Abstract
Background: Echinocandins are an important class of antifungal agents in the treatment of invasive candidiasis. However, little is known about the metabolomic effects of echinocandins on Candida. We therefore performed LC–high-resolution MS (LC-HRMS)-based metabolomics profiling of the response of Candida albicans cells to increasing concentrations of micafungin to determine the metabolic response of Candida to micafungin subinhibitory injury.
Methods: Isolates of C. albicans were cultured on nitrocellulose filters to mid-logarithmic phase of growth and micafungin (0–0.25 mg/L) was added. At mid-logarithmic phase, replicates were metabolically quenched. Intracellular metabolites were analysed by LC-HRMS. Changes in pool sizes of individual metabolites were analysed by Student’s t-test adjusted for multiple hypothesis testing by Benjamini–Hochberg correction. Metabolites were ascribed by the Kyoto Encyclopedia of Genes and Genomes (KEGG) metabolic pathways database.
Results: Among 3446 detected metabolites, 204 were identified by comparison against pure standard or comparison against a library of mass-retention-time pairs. Fifty had significantly altered abundances in response to increasing micafungin concentrations. Pool sizes of amino acids, nucleic acids and polyamine metabolism were significantly increased at subinhibitory concentrations, while exposure to inhibitory concentrations resulted in a precipitous decrease consistent with fungicidal activity.
Conclusions: Micafungin induces a re-routing of metabolic pathways inhibiting protein synthesis and cell replication. These results shed light on new mechanisms of action of echinocandins.
Introduction
Candida species are part of the normal microbiota of the oral mucosa, gastrointestinal tract and vagina and are responsible for different infections, ranging from mucocutaneous diseases to disseminated candidiasis.1 Critically ill and immunocompromised patients are particularly susceptible to invasive, life-threatening Candida infections.2–4
Echinocandins represent the newest class of antifungals and act by inhibiting the production of (1→3)-β-d-glucan, an essential component of the fungal cell wall.5 While considerable progress has been achieved in understanding the mechanisms of action of echinocandins, little is known about their effects on the metabolic activity of Candida spp. We therefore performed LC–high resolution MS (LC-HRMS)-based metabolomics profiling of the response of Candida albicans cells to increasing concentrations of micafungin to determine the metabolic response of Candida to micafungin subinhibitory injury.
Materials and methods
Organism
C. albicans (strain SC5314/ATCC MYA-2876) was used.6,7 Micafungin MIC was determined according to the CLSI M27-A3 method. The MIC was the lowest drug concentration at which a prominent decrease in turbidity was observed (MIC 0.25 mg/L).8
Sample preparation and LC-HRMS
Sample preparation and LC-HRMS were performed as described in the supplementary Materials and methods (available as Supplementary data at JAC Online) using methods similar to those previously described.9–11
LC-HRMS data processing and analysis
Global metabolomics profiling was performed on a Thermo Q-Exactive Orbitrap mass spectrometer with a Dionex UHPLC and autosampler. All samples were analysed using positive and negative heated electrospray ionization with a mass resolution of 70 000 at m/z 200 as separate injections (2 μL for positive and 4 μL for negative). Metabolites were searched by chemical formula and by molecular feature. Identities of specific metabolites were confirmed against pure chemical standards (where available) by molecular mass (mass tolerance <0.01 Da) and retention times. MZmine (a metabolomics data processing program) was used to identify features, deisotope, align features and perform gap filling to fill in any features that may have been missed in the first alignment algorithm (see Figure 1 legend).
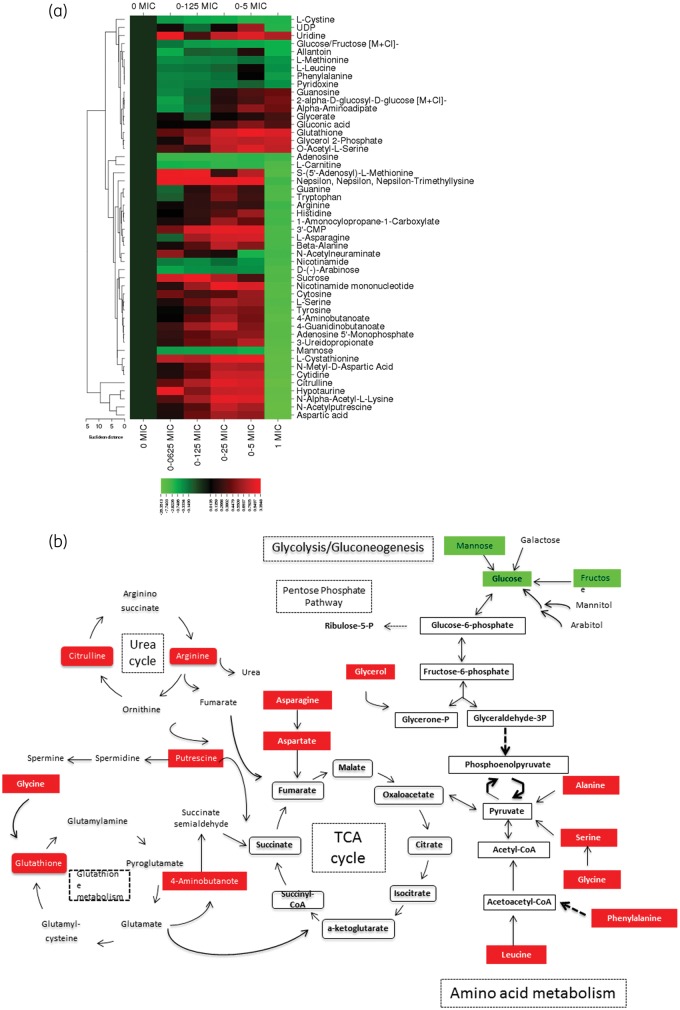
Figure 1.
(a) Heat map of altered metabolites in C. albicans in response to increasing micafungin concentrations. The heat map shows the 50 metabolites whose abundance was significantly altered in response to increasing micafungin concentrations from 0 mg/L (0 × MIC for this strain) to 0.25 mg/L (1 × MIC for this strain). Changes in abundance of each metabolite are indicated by colour coding, with red indicative of an increase in intracellular abundance and green indicative of a decrease relative to the baseline (as defined by the abundance in untreated C. albicans). Normalized levels for each metabolite across replicates from independent experiments were generated by dividing the adjusted abundance for each by the average adjusted abundance. A heat map was created using CIMminer software after log(2) transformation of the average normalized abundance of each metabolite (average of replicates across independent experiments). The metabolites that are depicted in the heat map are those with an adjusted P value ≤0.5 after correction for multiple hypothesis testing (Benjamini–Hochberg correction). (b) Schematic overview of the key molecules of C. albicans metabolism that are affected during treatment with micafungin at subinhibitory concentrations (red indicates increased abundances and green decreased abundances at subinhibitory concentrations of micafungin). Unless otherwise cited, metabolites were ascribed to pathways by comparison against the KEGG (Kyoto Encyclopedia of Genes and Genomes) Database. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Statistical analyses
As adjusted ion counts plotted using an Excel Microsoft® database demonstrated a clear relationship with a λ value approaching zero, log transformation was used. After log transformation of the raw data, changes in pool sizes of individual metabolites were verified by Student’s t-test adjusted for multiple hypothesis testing by Benjamini–Hochberg correction.12
Results
An untargeted metabolomics analysis was used to identify the metabolites that were differentially produced between micafungin-treated and control untreated C. albicans cells. Among the 3446 features detected from all measurements, 204 could be identified by name and molecular formula (Table S1).
As depicted in the heat map (Figure 1), the abundance of the majority of identified amino acids was abruptly decreased at the inhibitory concentration (MIC 0.25 mg/L), while at the subinhibitory concentrations (ranging from 0.0625 × MIC to 0.5 × MIC) there was a noted concentration–response accumulation ranging from 2- to 17-fold. This pattern was observed in 22 of the 26 identified amino acids [l-cystine, l-methionine, l-leucine, phenylalanine, carnitine, S-(5′-adenosyl)-l-methionine, trimethyllysine, tryptophan, l-asparagine, β-alanine, arginine, 4-aminobutanoate, histidine, guanidinobutanoate, l-cystathionine, N-methyl-d-aspartic acid, l-serine, tyrosine, citrulline, hypotaurine, N-α-acetyl-l-lysine and aspartic acid]. The most prominent decreases were noted with citrulline, aspartic acid, N-α-d-aspartic acid and N-α-acetyl-l-lysine, which were 20-, 18-, 10- and 14-fold diminished, respectively (P < 0.05). A similar trend was noted with the polyamine, N-acetylputrescine, where abundance was moderately increased, by 0.7-fold, at subinhibitory micafungin concentrations, while exposure to inhibitory concentrations (at MIC) resulted in a noticeable 20-fold decrease in the intracellular polyamine.
A similar concentration-dependent pattern of increasing nucleic acids (e.g. guanine, cytosine and cytidine) occurred at subinhibitory concentrations followed by a precipitous decrease at MIC. The abundance of cytosine, adenosine-5-monophosphate (AMP), cytidine 3′-monophosphate (3′CMP) and uridine diphosphate (UDP) was decreased at the MIC by 8-, 8-, 5- and 2-fold, respectively (P < 0.05; Figure 1).
The impact of micafungin upon central carbon metabolism varied depending on the metabolite. The abundances of four pivotal intermediates of glycolysis/gluconeogenesis and the pentose phosphate pathway, namely 2-α-d-glucosyl-d-glucose, gluconic acid, glycerol-2-phosphate and glycerate, were significantly increased in the micafungin-treated organisms (Figure 1). However, the increase was moderate, ranging from 0.3- to 0.7-fold, while the most prominent increase (0.7-fold) was noted for glycerol-2-phosphate. By comparison, for glucose/fructose, d-(–)-arabinose, sucrose and mannose, micafungin exerted a concentration–response negative effect causing a significant 2-, 7-, 6- and 9-fold decrease, respectively.
Intracellular glutathione was increased in micafungin-treated C. albicans cells. The increase in intracellular pool sizes of glutathione in response to micafungin treatment followed a concentration–response trend, ranging from an increase of 0.4-fold at the lowest concentration to 0.9-fold increase at the highest concentration tested (MIC).
Exposure of C. albicans across a range of tested micafungin concentrations did not significantly affect the abundance of intracellular lipids. By comparison, significant differences were observed in the abundance of amino acids, nucleic acids and central carbon metabolites.
Discussion
The main goal of this study was to investigate the metabolomic profile of C. albicans in response to increasing concentrations of micafungin. Based on LC-HRMS data, 50 metabolites were found to be differentially expressed between micafungin-treated and untreated C. albicans cells to a significant degree. Specific subsets of metabolites, including amino acids, nucleic acids, polyamines and intermediates of central carbon metabolism and glutathione metabolism, were observed (Figure 1).
Echinocandins inhibit (1→3)-β-d-glucan synthetase. As an important polysaccharide component of the cell wall, (1→3)-β-d-glucan is largely responsible for the strength and integrity of the cell wall. In this regard, β-glucan destruction prevents resistance to osmotic forces, which leads to cell lysis.13 However, the metabolic adaptations of C. albicans to echinocandins and the damage caused to the organism by echinocandins are unknown and this is the first comprehensive analysis.
The increase in pool sizes of almost all amino acids and nucleic acids in response to subinhibitory concentrations of micafungin is most likely multifactorial. First, there may be an adaptive response to changes in membrane permeability. Since amino acids and nucleic acids are essential nutrients for protein synthesis and cell proliferation, respectively, their accumulation may also be explained by the decreased energy metabolism for all the transmembrane proteins. As protein synthesis decreases, the activity of enzymes driving key metabolic pathways is also reduced. Moreover, many amino acids, can be metabolized into tricarboxylic acid (TCA) cycle intermediates and enter this cycle. Reduced TCA cycle activity would affect the consumption of the corresponding amino acids (Figure 1b). The increased amino acid levels during exposure to subinhibitory micafungin concentrations are consistent with a reduced consumption of amino acids due to inhibited TCA cycle activity. Eventually, when the concentration of micafungin reaches its fungicidal concentrations at the MIC, the osmotically fragile spheroplast is lysed, the intracellular metabolites efflux and their abundance is abruptly decreased (Figure 1). Thus, we hypothesize that amino acids initially accumulate, as they are utilized neither in the TCA cycle nor in the protein synthesis pathways during subinhibitory exposure to micafungin. Moreover, amino acids are rapidly depleted by efflux from the osmotically damaged Candida cell membrane at the fungicidal MIC of micafungin.
The same trend is followed by polyamines (N-acetylputrescine) and citrulline. Polyamine accumulation is a known bacterial stress response as it provides enhanced scavenging of intracellular reactive oxygen species (ROS) and thus enables the cell temporarily to survive the antibiotic effects.14–16 Since the intracellular arginine, S-5′-adenosyl-l-methionine, citrulline and N-acetylputrescine levels were stimulated at the subinhibitory concentrations of micafungin and, of note, decreased at lethal concentrations of the drug, we might speculate that polyamines regulate the fungal cell’s susceptibility to micafungin by providing more scavenging capacity against ROS. This hypothesis is consistent with results from other studies showing that polyamines play an important role in protecting C. albicans cells from amphotericin B killing. The mechanism by which polyamines protect C. albicans cells from antifungal killing may be related to attenuation of ROS accumulation.17
Glutathione is an antioxidant that plays a crucial role in cellular detoxification against damaging compounds. Our results show that the synthesis of intracellular glutathione is increased and this might explain a complex defensive response of C. albicans elicited by the fungicidal action of micafungin. Increased glutathione levels indicate an increased capacity for oxidative stress defence in micafungin-treated cells. The protective intracellular accumulation of glutathione and polyamines may constitute mechanisms involved in the resistance of C. albicans to micafungin.
These experiments showed that the changes in metabolite pools of C. albicans in response to micafungin are class specific. In particular, our previous experiments showed that exposure of C. albicans to fluconazole was associated with increased abundances of intermediates of central carbon metabolites and decreased pool sizes of intermediates of amino acid synthesis. In addition, fluconazole promoted the accumulation of mevalonate, while on the contrary micafungin did not have any effect on the sterol biosynthetic pathway.18 The metabolomic study of the effects of antifungal agents against C. albicans complements classical time–kill assays by further providing powerful biochemical insights into the mechanisms of the antifungal agents and further elucidating their fungicidal versus fungistatic properties.
In conclusion, our study demonstrates that the effects of micafungin on the metabolic state of C. albicans are concentration dependent and class specific. In addition, micafungin induces selective stress responses to C. albicans cells. These studies further shed new light on the mechanisms of the fungicidal effects of echinocandins, providing the framework for newer antifungal development.
Funding
The research project was implemented within the framework of the Action ‘Supporting Postdoctoral Researchers’ of the Operational Program ‘Education and Lifelong Learning’ (Action’s Beneficiary: General Secretariat for Research and Technology) and is co-financed by the European Social Fund (ESF) and the Greek State [LS6 (3309) No 38281/2012].
This study was supported by an ESPID Fellowship Award 2014 to A. K., a SECIM pilot and feasibility Award (NIH Grant #U24 DK097209) to T. J. W. Clinical Trial Registration, research scholar awards to T. J. W. from Save Our Sick Children (SOS) Foundation and the Sharp Family Foundation.
Transparency declarations
E. R. has received research grant support from Pfizer, Gilead and Merck, has served as a consultant to Gilead, Astellas Gilead, Cephalon and Pfizer, and has been in the speakers’ bureau of Merck, Pfizer, Gilead and Astellas. T. J. W. receives research grants to Weill Cornell Medical Center for experimental and clinical antimicrobial pharmacotherapeutics, new diagnostic systems and strategies for augmentation of host defence against life-threatening infections in immunocompromised children and adult patients from Astellas, Novartis, Merck, ContraFect, Cubist and Pfizer, and has served as a consultant to Astellas, ContraFect, iCo, Novartis, Pfizer, Methylgene, SigmaTau and Trius. All other authors: none to declare.
Supplementary Material
References
- 1. Kullberg BJ, Arendrup MC. Invasive candidiasis. N Engl J Med 2015; 373: 1445–56. [DOI] [PubMed] [Google Scholar]
- 2. Mean M, Marchetti O, Calandra T. Bench-to-bedside review: Candida infections in the intensive care unit. Crit Care 2008; 12: 204.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Achkar JM, Fries BC. Candida infections of the genitourinary tract. Clin Microbiol Rev 2010; 23: 253–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Delaloye J, Calandra T. Invasive candidiasis as a cause of sepsis in the critically ill patient. Virulence 2014; 5: 161–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ghannoum MA, Rice LB. Antifungal agents: mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clin Microbiol Rev 1999; 12: 501–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gillum AM, Tsay EY, Kirsch DR. Isolation of the Candida albicans gene for orotidine-5'-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet 1984; 198: 179–82. [DOI] [PubMed] [Google Scholar]
- 7. Odds FC, Brown AJ, Gow NA. Candida albicans genome sequence: a platform for genomics in the absence of genetics. Genome Biol 2004; 5: 230.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clinical and Laboratory Standards Institute. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts—Third Edition: Approved Standard M27-A3. CLSI, Wayne, PA, USA, 2008. [Google Scholar]
- 9. de Carvalho LP, Fischer SM, Marrero J. et al. Metabolomics of Mycobacterium tuberculosis reveals compartmentalized co-catabolism of carbon substrates. Chem Biol 2010; 17: 1122–31. [DOI] [PubMed] [Google Scholar]
- 10. Pesek JJ, Matyska MT, Loo JA. et al. Analysis of hydrophilic metabolites in physiological fluids by HPLC-MS using a silica hydride-based stationary phase. J Sep Sci 2009; 32: 2200–8. [DOI] [PubMed] [Google Scholar]
- 11. Ulmer CZ, Yost RA, Chen J. et al. Liquid chromatography-mass spectrometry metabolic and lipidomic sample preparation workflow for suspension-cultured mammalian cells using Jurkat T lymphocyte cells. J Proteomics Bioinform 2015; 8: 126–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med 1990; 9: 811–8. [DOI] [PubMed] [Google Scholar]
- 13. Nett JE, Andes DR. Antifungal agents: spectrum of activity, pharmacology, and clinical indications. Infect Dis Clin North Am 2016; 30: 51–83. [DOI] [PubMed] [Google Scholar]
- 14. Johnson L, Mulcahy H, Kanevets U. et al. Surface-localized spermidine protects the Pseudomonas aeruginosa outer membrane from antibiotic treatment and oxidative stress. J Bacteriol 2012; 194: 813–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kohanski MA, Dwyer DJ, Hayete B. et al. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 2007; 130: 797–810. [DOI] [PubMed] [Google Scholar]
- 16. Tkachenko AG, Akhova AV, Shumkov MS. et al. Polyamines reduce oxidative stress in Escherichia coli cells exposed to bactericidal antibiotics. Res Microbiol 2012; 163: 83–91. [DOI] [PubMed] [Google Scholar]
- 17. Cao Y, Zhu Z, Chen X. et al. Effect of amphotericin B on the metabolic profiles of Candida albicans. J Proteome Res 2013; 12: 2921–32. [DOI] [PubMed] [Google Scholar]
- 18. Katragkou A, Alexander EL, Eoh H. et al. Effects of fluconazole on the metabolomic profile of Candida albicans. J Antimicrob Chemother 2016; 71: 635–40. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.