Abstract
Background
Antimicrobial-resistant Neisseria gonorrhoeae is a major threat to public health. No studies to date have examined the genomic epidemiology of gonorrhoea in the Western Pacific Region, where the incidence of gonorrhoea is particularly high.
Methods
A population-level study of N. gonorrhoeae in New Zealand (October 2014 to May 2015). Comprehensive susceptibility testing and WGS data were obtained for 398 isolates. Relatedness was inferred using phylogenetic trees, and pairwise core SNPs. Mutations and genes known to be associated with resistance were identified, and correlated with phenotype.
Results
Eleven clusters were identified. In six of these clusters, >25% of isolates were from females, while in eight of them, >15% of isolates were from females. Drug resistance was common; 98%, 32% and 68% of isolates were non-susceptible to penicillin, ciprofloxacin and tetracycline, respectively. Elevated MICs to extended-spectrum cephalosporins (ESCs) were observed in 3.5% of isolates (cefixime MICs ≥ 0.12 mg/L, ceftriaxone MICs ≥ 0.06 mg/L). Only nine isolates had penA XXXIV genotypes, three of which had decreased susceptibility to ESCs (MIC = 0.12 mg/L). Azithromycin non-susceptibility was identified in 43 isolates (10.8%); two of these isolates had 23S mutations (C2611T, 4/4 alleles), while all had mutations in mtrR or its promoter.
Conclusions
The high proportion of females in clusters suggests transmission is not exclusively among MSM in New Zealand; re-assessment of risk factors for transmission may be warranted in this context. As elevated MICs of ESCs and/or azithromycin were found in closely related strains, targeted public health interventions to halt transmission are urgently needed.
Introduction
Gonorrhoea is the second most common sexually transmitted infection. In 2012, approximately 78 million incident cases were diagnosed worldwide among adolescents and adults aged 15–49 years.1 Untreated, gonorrhoea can cause pelvic inflammatory disease, ectopic pregnancy and/or infertility in women, and urethritis and infertility in men.2 As there are strains of Neisseria gonorrhoeae that are resistant to nearly all available antimicrobial agents, untreatable gonorrhoea represents an imminent global health threat. This was recently highlighted by a WHO report;3 as such, rapid diagnosis and appropriate antibiotic therapy are essential.
Over the past decade, diagnosis of gonorrhoea has moved from culture-based methods to rapid nucleic acid amplification tests (NAATs)4. While NAATs offer increased diagnostic sensitivity compared with bacterial culture,4 current high-throughput commercial platforms do not enable molecular epidemiological investigation, or prediction of phenotypic drug resistance. Accordingly, population-level studies using bacterial cultures are crucial to understanding the local molecular epidemiology of gonorrhoea, monitoring of resistance patterns and informing treatment guidelines.
Several recent studies have used WGS to provide insight into the epidemiology of gonorrhoea and mechanisms of drug resistance.5–8 To date, however, no WGS-based studies have been published from the WHO Western Pacific Region (WPR). This region had the highest reported number of incident cases of gonorrhoea in the world in 2012.9 As resistance to many drug classes [e.g. penicillins, quinolones, and more recently, extended-spectrum cephalosporins (ESCs)] appears to have emerged in this region before disseminating globally,10 a current assessment of local strain diversity and underlying resistance mutations is warranted. Herein, we help address this gap by providing a detailed genomic investigation of N. gonorrhoeae in New Zealand, an island nation in the South West Pacific.
Materials and methods
Ethics
This work was approved by the Health and Disability Ethics Committee of New Zealand. Data were collected as part of public health surveillance, and individual patient consent was not required. All identifiable patient-level data were anonymized.
Study design
Between 27 October 2014 and 29 May 2015, a population-level survey of antimicrobial susceptibility of N. gonorrhoeae was conducted in New Zealand. All N. gonorrhoeae isolates cultured and corresponding clinical data (age, sex, anatomical site of isolation) were provided by diagnostic laboratories.
Phenotypic susceptibility
Antimicrobial susceptibility testing (AST) to azithromycin, cefixime, ceftriaxone, ciprofloxacin, ertapenem, gentamicin, penicillin, spectinomycin and tetracycline was performed at the Antimicrobial Reference Laboratory at the Institute for Environmental Science and Research (ESR) in New Zealand, using the agar dilution method according to CLSI guidelines.11 MICs were interpreted according to CLSI breakpoints11 where available; otherwise, EUCAST breakpoints12 were used. Both the N. gonorrhoeae drug-susceptible strain ATCC 49226 and strain NCTC 13479 (WHO K), which has decreased susceptibility to ceftriaxone, were used for quality control.
MDR has previously been defined as resistance to at least one category I drug (ESCs or spectinomycin) plus two category II drugs (including penicillins, fluoroquinolones, azithromycin, aminoglycosides and carbapenems),10 whereas XDR has been defined as resistance to two or more category I drugs, and at least three category II drugs. We employed these definitions, but excluded aminoglycosides and carbapenems, as no MIC thresholds currently exist for N. gonorrhoeae for these drugs. We also defined as ‘pre-MDR’ isolates that could become MDR should resistance develop to a single additional class of drug.
DNA extraction and WGS
The first sample collected per patient typically underwent AST and was subsequently forwarded for sequencing [in two cases (0.5%), a patient’s second sample was submitted]. DNA extraction and WGS were performed at the Microbiological Diagnostic Unit Public Health Laboratory, The University of Melbourne. WGS was performed using Illumina NextSeq (error rate < 1%13) with 150 bp paired-end reads. Reads are available on the National Center for Biotechnology Information (NCBI) Sequence Read Archive (BioProject PRJNA394216; Table S1, available as Supplementary data at JAC Online).
Bioinformatics
Sequences were analysed using the Nullarbor pipeline (T. Seemann, available at: https://github.com/tseemann/nullarbor), described in depth in the Supplementary Methods. In brief, WGS data quality was assessed, reads were aligned to the most closely related reference genome (NCCP11945, NCBI Accession CP001050; Supplementary Results and Table S2) and SNPs were identified. De novo assembly was performed using SPAdes (v.3.9.0).14 See the Supplementary Results and Table S3 for detailed assembly statistics.
Population structure and epidemiology
Recombination was detected using Gubbins (v.2.2.0–1),15 and population structure was investigated using hierarchical Bayesian analysis (v.6.0).16 A recombination-adjusted maximum likelihood tree was produced with IQ-TREE (v.1.4.3),17 with the lowest Bayesian Information Criterion used to select the model of nucleotide substitution. Confidence in phylogenies was assessed via the ultra-fast bootstrap approximation with 10 000 iterations.18
To provide phylogeographic context for New Zealand isolates, we included available international N. gonorrhoeae sequences from NCBI. The same quality control exclusion criteria were applied as previously (Table S4); a total of 2442 international sequences were included, representing the UK, Canada, Chile and the USA.5–8,19,20
Conventional genotyping
N. gonorrhoeae multi-antigen sequence typing (NG-MAST) was performed using a previously developed in silico typing tool.21 Novel NG-MAST types were inferred as described in the Supplementary Methods.
Antimicrobial resistance
Chromosomal mutations were investigated by re-aligning reads to a drug-susceptible reference (FA1090, NCBI accession number NC_002946.2),22 and calling variants with respect to this reference. Assembled contigs were screened for acquired antibiotic resistance genes, as well as these mutations (Supplementary Methods).
Statistics
All statistical analyses were performed in Stata (v.14.2, College Station, TX, USA: StataCorp). Non-normal distributions were compared using the Mann–Whitney test. Geometric means and 95% CIs were calculated using the ‘ameans’ command. Multivariate logistic regression was used to evaluate the association between resistance mutations and phenotype. See the Supplementary Methods for further detail.
Results
There were 425 patients with at least one positive culture for N. gonorrhoeae during the study period. Although ten patients had two positive cultures, only three of these pairs were from different anatomical sites, all of which had the same susceptibility profile. Overall, 295/425 (69%) of patients were from the Auckland region, the largest urban centre in New Zealand. Given that Auckland isolates were over-represented in this dataset, these were randomly down-sampled, such that a total of 400 isolates (from 400 unique patients across four major geographical regions) were selected for sequencing (Figure S1). High-quality WGS data were obtained for 398 (99.5%) of these isolates (see Supplementary Results). Most isolates were from males (81%); 64.6% of these were urethral, 16.8% were anorectal, and 13.7% were penile. Only 2.8% of isolates were throat or pharyngeal. Among females, 90% of isolates were from cervical or vaginal specimens.
Population structure and epidemiology
Predicted recombination events are shown in Figure S2. Prior to adjusting for recombination, 31 492 core SNP positions were identified. Adjustment for recombination reduced this to 4529 core SNP positions, with a median pairwise core SNP distance of 581 between isolates (IQR 473–632).
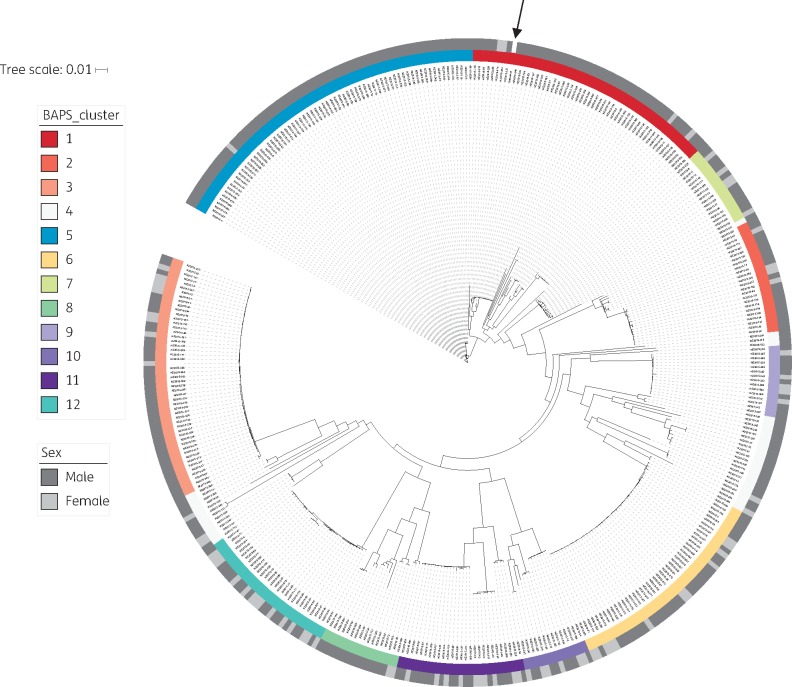
The recombination-adjusted maximum likelihood tree for New Zealand is shown in Figure 1, with bootstrap support values provided in Figure S3. Eleven major Bayesian Analysis of Population Structure (BAPS) clusters were identified, which were largely in agreement with the inferred phylogeny. An additional cluster (BAPS-4) artificially binned the most divergent isolates together; this is a known limitation of BAPS and does not represent a biologically meaningful grouping.16 As such, this cluster was excluded from further analysis.
Figure 1.
Population structure of Neisseria gonorrhoeae in New Zealand, 2014–15. High-quality WGS data were obtained for 398 of 400 New Zealand isolates (99.5%). Genome coverage to the NCCP11945 reference was 97% (SD 1.1%), with median depth of coverage of 96× (IQR 87–108). Recombination was identified and masked using Gubbins.15 A subsequent phylogenetic tree was built based on 4529 total core SNP positions (with 3291 informative sites) using the maximum likelihood method via IQ-TREE.17 A General Time Reversible model of nucleotide substitution with four gamma categories was selected based on the lowest Bayesian Information Criterion, with an ascertainment bias correction to account for only SNP sites included in the input. Ten-thousand ultra-fast bootstrap replicates were performed to assess uncertainty in tree topology; for clarity, these are shown in Figure S3. Bayesian Analysis of Population Structure (BAPS) was used to identify clusters; only the first level of clustering is shown. Note that in BAPS-4 the most divergent isolates are grouped together; this does not represent a biologically meaningful cluster. The sex of the patients is also shown. The reference genome is indicated with an arrow.
All clusters included isolates from both sexes, with females representing > 15% of isolates in eight of the BAPS groups, and >25% of isolates in six of the BAPS groups (Table S5). The median pairwise core SNPs within BAPS clusters was 13 (IQR 3–84), compared with 597 (IQR 499–633) pairwise core SNPs between isolates from different clusters (P < 0.0001). BAPS-3, -6 and -9 in particular were highly clonal, with median pairwise core SNP distance between isolates of 5 (IQR 3–6) within BAPS-3; 2 (IQR 1–5) in BAPS-6; and 3 (IQR 2–5) in BAPS-9. Though inconclusive without epidemiological data on contact, such closely related isolates may belong to the same transmission network; this is consistent with the transmission nomogram proposed by De Silva et al.20 which suggests that two N. gonorrhoeae isolates involved in transmission may have up to 11 SNPs between them if they are sampled 6 months apart. Overall, 84% of New Zealand isolates had ≥ 1 other sample within this distance based on the core genome, while 31% of isolates had at least one other isolate within 0 SNPs.
In the context of global N. gonorrhoeae strains (Figure S4), New Zealand isolates were often interspersed with those from Canada, the UK, Chile and/or the USA. This supports the idea of global dissemination of N. gonorrhoeae strains, in accordance with the WGS-based study from Ezewudo et al.19
Conventional genotyping
Fifty-seven known NG-MAST types were detected in New Zealand. One-hundred and thirty-five isolates (33.9%) did not have a known NG-MAST type; 59 new types were identified, with the most common of these accounting for 14 isolates. Types 4186 (11.3%), 2400 (10.8%) and 9368 (6.8%) were most frequently represented, overall. NG-MAST types were distributed across the tree and were not necessarily predictive of relatedness as identified by WGS (Figure S5).
NG-MAST was also determined for global N. gonorrhoeae strains (Table S6). Compared to New Zealand, a higher proportion of these had known NG-MAST types (66.1% versus 91.0%, respectively; P < 0.0005), reinforcing the need to better catalogue and characterize strains from the WPR.
Antimicrobial resistance
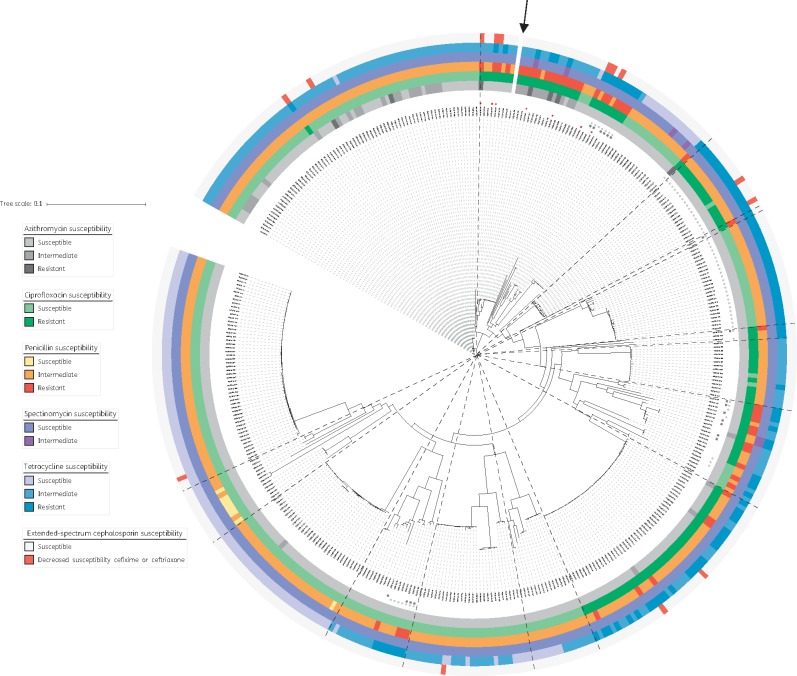
Phenotypic AST data were available for all New Zealand isolates (Figure 2). No MDR or XDR was identified. However, seven isolates had either decreased susceptibility to ESCs or spectinomycin (category I drugs), plus resistance to two or more category II drugs – marking them as ‘pre-MDR’ (Figure 2). One of these isolates had the highest ertapenem MIC detected (0.12 mg/L; Table 1). While six isolates had high gentamicin MICs of 16 mg/L (Table 1), none had resistance to category I drugs, and only two had resistance to a second category II drug (ciprofloxacin).
Figure 2.
Antimicrobial resistance among Neisseria gonorrhoeae isolates in New Zealand, 2014–15. Maximum likelihood tree of all 398 New Zealand isolates, after adjusting for recombination with Gubbins.15 Classifications based on MIC thresholds from reference 11, or if unavailable from reference 12. For azithromycin, resistance corresponds to MIC >0.5 mg/L, intermediate susceptibility corresponds to MIC of 0.5 mg/L and susceptible corresponds to MIC ≤0.25 mg/L. For ciprofloxacin, resistance corresponds to MIC ≥1 mg/L, intermediate is 0.12–0.5 mg/L and susceptible is ≤ 0.06 mg/L. For penicillin, resistance corresponds to MIC ≥ 2 mg/L, intermediate susceptibility is 0.12–1 mg/L and susceptible is ≤ 0.06 mg/L. For spectinomycin, resistance corresponds to MIC ≥128 mg/L, intermediate susceptibility is 64 mg/L and susceptible is ≤ 32 mg/L. For tetracycline, resistance corresponds to MIC ≥2 mg/L, intermediate susceptibility is 0.5–1 mg/L and susceptible is ≤ 0.25 mg/L. For the extended-spectrum cephalosporins, cefixime and ceftriaxone: decreased susceptibility corresponds to MICs of ≥ 0.12 mg/L and ≥0.06 mg/L, respectively. Isolates with the blaTEM-1 gene, associated with resistance to penicillins, are indicated with black squares, while isolates with the tetM gene are indicated with light grey circles. Isolates classified as pre-MDR are indicated in the inner ring with red stars. The BAPS groups identified (as shown in Figure 1) have been overlaid using dashed lines. The reference genome is indicated with an arrow.
Table 1.
MICs of drugs tested, by class
| MIC (mg/L) |
|||||
|---|---|---|---|---|---|
| Drug class | Drug | median | IQR | minimum | maximum |
| Aminocyclitol | spectinomycin | 32 | 16–32 | 4 | 64 |
| Aminoglycosides | gentamicina,b | 8 | 4–8 | 1 | 16c |
| Carbapenems | ertapenema,d | 0.016 | 0.008–0.016 | 0.002 | 0.12e |
| Cephalosporins | cefixime | 0.008 | 0.008–0.016 | 0.002 | 0.12 |
| ceftriaxone | 0.004 | 0.004–0.008 | 0.002 | 0.12 | |
| Macrolides | azithromycin | 0.25 | 0.06–0.25 | 0.03 | 8 |
| Penicillins | penicillin | 0.5 | 0.25–0.5 | 0.03 | 16 |
| Tetracyclines | tetracycline | 0.5 | 0.25–2 | 0.03 | 32 |
| Quinolones | ciprofloxacin | 0.004 | 0.002–4 | 0.002 | 8 |
No MIC thresholds are available for gentamicin or ertapenem.
One isolate was missing an MIC for this drug.
Six isolates had gentamicin MICs of 16 mg/L.
As per EUCAST, there is insufficient evidence for treatment using carbapenems.12
One isolate had an ertapenem MIC of 0.12 mg/L.
Phenotypic data, along with genes/mutations investigated in this study, have been provided for each isolate in Supplementary Dataset 1.
Penicillin
Most isolates had intermediate susceptibility to penicillin (n = 341, 85.7%; Table 2), although 49 (12.3%) were resistant. Twenty-six isolates had MICs > 2 mg/L, 19 of which had the blaTEM-1B gene,23 consistent with penicillinase-producing N. gonorrhoeae. MICs were significantly greater in those with blaTEM-1B than those without [geometric mean 14.9 mg/L (95% CI 12.8–17.3) versus 0.5 mg/L (95% CI 0.4–0.5)], and all isolates with blaTEM-1 tested positive for β-lactamase production on phenotypic testing. Importantly, no isolates were identified with the blaTEM-135 gene, which only requires a single additional SNP to produce an extended-spectrum β-lactamase.24
Table 2.
Main resistance-associated mutations from the literature and corresponding phenotype
| Drug | Gene (protein) | Mutation | Reference | Isolates with this gene/mutation | Intermediate susceptibility, n (%) | Resistant, n (%) |
|---|---|---|---|---|---|---|
| Penicillin | blaTEM-1B | — | 24 | 19 | 0 (0) | 19 (100) |
| blaTEM-135 | — | 24 | 0 | — | ||
| mtrR promoter | −35, a deletion in the 13 bp inverted repeat | 39,40 | 104 | 72 (69.2) | 32 (30.8) | |
| mtrR (MtrR) | Ala-39→Thr | 41 | 195 | 177 (90.8) | 14 (7.2) | |
| Gly-45→Asp | 41 | 42 | 35 (83.3) | 6 (14.3) | ||
| ponA (PBP1) | Leu-421→Pro | 42 | 133 | 96 (72.2) | 37 (27.8) | |
| penB (PorB1b) | Gly-120→Lys | 43 | 68 | 51 (75.0) | 17 (25.0) | |
| Gly-120→Lys & Ala-121→Asp | 43 | 54 | 41 (75.9) | 13 (24.1) | ||
| Gly-120→Lys & Ala-121→Asn | 44 | 14 | 10 (71.4) | 4 (28.6) | ||
| Gly-120→Asp | 43,44 | 0 | — | — | ||
| pilQ (PilQ) | Glu-666→Lys | 23 | 0 | — | — | |
| Total (N = 398) | 341 (85.7) | 49 (12.3) | ||||
| Ciprofloxacin | gyrA (GyrA) | Ser-91→Phe | 23,45 | 126 | 0 (0) | 124 (98.41) |
| Asp-95→Asn | 23,44 | 1 | 0 (0) | 0 (0) | ||
| Asp-95→Gly | 23,44 | 89 | 0 (0) | 89 (100) | ||
| Asp-95→Ala | 44 | 37 | 0 (0) | 35 (94.6) | ||
| Asp-95→Tyr | 44 | 0 | — | — | ||
| parC (ParC) | Asp-86→Asn | 23,45 | 10 | 0 (0) | 10 (100) | |
| Ser-87→Ile | 44 | 1 | 0 (0) | 1 (100) | ||
| Ser-87→Asn | 44 | 6 | 0 (0) | 6 (100) | ||
| Ser-87→Arg | 44 | 42 | 0 (0) | 40 (95.2) | ||
| Ser-88→Pro | 23 | 0 | — | — | ||
| Glu-91→Lys | 23 | 2 | 0 (0) | 2 (100) | ||
| parE (ParE) | Gly-410→Val | 44 | 0 | — | — | |
| norM promoter | C→T SNP in − 35 hexamer | 46 | 0 | — | — | |
| norM (NorM) | A→G SNP in the ribosomal binding site | 46 | 0 | — | — | |
| Total (N = 398) | 126 (31.7) | |||||
| Tetracycline | rpsJ (ribosomal protein S10) | Val-57→Met | 47 | 278 | 166 (59.7) | 106 (38.1) |
| mtrR promoter | −35 A deletion in the 13 bp inverted repeat | 40 | as above | 40 (38.5) | 56 (53.9) | |
| mtrR (MtrR) | Ala-39→Thr | 41 | as above | 87 (44.6) | 42 (21.5) | |
| Gly-45→Asp | 41 | as above | 2 (4.8) | 10 (23.8) | ||
| penB (PorB1b) | Gly-120→Lys | 43 | as above | 24 (35.3) | 44 (64.7) | |
| Gly-120→Lys & Ala-121→Asp | 43 | as above | 20 (37.0) | 34 (63.0) | ||
| Gly-120→Lys & Ala-121→Asn | 44 | as above | 4 (28.6) | 10 (71.4) | ||
| Gly-120→Asp | 43,44 | as above | — | — | ||
| pilQ (PilQ) | Glu-666→Lys | 23 | 0 | — | — | |
| tetM | — | 48 | 66 | 0 (0) | 66 (100) | |
| Total (N = 398) | 166 (41.7) | 106 (26.6) | ||||
| Spectinomycin | 16S | C1192U | 49 | 0 | — | — |
| rpsE (30S ribosomal protein S5) | Thr-24→Pro | 50 | 0 | — | — | |
| Val-27 deletion | 51 | 0 | — | — | ||
| Lys-28→Glu | 51 | 0 | — | — | ||
| Total (N = 398) | 6 (1.5) | |||||
| Azithromycin | 23S | A2059G | 52,53 | 0a | — | — |
| C2611T | 52,53 | 2a | 0 (0) | 2 (100) | ||
| mtrR promoter | −35, a deletion in the 13 bp inverted repeat | 54 | as above | 16 (15.4) | 6 (5.8) | |
| mtrR (MtrR) | Ala-39→Thr | 41 | as above | 19 (9.7) | 1 (1.0) | |
| Gly-45→Asp | 41,54 | as above | 1 (2.4) | 0 (0) | ||
| ermB | — | 55 | 0 | — | — | |
| ermC | — | 56 | 0 | — | — | |
| ermF | — | 55 | 0 | — | — | |
| macAB promoter | −10 hexamer sequence (TAGAAT→TATAAT) | 57 | 0 | — | — | |
| mef | — | 56 | 0 | — | — | |
| Total (N = 398) | 36 (9.1) | 7 (1.8) |
Some mutations shown may not be independently associated with resistance. MIC breakpoints were from reference 11, or if unavailable from reference 12. For azithromycin, resistance corresponds to MIC > 0.5 mg/L, intermediate susceptibility corresponds to MIC of 0.5 mg/L and susceptible corresponds to MIC ≤0.25 mg/L. For ciprofloxacin, resistance corresponds to MIC ≥1 mg/L, intermediate is 0.12–0.5 mg/L and susceptible is ≤ 0.06 mg/L. For penicillin, resistance corresponds to MIC ≥ 2 mg/L, intermediate susceptibility is 0.12–1 mg/L and susceptible is ≤ 0.06 mg/L. For spectinomycin, resistance corresponds to MIC ≥128 mg/L, intermediate susceptibility is 64 mg/L and susceptible is ≤ 32 mg/L. For tetracycline, resistance corresponds to MIC ≥2 mg/L, intermediate susceptibility is 0.5–1 mg/L and susceptible is ≤ 0.25 mg/L. ESCs (i.e. cefixime and ceftriaxone) are not shown above as only decreased susceptibility was detected; this corresponds to MICs of ≥ 0.12 mg/L and ≥0.06 mg/L, respectively.
All four alleles of 23S were inspected; none had the A2059G mutation, while 4 of 4 had the C2611T mutation.
In addition to acquired genes, penicillin resistance can be mediated by chromosomal point mutations. Mutations in the MtrR repressor (Ala-39→Thr or Gly-45→Asp), which regulates the MtrCDE efflux pump, were identified in 236 isolates. Only one isolate possessed both mutations. Seven others had either the Ala-39→Thr or Gly-45→Asp mutation along with an [A] deletion in −13 bp inverted repeat of the mtrR promoter. This deletion was identified in an additional 97 isolates, and was strongly associated with resistance (Table S7). Other chromosomal mutations identified and associated with resistance on univariate analysis include the penicillin-binding protein 1 (PBP1) L421P mutation, encoded by the ponA gene, and the PorB1b Gly-120→Lys mutation. All 133 isolates with the PBP1 mutation had intermediate or resistant phenotypes. This was also true for the 54 isolates with PorB1b Gly-120→Lys and Ala-121→Asp mutations, 98% of which possessed a concomitant mutation in either mtrR or its promoter.
We evaluated whether the associations between different mutations and penicillin resistance were due to confounding using multivariate logistic regression. Given the strong association of blaTEM-1B with resistance, we restricted this analysis to isolates without this gene, and found that only the mtrR promoter [A] deletion and PBP1 Leu-421→Pro were independently associated with the outcome (Table S7).
Ciprofloxacin
Approximately one-third of isolates were resistant to ciprofloxacin (n = 126). One-hundred and twenty-four of these resistant isolates had the GyrA Ser-91→Phe mutation (Table 2) plus either GyrA Asp-95→Gly (71.8%) or Asp-95→Ala (28.2%) mutations. Ten isolates with the GyrA Ser-91→Phe also had an Asp-86→Asn mutation in the QRDR. parC mutations were identified in 59 isolates; 57 of these were resistant (96.6%) and all had the GyrA Ser-91→Phe mutation. Interestingly, two isolates were phenotypically resistant, with an MIC of 8 mg/L, but had no known resistance-conferring mutations. Both had novel missense variants in gyrA, parC and parE; however, these mutations were also present in susceptible isolates.
Tetracycline
One-hundred and sixty-six isolates had intermediate susceptibility to tetracycline (41.7%), and another 106 were resistant (26.6%). The tetM gene was identified in 66 isolates (62.3%, all of which were resistant), and in 62 of the 63 isolates with MICs ≥ 16 mg/L, marking these as high-level, plasmid-mediated tetracycline-resistant N. gonorrhoeae. Among isolates without tetM, the mtrR promoter deletion was present in 40 of 166 with an intermediate phenotype (24.1%), and 35 of the 40 with resistance (87.5%). In contrast, MtrR Ala-39→Thr and Gly-45→Asp mutations appeared negatively associated with resistance to tetracycline (Table 2). PorB1b Gly-120→Lys and Ala-121→Asp mutations were present only in intermediate or resistant isolates. The ribosomal protein S10 Val-57→Met mutation, in contrast, was found in both non-susceptible and susceptible isolates.
Spectinomycin
Six isolates had intermediate susceptibility to spectinomycin (Table 2), with MICs of 64 mg/L. No known resistance-conferring mutations were identified; while two of six had a novel SNP in rpsE, identical mutations were identified in 20 other isolates that were phenotypically susceptible.
Cefixime and ceftriaxone
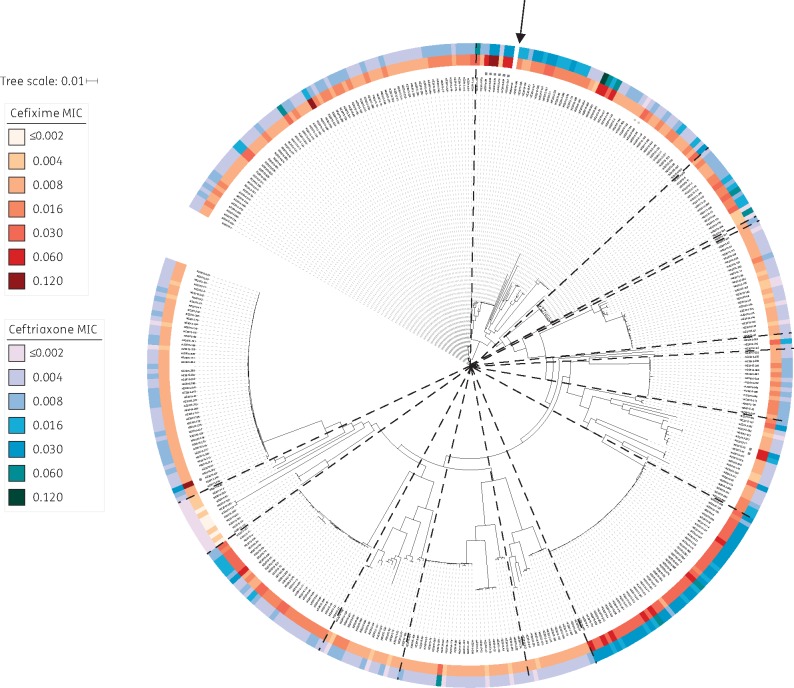
There was no outright resistance to either ESC. Decreased susceptibility (DS) to cefixime (MIC ≥ 0.12 mg/L) or ceftriaxone (MIC ≥ 0.06 mg/L) was noted in 14 isolates (3.5%; 4 cefixime, 10 ceftriaxone), and MICs were moderately correlated (R = 0.6). Within the New Zealand N. gonorrhoeae population, we identified several clusters of isolates with raised MICs to both ESCs (Figure 3), suggestive of de novo acquisition of reduced ESC susceptibility, with subsequent transmission within clusters.
Figure 3.
MICs (mg/L) of extended-spectrum cephalosporins among Neisseria gonorrhoeae isolates in New Zealand. Maximum likelihood tree of all 398 New Zealand isolates, after adjusting for recombination with Gubbins.15 MICs of cefixime and ceftriaxone are shown. Isolates with mosaic penA XXXIV alleles are indicated with black stars, while those with mosaic-like penA alleles as described in text are indicated with grey stars. The BAPS groups identified (as shown in Figure 1) have been overlaid using dashed lines. The reference genome is indicated with an arrow.
Nine isolates had the mosaic penA XXXIV allele (Figure 3), but only three had DS to cefixime, and none had DS to ceftriaxone. As all had the Gly-545→Ser, Ile-312→Met, Val-316→Thr25 and Asn-512→Tyr mutations in penicillin-binding protein 2 (PBP2) associated with resistance to ESCs,26 this suggests that other mutations may be necessary for resistance, either independently or via interaction with these SNPs. An additional two isolates had mosaic-like alleles (lacking Gly-545→Ser and Asn-512→Tyr mutations in PBP2); neither had DS to either drug.
The non-mosaic PBP2 Ala-501→Val mutation, associated with resistance to ESCs,27 was identified in six isolates; three had DS to ceftriaxone, with MICs ranging from 0.06 to 0.12 mg/L. Forty-five other isolates had a PBP2 Ala-501→Thr mutation; seven of these also had DS to ceftriaxone. No A501 mutations were associated with DS to cefixime.
A single isolate (NZ2015–369) had unexplained DS to cefixime. This isolate had no known mutations in MtrR (other than Ala-39→Thr) or PorB1b, and lacked the Glu-666→Lys mutation in PilQ.
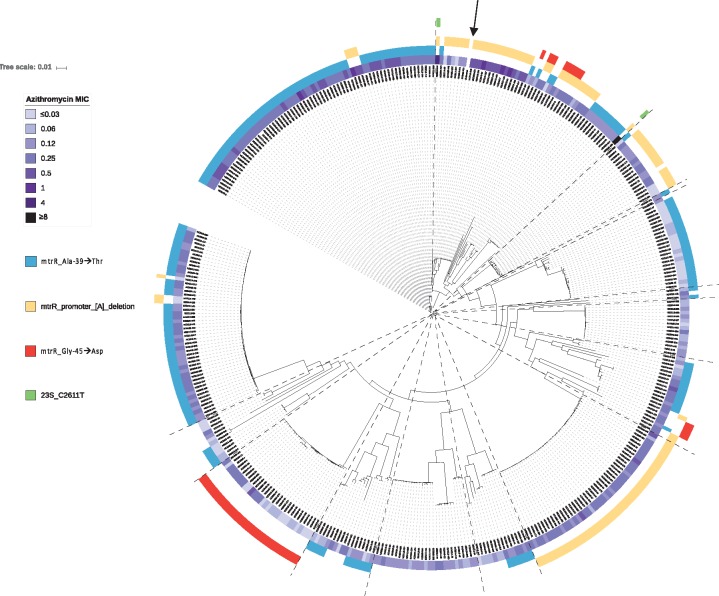
Azithromycin
Thirty-six isolates had intermediate susceptibility to azithromycin, and seven isolates were resistant (Table 2). No acquired azithromycin resistance genes (e.g. ermB, ermC, ermF or mef) were detected. All isolates, however, had at least one mutation associated with resistance (Figure 4). Two unrelated isolates had 23S mutations, with the C2611T mutation present in all four alleles. This mutation is known to confer low to moderate levels of resistance,28 and corresponding MICs were 4 and ≥ 8 mg/L. Both isolates also had the mtrR promoter [A] deletion. Excluding isolates with 23S mutations, the mtrR promoter [A] deletion was associated with higher azithromycin MICs [geometric mean 0.2 mg/L (95% CI 0.2–0.2) versus 0.1 mg/L (95% CI 0.1–0.1) for isolates without the deletion]. This deletion was found in multiple distinct clusters in the phylogeny, and was also shared within clusters among highly related isolates (Figure 4). This suggests that azithromycin MICs may be increasing due to transmission of isolates already possessing mutations associated with resistance, as well as their de novo acquisition of resistance-conferring mutations.
Figure 4.
Mechanisms of azithromycin resistance in New Zealand. Maximum likelihood tree of all 398 New Zealand isolates, after adjusting for recombination with Gubbins.15 Azithromycin MICs in mg/L are shown, along with corresponding mutations known to be associated with phenotypic resistance. The BAPS groups identified (as shown in Figure 1) have been overlaid using dashed lines. The reference genome is indicated with an arrow.
No promoter mutations were identified in macAB, a macrolide transport protein associated with resistance.
Discussion
Herein, to our knowledge, we provide the first in-depth WGS-based analysis of N. gonorrhoeae epidemiology and antimicrobial resistance in the WPR. In this study, we have shown that resistance to antimicrobials is highly prevalent in New Zealand. Although outright resistance to ESCs was not detected, decreased susceptibility to cefixime and ceftriaxone was identified in ∼4% of samples. As ESCs represent a cornerstone of current empirical treatment for gonorrhoea, this finding is not only relevant for regional public health programmes, but global gonorrhoea control as well.
Gonorrhoea incidence has typically been highest among MSM, with most transmission thought to occur within this population.7,29 In accordance with this, a recent Australian study by Trembizki et al.30 suggested that most strains were associated with male-dominated clusters. In New Zealand, however, we found that most clusters identified had a high proportion of isolates from both male and female patients; several of these clusters were highly clonal, suggestive of ‘bridging’ transmission across heterosexual and/or bisexual populations, rather than exclusively between MSM.
N. gonorrhoeae has rapidly acquired resistance to numerous antibiotics;2 New Zealand N. gonorrhoeae—in that sense—is no different, with resistance to penicillins, fluoroquinolones (ciprofloxacin) and tetracyclines all highly prevalent. While we did not detect outright resistance to ESCs, DS to both cefixime and ceftriaxone were identified. Consistent with the findings of Grad et al.,8 isolates with elevated MICs to cefixime and ceftriaxone were largely clonal, suggesting that DS to ESCs may be partly mediated by transmission of already-resistant strains in this context. The mosaic penA XXXIV allele was present in 2.3% of isolates; as this allele appeared in several different clusters in the phylogeny, this suggests novel introductions of penA XXXIV allele have occurred, either through undetected transmission or horizontal gene transfer across lineages. Similar to Australia,30 only approximately one-third of these isolates displayed decreased susceptibility to either ESC tested, indicating that additional mutations are needed to confer phenotypic resistance.8 The broad range of resistance mechanisms associated with reduced susceptibility to ESCs highlights the potential limitations of PCR-based detection of ESC resistance, and underlines the importance of combined genotypic/phenotypic studies for understanding antimicrobial resistance in N. gonorrhoeae.
Decreased susceptibility to ESCs has been shown to co-evolve with azithromycin resistance.28 Overall, this was not apparent in our dataset, potentially due to the small numbers of isolates with DS to ESCs; most isolates with higher MICs to cefixime or ceftriaxone were susceptible to azithromycin. This is consistent with the findings of the USA CDC’s Gonococcal Isolate Surveillance Project,31 although an outbreak involving azithromycin-resistant isolates with DS to ESCs was detected in 2016.32 Considering azithromycin, no isolates were identified with the A2059G mutation in the 23S rRNA, which confers high-level resistance (>256 mg/L), and has recently been reported in Australia.33 Despite this, with 10.8% of isolates overall having azithromycin MICs of 0.5 mg/L or greater, resistance to this drug is still concerning. Current guidelines recommend empirical therapy with antimicrobials that ‘are at least 95% effective’, based on local prevalence of drug resistance;4,34 although azithromycin is given as dual therapy with ESCs, the high proportion of isolates with azithromycin non-susceptibility suggests its time as an effective antimicrobial against gonorrhoea may be limited.
This study has several key strengths. By including all culture-positive N. gonorrhoeae samples over the study period, we have provided a comprehensive snapshot of antimicrobial resistance in New Zealand. Coupled with WGS, this has facilitated a detailed investigation of local N. gonorrhoeae population structure and underlying resistance-conferring genes/mutations. To our knowledge, this study is the first to apply WGS to describe the molecular epidemiology of N. gonorrhoeae in the WPR, where drug resistance is thought to be greatest. Unlike most studies that have applied WGS to gonorrhoea outside this region,5–8 our samples were collected irrespective of susceptibility, increasing the accuracy of prevalence estimates for resistance and concomitant mutations. Sampling was also not biased by sex, allowing us to investigate genetic relatedness between isolates from male and female patients. Finally, while AST is typically only provided at the aggregate level, we have shared both WGS and corresponding phenotypic data (including MIC) per isolate, to facilitate future studies on gonorrhoea resistance and global dissemination of such strains.
There are several limitations to this work. Firstly, epidemiological data describing contact between cases was not available; therefore, direct person-to-person transmission events could not be inferred. Secondly, since 2008, NAATs have become the primary method of diagnosing N. gonorrhoeae in New Zealand,35 due to higher sensitivity and inherent difficulty in maintaining gonococcal viability for culture during transport and storage.4 Without cultures, no sample is available for AST and WGS. Thus, while all positive cultures in New Zealand were obtained, the overall sampling fraction was approximately 27% [calculated using the N. gonorrhoeae incidence reported in 2014 (70 per 100 000)36 and population denominator from the New Zealand 2013 Census, available at http://www.stats.govt.nz/]. This is consistent with previous molecular epidemiology-based studies of gonorrhoea, as the use of NAATs in lieu of culture-based diagnostics has increased in most settings and/or studies have relied on sentinel surveillance data.7,8,30 Unfortunately, such sampling limits the epidemiological inferences that can be drawn, as many source/secondary cases may not be included. In addition to this, as women are often asymptomatic,37 they may be less likely to be diagnosed than men. Therefore, our analysis may under-estimate the number of women involved in potential transmission networks.
In summary, to our knowledge, we present the first population-level WGS-based analysis of N. gonorrhoeae from the WPR, and provide a genomic framework for future studies from this region. Moreover, our comprehensive genotypic–phenotypic analysis highlights the ongoing need to identify resistance mechanisms in N. gonorrhoeae. WGS-based studies offer the opportunity to investigate these mechanisms in detail. To facilitate this understanding, open sharing of genomic data with corresponding phenotypic data per isolate, as is being done for other pathogens (e.g. Mycobacterium tuberculosis, with ReSeqTB38), is essential.
Supplementary Material
Acknowledgements
We thank the Antimicrobial Reference Laboratory at the Institute for Environmental Science and Research in Wellington, New Zealand, for performing the AST, and the Microbiological Diagnostic Public Health Laboratory in Melbourne, Australia, for performing the DNA extraction and WGS. We also thank the regional laboratories across New Zealand for their provision of isolates, and the corresponding clinical data.
Funding
R. S. L. is supported by a Fellowship from the Canadian Institutes of Health Research (Funding Reference Number 152448). B. P. H. has a Practitioner Fellowship from the National Health and Medical Research Council, Australia (GNT1105905). J. C. K. is supported by a postgraduate scholarship from the National Health and Medical Research Council (GNT1074824). D. A. W. is supported by an Early Career Fellowship, also from the National Health and Medical Research Council (GNT1123854). This study was funded by the Ministry of Health of New Zealand. The Microbiological Diagnostic Unit Public Health Laboratory is funded by the Victorian Government, Australia.
Transparency declarations
None to declare.
Author contributions
R. S. L. designed and performed analyses, interpreted results, and wrote the first draft of the manuscript. T. S. provided input into the bioinformatics analysis, contributed bioinformatics tools and helped interpret results. H. H. helped plan and conduct the survey in 2014–15. J. C. K. and A. G. da S. provided input into the bioinformatics and phylogenetic analyses, contributed bioinformatics tools and helped interpret results. G. P. C. performed laboratory work and helped interpret results. D. M. B. assisted with the analysis and helped interpret results. K. H. D. and R. W. also contributed to planning and conducted the survey in 2014–15. T. P. S. and B. P. H. advised on analyses and helped interpret results. D. A. W. led the population-level study in 2014–15, conceived the current study, helped design the analyses and contributed to writing the manuscript. All authors critically reviewed the manuscript for content.
Supplementary data
Supplementary Methods, Results, Figures S1 to S5, Tables S1 to S7 and a dataset are available at JAC Online.
References
- 1. World Health Organization. WHO Guidelines for the Treatment of Neisseria gonorrhoeae .World Health Organization, Geneva, Switzerland, 2016. http://www.who.int/reproductivehealth/publications/rtis/gonorrhoea-treatment-guidelines/en/. [PubMed] [Google Scholar]
- 2. Unemo M. Current and future antimicrobial treatment of gonorrhea—the rapidly evolving Neisseria gonorrhoeae continues to challenge. BMC Infect Dis 2015; 15: 364.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wi T, Lahra MM, Ndowa F. et al. Antimicrobial resistance in Neisseria gonorrhoeae: global surveillance and a call for international collaborative action. PLoS Med 2017; 14: e1002344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Whiley DM, Tapsall JW, Sloots TP.. Nucleic acid amplification testing for Neisseria gonorrhoeae: an ongoing challenge. J Mol Diagn 2006; 8: 3–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Demczuk W, Lynch T, Martin I. et al. Whole-genome phylogenomic heterogeneity of Neisseria gonorrhoeae isolates with decreased cephalosporin susceptibility collected in Canada between 1989 and 2013. J Clin Microbiol 2014; 53: 191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Demczuk W, Martin I, Peterson S. et al. Genomic epidemiology and molecular resistance mechanisms of azithromycin-resistant Neisseria gonorrhoeae in Canada from 1997 to 2014. J Clin Microbiol 2016; 54: 1304–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Grad YH, Kirkcaldy RD, Trees D. et al. Genomic epidemiology of Neisseria gonorrhoeae with reduced susceptibility to cefixime in the USA: a retrospective observational study. Lancet Infect Dis 2014; 14: 220–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Grad YH, Harris SR, Kirkcaldy RD. et al. Genomic epidemiology of gonococcal resistance to extended-spectrum cephalosporins, macrolides, and fluoroquinolones in the United States, 2000-2013. J Infect Dis 2016; 214: 1579–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Newman L, Rowley J, Vander Hoorn S. et al. Global estimates of the prevalence and incidence of four curable sexually transmitted infections in 2012 based on systematic review and global reporting. PLoS One 2015; 10: e0143304.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tapsall JW, Ndowa F, Lewis DA. et al. Meeting the public health challenge of multidrug- and extensively drug-resistant Neisseria gonorrhoeae. Expert Rev Anti Infect Ther 2009; 7: 821–34. [DOI] [PubMed] [Google Scholar]
- 11. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-sixth Informational Supplement M100-S26. CLSI, Wayne, PA, USA, 2016. [Google Scholar]
- 12. EUCAST. Breakpoint tables for interpretation of MICs and zone diameters, version 7.0. 2017. http://www.eucast.org/.
- 13. Goodwin S, McPherson JD, McCombie WR.. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet 2016; 17: 333–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bankevich A, Nurk S, Antipov D. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012; 19: 455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Croucher NJ, Page AJ, Connor TR. et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res 2015; 43: e15.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cheng L, Connor TR, Siren J. et al. Hierarchical and spatially explicit clustering of DNA sequences with BAPS software. Mol Biol Evol 2013; 30: 1224–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nguyen LT, Schmidt HA, von Haeseler A. et al. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 2015; 32: 268–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Minh BQ, Nguyen MA, von Haeseler A.. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol 2013; 30: 1188–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ezewudo MN, Joseph SJ, Castillo-Ramirez S. et al. Population structure of Neisseria gonorrhoeae based on whole genome data and its relationship with antibiotic resistance. PeerJ 2015; 3: e806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. De Silva D, Peters J, Cole K. et al. Whole-genome sequencing to determine transmission of Neisseria gonorrhoeae: an observational study. Lancet Infect Dis 2016; 16: 1295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kwong JC, Goncalves da Silva A, Dyet K. et al. NGMASTER: in silico multi-antigen sequence typing for Neisseria gonorrhoeae. Microbial Genomics 2016; 2: e000076.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. arXiv 2012: 1207.3907v1202.
- 23. Unemo M, Shafer WM.. Antimicrobial resistance in Neisseria gonorrhoeae in the 21st century: past, evolution, and future. Clin Microbiol Rev 2014; 27: 587–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Muhammad IDG, Dillon JR, Johansson A. et al. Characterisation of blaTEM genes and types of β-lactamase plasmids in Neisseria gonorrhoeae – the prevalent and conserved blaTEM-135 has not recently evolved and existed in the Toronto plasmid from the origin. BMC Infect Dis 2014; 14: 454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Takahata S, Senju N, Osaki Y. et al. Amino acid substitutions in mosaic penicillin-binding protein 2 associated with reduced susceptibility to cefixime in clinical isolates of Neisseria gonorrhoeae. Antimicrob Agents Chemother 2006; 50: 3638–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tomberg J, Unemo M, Davies C. et al. Molecular and structural analysis of mosaic variants of penicillin-binding protein 2 conferring decreased susceptibility to expanded-spectrum cephalosporins in Neisseria gonorrhoeae: role of epistatic mutations. Biochemistry 2010; 49: 8062–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Unemo M, Golparian D, Nicholas R. et al. High-level cefixime- and ceftriaxone-resistant Neisseria gonorrhoeae in France: novel penA mosaic allele in a successful international clone causes treatment failure. Antimicrob Agents Chemother 2012; 56: 1273–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Allen VSC, Melano RG.. Azithromycin resistance is coevolving with reduced susceptibility to cephalosporins in Neisseria gonorrhoeae in Ontario, Canada. Antimicrob Agents Chemother 2014; 58: 2528–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fairley CK, Hocking JS, Zhang L. et al. Frequent transmission of gonorrhea in men who have sex with men. Emerg Infect Dis 2017; 23: 102–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Trembizki E, Wand H, Donovan B. et al. The molecular epidemiology and antimicrobial resistance of Neisseria gonorrhoeae in Australia: a nationwide cross-sectional study, 2012. Clin Infect Dis 2016; 63: 1591–8. [DOI] [PubMed] [Google Scholar]
- 31. Kirkcaldy RD, Hook EW 3rd, Soge OO. et al. Trends in Neisseria gonorrhoeae susceptibility to cephalosporins in the United States, 2006-2014. JAMA 2015; 314: 1869–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Papp JR, Abrams AJ, Nash E. et al. Azithromycin resistance and decreased ceftriaxone susceptibility in Neisseria gonorrhoeae, Hawaii, USA. Emerg Infect Dis 2017; 23: 830–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stevens K, Zaia A, Tawil S. et al. Neisseria gonorrhoeae isolates with high-level resistance to azithromycin in Australia. J Antimicrob Chemother 2015; 70: 1267–8. [DOI] [PubMed] [Google Scholar]
- 34. Allen VG, Melano RG.. Whole-genome sequencing—new tools for gonorrhoea control. Lancet Infect Dis 2016; 16: 1214–5. [DOI] [PubMed] [Google Scholar]
- 35. Gonorrhoea Guideline Writing Group on behalf of the New Zealand Sexual Health Society. New Zealand Guideline for the Management of Gonorrhoea, 2014 2014. http://www.nzshs.org.
- 36. The Institute of Environmental Science and Research Ltd. Sexually Transmitted Infections in New Zealand 2014 2015. https://surv.esr.cri.nz.
- 37. Goire N, Lahra MM, Chen M. et al. Molecular approaches to enhance surveillance of gonococcal antimicrobial resistance. Nat Rev Microbiol 2014; 12: 223–9. [DOI] [PubMed] [Google Scholar]
- 38. Starks AM, Aviles E, Cirillo DM. et al. Collaborative effort for a centralized worldwide tuberculosis relational sequencing data platform. Clin Infect Dis 2015; 61 Suppl 3: S141–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hagman KE, Shafer WM.. Transcriptional control of the mtr efflux pump system of Neisseria gonorrhoeae. J Bacteriol 1995; 177: 4162–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Veal WL, Nicholas RA, Shafer WM.. Overexpression of the MtrC-MtrD-MtrE efflux pump due to an mtrR mutation is required for chromosomally mediated penicillin resistance in Neisseria gonorrhoeae. J Bacter 2002; 184: 5619–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shafer WM, Balthazar JT, Hagman KE. et al. Missense mutations that alter the DNA-binding domain of the MtrR protein occur frequently in rectal isolates of Neisseria gonorrhoeae that are resistant to faecal lipids. Microbiology 1995; 141: 907–11. [DOI] [PubMed] [Google Scholar]
- 42. Ropp PA, Hu M, Olesky M, Nicholas RA.. Mutations in ponA, the gene encoding penicillin-binding protein 1, and a novel locus, penC, are required for high-level chromosomally mediated penicillin resistance in Neisseria gonorrhoeae. Antimicrob Agents Chemother 2002; 46: 769–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Olesky M, Hobbs M, Nicholas RA.. Identification and analysis of amino acid mutations in porin 1B that mediate intermediate-level resistance to penicillin and tetracycline in Neisseria gonorrhoeae. Antimicrob Agents Chemother 2002; 46: 2811–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Harrison OB, Clemence M, Dillard JP. et al. Genomic analyses of Neisseria gonorrhoeae reveal an association of the gonococcal genetic island with antimicrobial resistance. J Infect 2016; 73: 578–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Belland RJ, Morrison SG, Ison C. et al. Neisseria gonorrhoeae acquires mutations in analogous regions of gyrA and parC in fluoroquinolone-resistant isolates. Mol Microbiol 1994; 14: 271–380. [DOI] [PubMed] [Google Scholar]
- 46. Rouquette-Loughlin C, Dunham SA, Kuhn M. et al. The NorM efflux pump of Neisseria gonorrhoeae and Neisseria meningitidis recognizes antimicrobial cationic compounds. J Bacteriol 2003; 185: 1101–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hu M, Nandi S, Davies C. et al. High-level chromosomally mediated tetracycline resistance in Neisseria gonorrhoeae results from a point mutation in the rpsJ gene encoding ribosomal protein S10 in combination with the mtrR and penB resistance determinants. Antimicrob Agents Chemother 2005; 49: 4327–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Morse SA, Johnson SR, Biddle JW. et al. High-level tetracycline resistance in Neisseria gonorrhoeae is result of acquisition of streptococcal TetM determinant. Antimicrob Agents Chemother 1986; 30: 664–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Galimand M, Gerbaud G, Courvalin P.. Spectinomycin resistance in Neisseria spp. due to mutations in 16S rRNA. Antimicrob Agents Chemother 2000; 44: 1365–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ilina EN, Malakhova MV, Bodoev IN. et al. Mutation in ribosomal protein S5 leads to spectinomycin resistance in Neisseria gonorrhoeae. Front Microbiol 2013; 4: 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Unemo M, Golparian D, Skogen V. et al. Neisseria gonorrhoeae strain with high-level resistance to spectinomycin due to a novel resistance mechanism (mutated ribosomal protein S5) verified in Norway. Antimicrob Agents Chemother 2013; 57: 1057–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Galarza PG, Abad R, Canigia LF. et al. New mutation in 23S rRNA gene associated with high level of azithromycin resistance in Neisseria gonorrhoeae. Antimicrob Agents Chemother 2010; 54: 1652–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chisholm SA, Dave J, Ison CA.. High-level azithromycin resistance occurs in Neisseria gonorrhoeae as a result of a single point mutation in the 23S rRNA genes. Antimicrob Agents Chemother 2010; 54: 3812–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zarantonelli L, Borthagaray G, Lee E-H. et al. Decreased azithromycin susceptibility of Neisseria gonorrhoeae due to mtrR mutations. J Antimicrob Chemother 1999; 43: 2468–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Roberts MC, Chung WO, Roe D. et al. Erythromycin-resistant Neisseria gonorrhoeae and oral commensal Neisseria spp. carry known rRNA methylase genes. Antimicrob Agents Chemother 1999; 43: 1367–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cousin S, Whittington WLH, Roberts MC.. Acquired macrolide resistance genes in pathogenic Neisseria spp. isolated between 1940 and 1987. Antimicrob Agents Chemother 2003; 47: 3877–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rouquette-Loughlin CE, Balthazar JT. et al. Characterization of the MacA-MacB efflux system in Neisseria gonorrhoeae. J Antimicrob Chemother 2005; 56: 856–60. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.