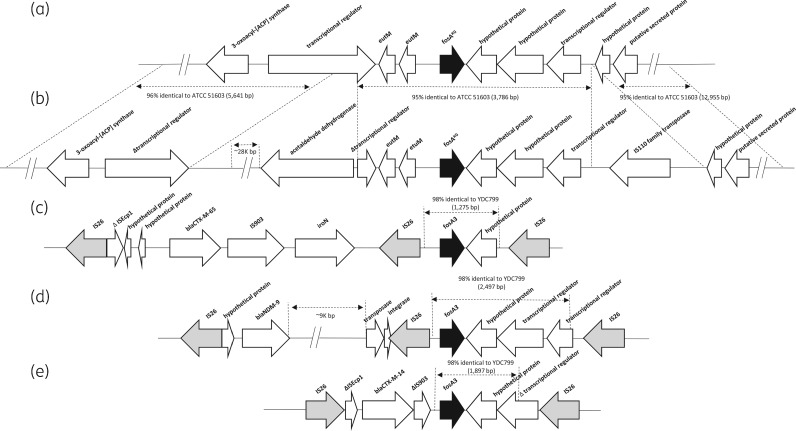
The 3.8 kb region containing fosAKG in K. georgiana YDC799 and ATCC 51603 shared 95% nucleotide identity and consisted of a truncated transcriptional regulator gene, eutM, etuM, fosAKG, two additional ORFs and a second transcription regulator gene (Figure 2). Although the upstream transcriptional regulator gene in K. georgiana YDC799 was truncated, it shared 95% identity with that of K. georgiana ATCC 51603. The chromosomal sequences of the two K. georgiana strains diverged beyond this 3.8 kb region containing fosAKG. In particular, an IS of the IS110 family, members of which share a highly conserved tetrad motif with reverse transcriptase,15 was located downstream of fosAKG in YDC799. In addition, the regions further upstream and downstream of fosAKG in YDC799 shared 96% (5641 bp) and 95% (12 955 bp) identity with those of K. georgiana ATCC 51603 at the nucleotide level, respectively.
Figure 2.
Genetic environment of FosA and the neighbouring regions in K. georgiana ATCC 51603 (a), K. georgiana YDC799 (b), E. coli YD472 (c), E. coli 5CRE51 (d) and E. coli 21TF (e).
The region downstream of fosAKG of the K. georgiana YDC799 chromosome shared 98% (1275 bp), 98% (2497 bp) and 98% (1897 bp) nucleotide identity with the corresponding regions downstream of plasmid-mediated fosA3 in E. coli YD472, E. coli 5CRE51 and E. coli 21TF, respectively (Figure 2). These high levels of identity were only interrupted by ISs (two tandem copies of IS26 for E. coli YDC472 and 5CRE51, and IS26 and a truncated IS903 in the case of E. coli 21TF). These data strongly suggest that K. georgiana served as the origin of fosA3 and that IS26 played a pivotal role in the acquisition of the plasmid-mediated fosA3 gene.13 Furthermore, truncation of the fosA3-containing regions by IS26 and other ISs at various locations suggests that mobilization of fosA3 may represent multiple independent events rather than a single event in the past.
Discussion
There is an increasing interest in using fosfomycin to treat infections due to MDR Gram-negative pathogens, in particular ESBL-producing E. coli. In this context, the emergence and dissemination of plasmid-mediated fosA in E. coli and other Enterobacteriaceae represents a major threat against its clinical utility. The origin of some plasmid-mediated fosA genes has been identified. For instance, fosATn2921 originated from the chromosome of Enterobacter cloacae, and fosA5 and fosA6 originated from the chromosome of Klebsiella pneumoniae.10,16 However, the origin of fosA3 has not been identified to date. Here, we provide evidence that fosA3 was likely mobilized from the chromosome of K. georgiana. It has previously been postulated that fosA3 may have originated from K. pneumoniae due to moderate sequence identity (∼78%) of the fosA genes themselves as well as downstream of fosA3 and chromosomal fosA of K. pneumoniae 342.4,13 However, the higher identity of fosAKG with fosA3 including their genetic environments (∼98%) suggests that the chromosome of K. georgiana is the more likely origin of fosA3.
While both K. georgiana strains used in this study were susceptible to fosfomycin, there was a significant difference in their MICs (32 mg/L for YDC799 versus 0.5 mg/L for ATCC 51603). The fosfomycin MICs for the transformants harbouring fosAKG-YDC799 and fosAKG-ATCC 51603 with the native promoter regions were over 1024 mg/L. While the reason for the discordance is unclear, we assume that fosAKG-ATCC 51603 may be poorly expressed in ATCC 51603, a hypothesis that is also supported by the lack of inhibition of FosA activity by phosphonoformate.
Kluyvera is a genus of the Enterobacteriaceae family and members of this genus are generally considered to be non-pathogenic although there are rare reports of them as human pathogens.17 Of interest, the β-lactamase gene blaKLUG-1 located on the chromosome of K. georgiana, including YDC799, has been identified as the origin of blaCTX-M-8, an ESBL gene frequently identified in E. coli in South America.18 Our findings therefore underscore the importance of Kluyvera spp. as a reservoir of resistance genes that can be transferred to Gram-negative pathogens and render them resistant to clinically important antimicrobial agents.
In summary, our study identified the origin of plasmid-mediated fosA3 as the chromosomal fosAKG gene in K. georgiana. While fosA3 is currently prevalent in East Asia, this finding suggests that its mobilization may occur elsewhere independently as fosfomycin use increases globally.
Nucleotide sequence accession number
The finished chromosome sequence of K. georgiana YDC799 was submitted to GenBank under accession number CP022114.1.
Funding
This study was supported by a research grant from the National Institutes of Health (grant number R21AI123747). The effort of Y. D. was also supported by R01AI104895.
Transparency declarations
Y. D. has served on advisory boards for Meiji, Allergan, Curetis, The Medicines Company and Roche. All other authors: none to declare.
References
- 1. Hendlin D, Stapley EO, Jackson M. et al. Phosphonomycin, a new antibiotic produced by strains of Streptomyces. Science 1969; 166: 122–3. [DOI] [PubMed] [Google Scholar]
- 2. Eschenburg S, Priestman M, Schonbrunn E.. Evidence that the fosfomycin target Cys115 in UDP-N-acetylglucosamine enolpyruvyl transferase (MurA) is essential for product release. J Biol Chem 2005; 280: 3757–63. [DOI] [PubMed] [Google Scholar]
- 3. Castaneda GA, Blazquez J, Rodriguez-Rojas A.. Molecular mechanisms and clinical impact of acquired and intrinsic fosfomycin resistance. Antibiotics 2013; 2: 217–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wachino J, Yamane K, Suzuki S. et al. Prevalence of fosfomycin resistance among CTX-M-producing Escherichia coli clinical isolates in Japan and identification of novel plasmid-mediated fosfomycin-modifying enzymes. Antimicrob Agents Chemother 2010; 54: 3061–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cao XL, Shen H, Xu YY. et al. High prevalence of fosfomycin resistance gene fosA3 in blaCTX-M-harbouring Escherichia coli from urine in a Chinese tertiary hospital during 2010-2014. Epidemiol Infect 2017; 145: 818–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Alrowais H, McElheny CL, Spychala CN. et al. Fosfomycin resistance in Escherichia coli, Pennsylvania, USA. Emerg Infect Dis 2015; 21: 2045–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Seventh Informational Supplement M100-S27. CLSI, Wayne, PA, USA, 2017. [Google Scholar]
- 8. Rigsby RE, Rife CL, Fillgrove KL. et al. Phosphonoformate: a minimal transition state analogue inhibitor of the fosfomycin resistance protein, FosA. Biochemistry 2004; 43: 13666–73. [DOI] [PubMed] [Google Scholar]
- 9. Nakamura G, Wachino J, Sato N. et al. Practical agar-based disk potentiation test for detection of fosfomycin-nonsusceptible Escherichia coli clinical isolates producing glutathione S-transferases. J Clin Microbiol 2014; 52: 3175–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Guo Q, Tomich AD, McElheny CL. et al. Glutathione-S-transferase FosA6 of Klebsiella pneumoniae origin conferring fosfomycin resistance in ESBL-producing Escherichia coli. J Antimicrob Chemother 2016; 71: 2460–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li JJ, Lee CS, Sheng JF. et al. Complete sequence of a conjugative incn plasmid harboring blaKPC-2, blaSHV-12, and qnrS1 from an Escherichia coli sequence type 648 strain. Antimicrob Agents Chemother 2014; 58: 6974–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Aziz RK, Bartels D, Best AA. et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 2008; 9: 75.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee SY, Park YJ, Yu JK. et al. Prevalence of acquired fosfomycin resistance among extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae clinical isolates in Korea and IS26-composite transposon surrounding fosA3. J Antimicrob Chemother 2012; 67: 2843–7. [DOI] [PubMed] [Google Scholar]
- 14. Zankari E, Hasman H, Cosentino S. et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 2012; 67: 2640–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mahillon J, Chandler M.. Insertion sequences. Microbiol Mol Biol Rev 1998; 62: 725–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ma Y, Xu X, Guo Q. et al. Characterization of fosA5, a new plasmid-mediated fosfomycin resistance gene in Escherichia coli. Lett Appl Microbiol 2015; 60: 259–64. [DOI] [PubMed] [Google Scholar]
- 17. Sarria JC, Vidal AM, Kimbrough RC 3rd. Infections caused by Kluyvera species in humans. Clin Infect Dis 2001; 33: E69–74. [DOI] [PubMed] [Google Scholar]
- 18. Poirel L, Kampfer P, Nordmann P.. Chromosome-encoded Ambler class A β-lactamase of Kluyvera georgiana, a probable progenitor of a subgroup of CTX-M extended-spectrum β-lactamases. Antimicrob Agents Chemother 2002; 46: 4038–40. [DOI] [PMC free article] [PubMed] [Google Scholar]