Abstract
Objectives
Test the performance of topical antimicrobial wound solutions against microbial biofilms using in vitro, ex vivo and in vivo model systems at clinically relevant exposure times.
Methods
Topical antimicrobial wound solutions were tested under three different conditions: (in vitro) 4% w/v Melaleuca oil, polyhexamethylene biguanide, chlorhexidine, povidone iodine and hypochlorous acid were tested at short duration exposure times for 15 min against 3 day mature biofilms of Staphylococcus aureus and Pseudomonas aeruginosa; (ex vivo) hypochlorous acid was tested in a porcine skin explant model with 12 cycles of 10 min exposure, over 24 h, against 3 day mature P. aeruginosa biofilms; and (in vivo) 4% w/v Melaleuca oil was applied for 15 min exposure, daily, for 7 days, in 10 patients with chronic non-healing diabetic foot ulcers complicated by biofilm.
Results
In vitro assessment demonstrated variable efficacy in reducing biofilms ranging from 0.5 log10 reductions to full eradication. Repeated instillation of hypochlorous acid in a porcine model achieved <1 log10 reduction (0.77 log10, P = 0.1). Application of 4% w/v Melaleuca oil in vivo resulted in no change to the total microbial load of diabetic foot ulcers complicated by biofilm (median log10 microbial load pre-treatment = 4.9 log10 versus 4.8 log10, P = 0.43).
Conclusions
Short durations of exposure to topical antimicrobial wound solutions commonly utilized by clinicians are ineffective against microbial biofilms, particularly when used in vivo. Wound solutions should not be used as a sole therapy and clinicians should consider multifaceted strategies that include sharp debridement as the gold standard.
Introduction
Chronic wounds are a serious cause of morbidity and mortality, and are associated with reduced patient health-related quality of life. The impacts on healthcare providers are reflected in the staggering cost of managing these wounds and associated comorbidities, with £5.3 billion attributed to UK National Health Service expenditure.1 Increasing evidence on the microorganisms involved in chronic wounds has identified that planktonic cells may not necessarily represent the phenotypic behaviour of microorganisms involved in chronic non-healing wounds. The focus has shifted towards the concept of microbial aggregates (biofilms), which differ markedly in their phenotypic behaviour and may contribute to the delayed healing of wounds.2 In addition, the ecology of chronic wounds explored through molecular DNA-based technologies (and not cultivation-based methods) has identified these wounds to be complicated by complex polymicrobial communities.3
Once established, complex biofilm communities often become highly tolerant to standard treatment and removal/eradication paradigms, yielding several hallmark features that distinguish biofilm phenotypes from those of planktonic counterparts. The most notable of these is a remarkable tolerance to antimicrobial agents,4,5 and host immune defences.6 The increasing awareness and promotion of the biofilm concept within the wound care arena has led to a dramatic rise in the use of topical antimicrobial solutions as part of wound care therapeutics.7
Unfortunately, the evidence for use of particular topical antimicrobials in the treatment of biofilm-associated wounds is based on in vitro methodologies that lack standardization and clinical relevance to their intended applications.8 For example, the anti-biofilm effects of wound solutions, for which outcomes are based on reductions in biofilm markers (i.e. biomass, cfu/mL, LIVE/DEAD® stain viability), have been reported at exposure times far greater than their intended use. Many wound care/device companies promote a 15 min exposure time for their respective antimicrobial solutions (seconds for irrigation solutions), yet the bulk of data for effectiveness of these products in vitro have only reported outcomes at 24 h exposure times.9–11 This has important consequences at the treatment level where clinicians often seek guidance from laboratory-based studies (owing to a lack of available in vivo data) in choosing the most relevant and effective agent to reduce microbial biofilms. Therefore, in vitro data based on greater exposure times may not reflect the most clinically appropriate outcomes for clinicians using these products at shorter exposure times. This is highlighted succinctly by Castaneda et al.,12 who showed that in an in vitro biofilm model, antimicrobial susceptibility increased with antimicrobial exposure time.
The present study was designed to explore whether shorter durations of exposure to antimicrobial wound solutions were effective against microbial biofilms: (i) in vitro against mature biofilms of Staphylococcus aureus and Pseudomonas aeruginosa; (ii) in an ex vivo porcine skin explant model against mature P. aeruginosa biofilms; and (iii) in vivo in 10 patients with chronic non-healing diabetic foot ulcers (DFUs).
Materials and methods
Bacteria
The biofilm-forming reference strains utilized in vitro were S. aureus [ATCC® 25923™ (MSSA)] and P. aeruginosa (ATCC® 25619™), and P. aeruginosa PA01 (ATCC® BAA-47™) was used in the ex vivo porcine skin explant model.
Antimicrobial wound solutions
The solutions examined, any incorporated antimicrobials/antiseptics and their respective manufacturers, were as follows: surfactant-based antiseptic solution with 4% w/v Melaleuca oil (SBMO; Woundaid® Woundwash; Mundipharma, Singapore); surfactant-based antimicrobial solution with polyhexamethylene biguanide (SBPHMB; Prontosan®; B. Braun Medical, Melsungen, Germany); superoxidized solution (SOS) containing sodium hypochlorite, hypochlorous acid, sodium chloride and oxidized water (Microcyn®; Oculus Technologies of Mexico); chlorhexidine (CHX) 4.5 mg/30 mL (0.015% w/v) and cetrimide 45 mg/30 mL (0.15% w/v) irrigation solution (Pfizer, New York, USA); povidone iodine antiseptic solution 10% w/v equivalent to 1% w/v available iodine (PVP-I; BETADINE®; Mundipharma, Singapore); NaCl 0.9% (Baxter, IL, USA).
The decision to use SOS for the ex vivo porcine explant model and SBMO for the human in vivo study was based on clinical relevance. Both the use and promotion of these ‘newer generation’ solutions with antimicrobial properties (as opposed to traditional antimicrobials of CHX and PVP-I) by clinicians and industry for action against wound biofilm has increased significantly over the last decade. They now represent the predominant products used for wound cleansing and debridement.
Experimental models
In vitro model
Biofilm, containing 107–108 cells/coupon of P. aeruginosa (ATCC 25619) and 106 cells/coupon of S. aureus ATCC 25923 was grown under shear (130 rpm) on polycarbonate coupons in a CDC biofilm reactor (BioSurface Technologies Corp., Bozeman, MT, USA) as previously described by our group,13 in 400 mL of 15 g/L (50%) tryptic soy broth (Sigma–Aldrich, St Louis, MO, USA) at 35 °C in batch phase for 48 h, followed by incubation in fresh medium (20% tryptic soy broth, 6 g/L) for a further 24 h. Coupons were washed in 10 mL PBS to remove loosely attached planktonic bacteria. Each coupon had 107–108P. aeruginosa or 106S. aureus. Five antiseptic treatments were tested (SBMO, SBPHMB, SOS, CHX, PVP-I); four coupons were exposed to each treatment condition for 15 min, while an additional four coupons were used as controls.
The numbers of bacterial colony forming units (cfu) per coupon were tested in triplicate by sonication in an ultrasonic bath (Soniclean; JMR, Australia) for 10 min with a sweeping frequency of 42–47 kHz at 20°C. The coupon was then vortexed for 2 min in 2 mL of PBS followed by a sequential 10-fold dilution and plate count. Pre- and post-exposure average cfu/coupon was expressed as log10. Bacterial cell viability pre- and post-exposure was also assessed using BacLight™ (LIVE/DEAD® Bacterial Viability Kit, 7012; Molecular Probes, Invitrogen, Carlsbad, CA, USA) in conjunction with confocal laser scanning microscopy (CLSM) and expressed as the percentage of viability as determined by Imaris (v8.4, ImarisXT, Bitplane). For CLSM, we used an inverted laser scanning confocal microscope (ZEISS LSM 880; Carl Zeiss Ltd, Herefordshire, UK) for all the samples, with oil-immersion lenses (63× and 100×) and acquisition parameters of: frame size, 1024 × 1024; speed, 6; averaging, 2; bit depth, 12.
Ex vivo porcine skin explant model
The ex vivo porcine skin explant biofilm model used in this study is previously described14 and a detailed description can be found in the Supplementary data (Part S1, available as Supplementary data at JAC Online). One pig was used to obtain all explants, which were freshly harvested, shaved, cleaned and inflicted with a partial thickness excision wound. Explants were then sterilized by first submerging the explants in PBS containing 0.6% hypochlorous acid and 0.5% Tween 80 for 5 min then transferring them to a chlorine gas chamber for 45 min, followed by submerging the explants again in PBS containing 0.6% hypochlorous acid and 0.5% Tween 80 for 5 min. The sterile explants were rinsed twice in sterile PBS then transferred into 150 mm diameter by 25 mm deep culture plates (176 cm2 surface area) (Corning 430599) containing 0.5% tryptic soy soft agar containing antibiotic (gentamicin at 50 mg/mL) to limit planktonic growth and promote biofilm growth on the explants. One hundred microlitres of P. aeruginosa PA01 (∼107–108 cfu/mL) was inoculated onto the explants and incubated for 3 days at 37 °C. Porcine explants were subjected to three test groups: (i) negative pressure wound therapy alone (control); (ii) negative pressure wound therapy with instillation therapy for 12 cycles of 10 min of soak/dwell with SOS, totalling 24 h for the experiment; and (iii) negative pressure with instillation therapy for 12 cycles of 10 min of soak/dwell with saline (NaCl 0.9%). After 24 h, six 8 mm biopsies were obtained from the porcine skin explant and processed for measurement of cfu/mL and scanning electron microscopy (SEM). For each test group, six experiments were established and the cfu was averaged over these.
In vivo clinical study
We used a combined molecular and microscopy approach described previously15 to better understand the effects of a topical antimicrobial solution against the microbial load and diversity of chronic non-healing DFUs complicated by biofilm (Supplementary data Part S2). Ten patients with chronic non-healing DFUs (and not on current antimicrobial therapy) were enrolled over a 6 month period from a tertiary referral hospital (Liverpool Hospital High Risk Foot Service, Liverpool, Sydney). Ethics approval for this study was granted by the South West Sydney Local Health District Research and Ethics Committee (HREC/14/LPOOL/487, SSA/14/LPOOL/489). Sterile gauze was soaked in SBMO and applied to the wound for 15 min, every day for 7 days. Sharp debridement of tissue was withheld over the 7 day treatment period, as this would likely have affected the primary outcome measure.16 Tissue punch biopsies were obtained from the wound edge for each participant after cleansing the wound with NaCl 0.9% pre- and post-treatment. These were subjected to quantitative PCR (qPCR) to determine the total microbial load, next generation DNA sequencing to explore the microbiome of chronic DFUs and the effects on microbial communities following topical antimicrobial therapy, SEM to visualize biofilm structures and fluorescent in situ hybridization in conjunction with CLSM to examine spatial organization of microbial aggregates.
Statistics
Mann–Whitney U-tests were used to assess differences between pre- and post-log10 cfu using the Statistical Package for Social Sciences Version 23 (SPSS Inc., Chicago, IL, USA). CLC genomics workbench version 8.5.1 in combination with the microbial genome-finishing module (CLC bio, Qiagen Aarhus, Denmark) was used to analyse DNA sequence data. QIIME was utilized to visually represent data. Analysis of variance and permutational analysis of variance were used for statistical analysis of alpha and beta diversity measures. Principal coordinates analysis plots with Bray–Curtis distances were used to assess how dissimilar microbial communities were pre- and post-treatment. Community richness of DFUs was presented using richness index reporting the number of unique operational taxonomic units (OTUs) in each wound sample. The Shannon–Weaver Index is an ecological measure of diversity that includes the number of unique microbial taxa and their relative evenness within each sample. For all comparisons and modelling, the level of significance was set at P < 0.05. Data are given as mean, median (±SD) and 95% CI.
Results
Antimicrobial efficacies of topical wound solutions against mature biofilms in vitro
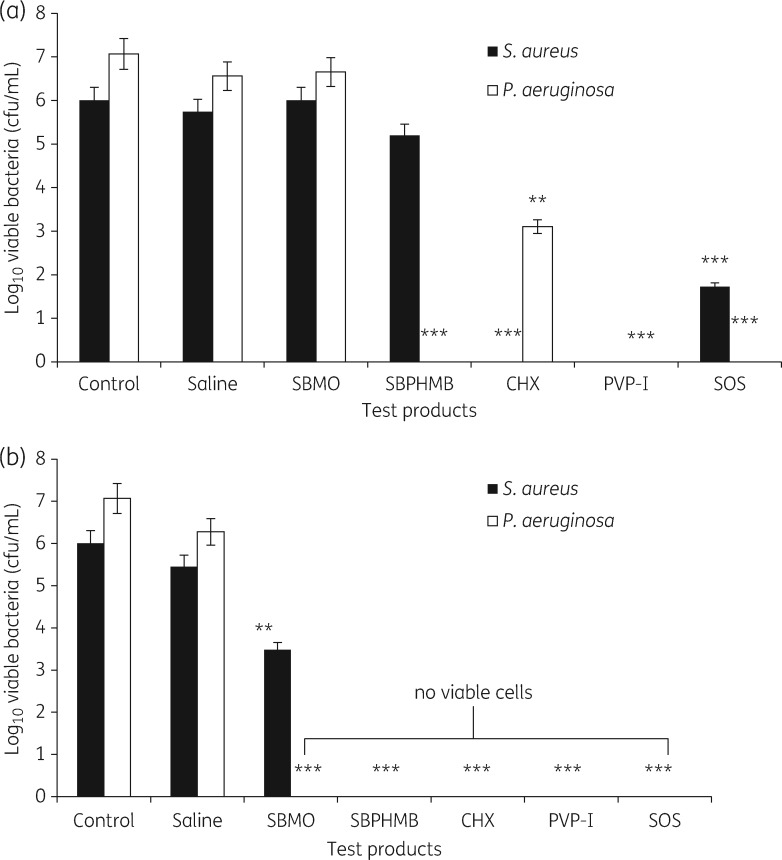
The effects of topical antimicrobial solutions on reductions in log10 cfu/coupon following treatments at 15 min and 24 h are shown in Figure 1(a and b). LIVE/DEAD® stain with CLSM and the percentage of red signal (dead/damaged cells) and green signal (viable cells) at 15 min exposures are noted in Supplementary data Part S3. At 15 min exposures PVP-I was the only solution to show complete and efficient killing of both S. aureus and P. aeruginosa biofilms (6 and 7 log10 reduction, P = 0.001). CHX was effective against S. aureus biofilms showing complete removal of all bacteria (6 log10 reduction, P = 0.001), and further demonstrated a 3.96 log10 cfu reduction against the P. aeruginosa biofilm (P = 0.01). In contrast, SOS demonstrated complete eradication of the P. aeruginosa biofilm (7 log10 reduction, P = 0.001) and a ≥4 log10 cfu/mL reduction against S. aureus (4.3 log10 reduction P = 0.01). No significant reduction in S. aureus counts was observed for treatment with SBPHMB (0.8 log10 reduction); however, it was highly effective against P. aeruginosa biofilm showing complete eradication (7 log10 reduction, P = 0.01). Treatment with SBMO was ineffective against both S. aureus and P. aeruginosa biofilm. In contrast, treatment of biofilm with topical antimicrobials for 24 h exposure showed complete and efficient killing of biofilm, except for SBMO, which failed to eradicate S. aureus (but still achieved a ≥2.5 log10 cfu/coupon).
Figure 1.
Effect of test products on bacterial viability. Bars represent means of logarithms of colony-forming units of viable biofilm cells after (a) 15 min exposure and (b) after 24 h exposure. Error bars represent the standard error of the means from three coupons (**P < 0.01, ***P < 0.001, no viable cells).
Antimicrobial efficacy of SOS against mature biofilms in an ex vivo porcine skin explant model
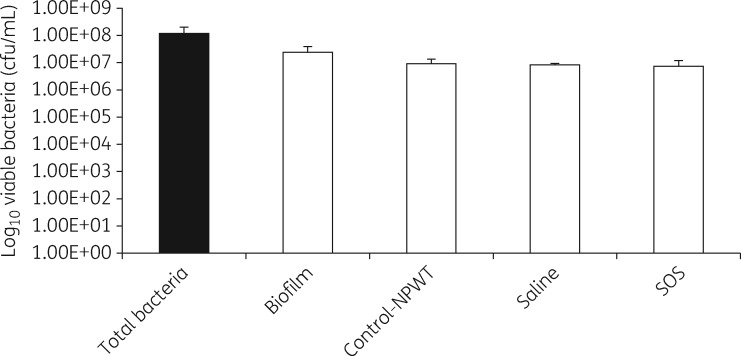
Levels of P. aeruginosa PA01 viable bacteria after 12 cycles of negative pressure therapy and instillation of saline or SOS are shown in Figure 2. The total bacterial bioburden (planktonic + biofilm) growing on the porcine skin explant was 8.0 log10 cfu/mL, of which 7.1 log10 cfu/mL were biofilm bacteria, as defined by being tolerant to incubation in 50× MIC gentamicin for 24 h at 37 °C. When porcine skin explants with mature P. aeruginosa PA01 biofilm were exposed to 12 cycles of negative pressure wound therapy alone without instillation of any solution (control for negative pressure wound therapy), which is equivalent to ‘pulsed or intermittent negative pressure wound therapy’, the level of P. aeruginosa PA01 cfu was reduced to 6.9 log10 cfu/mL. When the porcine skin explants were treated with negative pressure wound therapy with 12 cycles of instillation with saline with a 10 min exposure time, the level of P. aeruginosa PA01 bacteria was the same (6.9 log10 cfu/mL). Changing the instillation solution to SOS and using the same 12 cycles of instillation, the level of P. aeruginosa PAO1 bacteria was essentially the same as with saline instillation, with 6.8 log10 cfu/mL surviving the instillation treatment. In contrast, planktonic and biofilm bacteria were completely eradicated using the in vitro CDC biofilm reactor laboratory test.
Figure 2.
Treatment of porcine skin explants. 108 cfu of P. aeruginosa PA01 was inoculated onto porcine skin explants and after 3 days of growth at 37 °C, the average cfu of viable total bacteria or biofilm bacteria present before or after 12 cycles of 10 min instillations with saline or SOS solutions or only NPWT without instillation are shown. NPWT, negative pressure wound therapy.
As shown in Supplementary data Part S4, SEM of the wound area in the porcine skin explants demonstrated very thick continuous biofilm on untreated explants (panel A). SEM of explants treated with saline instillation (panel C) or explants treated only with negative pressure and no instillation (panel D) showed a reduction in biofilm structures, but substantial amounts of attached bacteria were still present. Explants treated with SOS instillation (panel B) also showed a reduction in the biofilm structure but persistence of attached bacteria.
The effect of SBMO against the microbial load and diversity of DFUs complicated by biofilm in vivo
Ten patients with chronic non-healing DFUs were enrolled. A total of 1306086 high-quality DNA sequences were generated (before = 623117, after = 682969), with a median of 61132 per sample-level data (range = 5702–168421). The OTUs identified 1976 unique taxa of which low-abundance OTUs were removed (<0.1%), leaving 124 OTUs for further analysis.
Confirmation of the presence or absence of biofilms in each DFU
Biofilms were visualized and confirmed in all 10 participants using SEM (Supplementary data Part S5). Biofilm architecture was graded using an arbitrary sliding scale from a score of 5 (heavy biofilm) to 0 (no biofilm) as previously reported.17 The median value of DFU biofilm architecture reduced from 4 pre-treatment (large microcolonies ∼100 cells, and a continuous film/matrix) to 3 post-treatment (large microcolonies ∼100 cells).
Microbial load of chronic non-healing DFUs complicated by biofilm
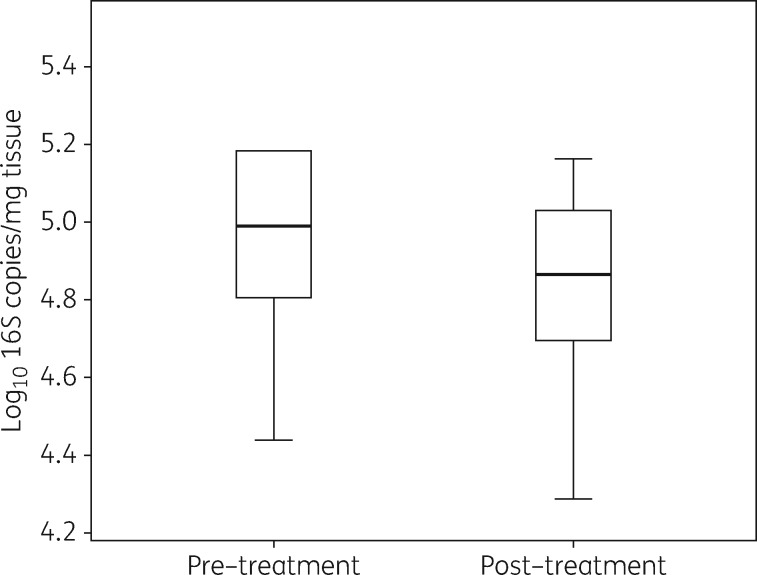
The application of SBMO for 15 min exposure daily, for 7 days, resulted in no change to the total microbial load (Figure 3) (median log10 microbial load pre-treatment = 4.9 log10 16S copies/mg of tissue, versus 4.8 log10 16S copies/mg of tissue, P = 0.43).
Figure 3.
Effects of SBMO pre- and post-treatment of 10 chronic non-healing diabetic foot ulcers. Box-and-whisker plots show the median log10 16S copies/mg of tissue values for all 10 patients.
Analysis of community richness and diversity of chronic non-healing DFUs treated with SBMO
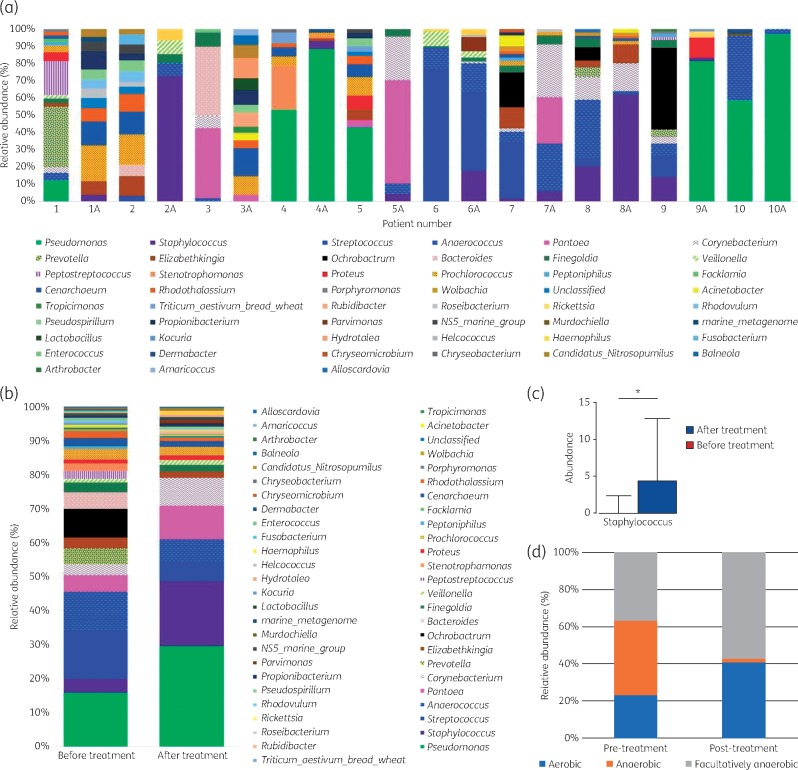
The most abundant OTUs contributing to >1% of the microorganisms within individual DFUs are shown in Figure 4(a); P. aeruginosa, S. aureus, Anaerococcus spp., Prevotella spp. and Streptococcus spp. were most commonly identified. The relative abundance of P. aeruginosa and S. aureus increased in all but one patient post-treatment with SBMO (Figure 4b), with pooled data from all samples identifying this to be statistically significant for the amount of Staphylococcus spp. DNA copies (P = 0.04). Only patient 9 seemed to experience a reduction in S. aureus levels (Supplementary data Part S6); however, a significant increase in P. aeruginosa was noted as a result (Figure 4a). Overall, there were increases in both aerobes and facultative microorganisms but these were reflected by a composite reduction in the relative abundance of anaerobic microorganisms (Figure 4d).
Figure 4.
Effects on microbial communities following treatment with SBMO. Pairwise comparisons of pre- and post-treatment (A) microbial communities at the genus level in microorganisms contributing >1% within each wound. Further analysis of pooled data depicts changes across all ten patients when all DNA copies are pooled and examined. (a) Relative abundance (%) for individual wound-level data pre- and post-treatment. (b) Pooled data (all DNA copies) from ten patients identifies the relative abundance (%) of microorganisms pre- and post-treatment. (c) Relative abundance of pooled sample data of Staphylococcus spp. DNA copies pre- and post-treatment identifies a statistically significant increase (P = 0.04). (d) Relative abundance (%) of pooled sample data detailing the aerotolerance of microorganisms. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Microorganisms contributing to >1% of microbial communities in individual DFUs and from pooled data were analysed by alpha and beta diversity measures. Chronic DFUs prior to treatment were rich and diverse, yet there were minimal changes to community richness (P = 0.3), diversity (P = 0.1) or community composition of DFUs post-treatment (P = 0.9) (Figure 5a–c).
Figure 5.
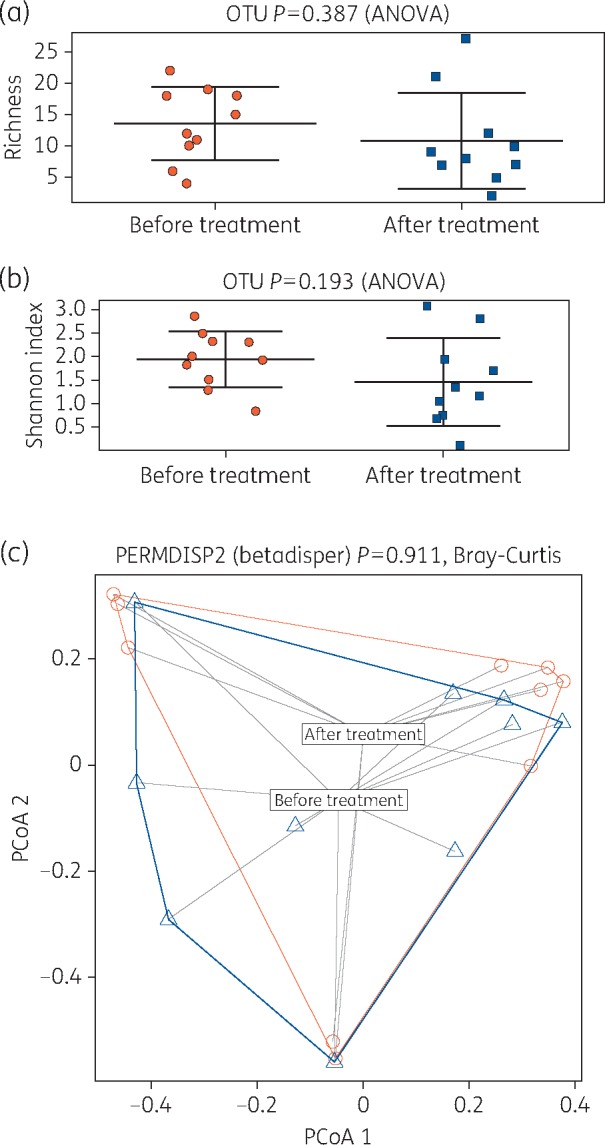
Alpha and beta diversity analysis pre- and post-treatment with SBMO. (a) The richness plot is a measure of the number of distinct or unique OTUs. These were reduced post-treatment but were non-significant. (b) The Shannon index is a measure of diversity that includes the number of unique microbial taxa and their relative evenness within each sample. Diversity of biofilm in diabetic foot ulcers post-treatment is reduced but non-significantly. (c) Principal coordinates analysis plots with Bray–Curtis distances between pre- and post-treatment samples identified that microbial communities are similar pre- and pre-treatment (blue triangles, pre-treatment; red circles, post-treatment). This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Discussion
Summary of key findings
We systematically tested the performance of topical antimicrobial solutions using short exposure times for in vitro and ex vivo models and an in vivo human trial. Our results suggest that the performance of these solutions is poor when challenged against mature biofilms using short exposure times that mimic real clinical use (i.e. 15 min application). Clinicians using topical antimicrobials to cleanse chronic wounds as a single therapy under the assumption of removing biofilm may therefore experience poor clinical outcomes. Clinicians should consider multifaceted strategies that include sharp debridement as the gold standard.16
What this study adds to the available evidence and new recommendations
There are many facets to the management of chronic wounds, with a large focus on managing wounds colonized or infected with either planktonic or biofilm phenotype microorganisms. While there is a plethora of data pertaining to the effectiveness of topical antimicrobials in vitro against both planktonic and sessile microorganisms, here we identify the inherent limitations of in vitro methodologies that fail to consider clinically relevant biofilm models when testing topical antimicrobials for use in wound care.9–11 In particular, in vitro models testing topical antimicrobial wound solutions have not considered the clinical applications of the products' intended use with regards to the time of exposure,11 and outcomes are often reported after 24 h exposure times. This does not reflect the typical clinical pattern of usage of these products or the ‘instructions for use’ explained in product inserts. Nor does the use of immature biofilms (early forming biofilm 24 h old) that have a less organized structure, a more active metabolism and a less pronounced stress response truly depict the complex, mature and highly tolerant biofilms identified in many chronic wounds.3,18
This may explain why some of the topical antimicrobials tested at clinically relevant times in this study performed poorly. Our in vitro model utilized two clinically relevant bacteria, P. aeruginosa and S. aureus, which have been noted as causes of delayed wound healing and as pathogens of infection.2,19 Testing the efficacy of solutions over a single 15 min exposure time in vitro, we identified great variability in test performances. In general, surfactant-based topical antimicrobials performed poorly (except for SBPHMB against P. aeruginosa) and were no more effective than normal saline (non-antimicrobial).
Traditional antiseptics such as CHX and PVP-I were highly effective, while new-generation solutions such as SOS were also highly efficacious. CHX is a cationic bisbiguanide with a broad-spectrum biocide that is active against both Gram-positive and -negative bacteria.20 Its primary action is against the negatively charged bacterial cell wall, leading to increased cell permeability resulting in cell death.20 The efficacy of CHX in reducing or eradicating single or multispecies biofilm has been demonstrated in vitro,21–23 with the combination of cetrimide and CHX producing enhanced antimicrobial activity (and anti-biofilm activity). One explanation for the effectiveness of CHX in vitro in this study may be the cationic surfactant properties of cetrimide, which has demonstrated the capacity to decrease the mechanical stability of biofilm (in addition to its proven bactericidal activity), but further work is required to elucidate further these effects in wound models.24
PVP-I also performed well in vitro, and as a broad-spectrum microbicide is capable of inactivating Gram-positive and -negative bacteria, bacterial spores, fungi, protozoa and several viruses.25 PVP enables the delivery of free iodine to a target cell membrane, where it destabilizes the structural components of cell membranes.25 It has demonstrated activity against biofilms in vitro;26,27 moreover, more recent in vitro data on the performance of a wound care-related PVP-I on multispecies biofilms using the CDC reactor have corroborated the results of this study.28
More recently, ‘newer generation’ topical solutions with antimicrobial properties such as SOS have been utilized as anti-biofilm therapies in wound care, even in the presence of a low evidence base. SOS contains as a primary ingredient hyperchlorous acid (which is not new generation), and only one in vitro study is available that used the concentrations of SOS found in current wound care solution formulations.10 Using a continuous flow tube reactor (to mimic the clinical scenario of a catheter) to grow mature 6-day-old P. aeruginosa PA01 biofilms, Sauer and colleagues10 utilized SOS at the same concentration (80 ppm) reported in this study, to achieve a 2.5 log10 reduction after 60 min exposure.
Our study identified that SOS could eradicate P. aeruginosa biofilms in addition to performing well against S. aureus biofilm. This was in contrast to the porcine skin model, which identified that SOS achieved only 0.77 log10 reduction against P. aeruginosa PA01 biofilms. Potential explanations to describe these results could be the two different strains of P. aeruginosa that were used for the study. The in vitro model utilized P. aeruginosa (ATCC® 25619™) and the porcine skin explant utilized P. aeruginosa (PA01, ATCC® BAA-47™). Sauer and colleagues also utilized P. aeruginosa (PA01, ATCC® BAA-47™). Interestingly, the use of the P. aeruginosa PA01 strain yielded results that identified a reduced effectiveness of SOS. It is possible that whilst our in vitro P. aeruginosa (ATCC® 25619™) strain readily formed a biofilm with the characteristic P. aeruginosa architecture, it did not develop a high-level biofilm-specific resistance,29 which may have arisen in the PA01 strain.
Other explanations for the different results observed for SOS in vitro versus the porcine skin model may be the surface the biofilms were formed on (i.e. the soft tissue dermal matrix of porcine skin, which more closely represented an actual wound bed compared with an abiotic polycarbonate disc). This may have contributed to alterations in microbial behaviour in response to the presence of biotic signals or organic material.30,31 Biofilms grown on biotic substrates or in vivo often do not display the morphological or architectural characteristics of those grown in vitro (e.g. mushroom structures and towers), which are important parameters that undoubtedly affect bacterial behaviour.32 Lastly unlike an abiotic surface, porcine skin has a striking similarity to human skin in terms of its structure and this is important given that microbial aggregates have been identified as not only forming on a wound surface, but also penetrating to deeper structures in a non-random distribution.33 In this scenario, any topical solution applied to a contact surface would have to penetrate a biofilm formed on that contact surface in addition to then penetrating between tissue cells. This in itself presents a greater challenge (than that already posed by biofilm tolerance mechanisms) and may contribute to the reduced effectiveness of topical antimicrobials.
Lastly, the performance of SBMO was tested on human tissue in an in vivo study of chronic non-healing DFUs. SBMO was applied daily for 15 min over a 7 day treatment period, with the results identifying no change in the total microbial load from tissue biopsies. Interestingly, our in vivo results identified a correlation between the poor performance of SBMO against P. aeruginosa and S. aureus that was also seen in vitro.
Next generation DNA sequencing was performed to understand the effects of SBMO on microbial communities in chronic non-healing DFUs. The relative abundances of both P. aeruginosa and S. aureus within the majority of DFUs increased post-treatment. Conversely, an overall reduction in the relative abundance of anaerobic microorganisms and low frequency taxa (microorganisms contributing <1% relative abundance) was noted; however, the total microbial loads within these wounds did not decrease. This potentially suggests that more dominant species such as Staphylococcus spp., or Pseudomonas spp., benefit from the increased nutrient availability caused by disruption to the microbial community (that resulted from removal of competing microorganisms),34 thus sustaining the microbial load within tissues.
Treatment with SBMO resulted in a reduction in the relative abundance of anaerobic microorganisms. Anaerobic microorganisms have been identified as part of polymicrobial communities cited for their involvement in delayed wound healing,35,36 as pathogens of infection in the diabetic foot37 and in biofilm production.38 In this instance reducing their numbers would seem like a positive step to reducing microorganisms with the potential to negatively impact the wound environment. Unfortunately, it is likely not this simple, particularly given the concomitant increases in pyogenic cocci (Staphylococcus spp.) and Gram-negative rods (P. aeruginosa), which are equal (if not greater) pathogens of infection.
To assess the overall effects of SBMO treatment on DFU microbiota (community richness, diversity, structure and composition) DNA sequence data were analysed using QIIME.39
Minimal reductions were seen in the number of OTUs (richness) and community diversity of chronic DFUs post-treatment. In a recent study by Loesche et al.,40 the temporal analysis of chronic DFUs found that patient samples that received systemic antimicrobial therapy had no alterations to species richness or diversity, and that antimicrobial exposure did not drive microbiota variation. Instead the data indicated that antimicrobial exposure disrupted the microbiota where antimicrobials were specifically directed to treat underlying wound infection. We found a similar pattern of events with our data, in that exposure to SBMO had some effects when we explored our samples individually. For example, sample 2 experienced a significant disruption to its microbiota whereby pre-treatment Staphylococcus spp. contributed <1% relative abundance; post-treatment this significantly increased to >65%. Similar patterns are seen across our data but it is not possible to infer if these changes would result in positive or negative effects to a wound. This intriguing aspect requires further correlation with longitudinal sampling that maps microbiota disruption to wound outcomes.
Our molecular-based data on the 16S gene, whilst informative in describing ‘who is there’, is unable to truly define ‘who is doing what’.41 In some wounds in which anaerobic microorganisms are acting synergistically with aerobic counterparts to increase pathogenicity or virulence in a chronic wound, their reduction may likely lead to positive effects. Conversely, and food for thought, any perturbations to the complex microflora seen within chronic wounds may lead to microbial dysbiosis. Of particular significance is the reduction in microbial diversity, which may directly contribute to pathogen selection and persistence.42 Longitudinal studies are required to determine whether the alterations to the microbial diversity of chronic non-infected wounds seen by using topical antimicrobials lead to future complications.
Limitations
The CDC biofilm reactor used in vitro was performed under flow allowing mature biofilms to form on the polycarbonate coupons; however, this abiotic surface does not reflect the complexity of human tissue and the absence of the host immune response. Secondly, most chronic wounds are contaminated with multiple species of bacteria3 and this study utilized single-species biofilms in vitro. That aside, our model tested clinically relevant exposure times against clinically relevant microorganisms involved in both chronic and infected wound types in screening the performance of topical antimicrobial solutions. qPCR was utilized to measure total microbial load in vivo;15 however, this method has limitations in its inability to differentiate live or dead bacteria. The log reductions noted in this study therefore represent the minimal response and we acknowledge that some of the bacteria detected by qPCR could be dead, resulting in a lower calculable efficacy.
Overall, the limitations in vitro were circumvented by the addition of an in vivo study. Costs to perform this study were a limiting factor in not being able to test a wider range of topical antimicrobials in vivo. Further studies incorporating a human in vivo design may be required to understand the efficacy of single products tested in the in vitro stage of this study against microbial biofilms. However, when taking the group of studies performed collectively, there is a strong correlation between exposure time and efficacy.
Conclusions
Polymicrobial communities forming biofilms in chronic wounds may have extended time periods to develop complex, highly tolerant communities that differ greatly from single-species biofilm models grown on polycarbonate coupons for 24–72 h. The discrepancies between the three different test parameters in this study raise an important question about in vitro testing for anti-biofilm therapeutics, in which results identifying potential effectiveness against biofilm differ markedly when the test parameters are changed. In vitro testing for anti-biofilm strategies could be used as a screening tool for identifying potential therapeutics that may perform well at the next stage of testing (i.e. when taken to animal models or to clinical studies). The effectiveness of an anti-biofilm therapeutic at this in vitro stage is however not absolute, yet for many medical device companies this is the only data available for use in the promotion of products. This highlights the limitations of clinicians relying solely on in vitro data. When using porcine explants and human in vivo tissue samples, our data are highly suggestive that the exposure time of topical antimicrobial wound solutions and irrigation solutions is too short and that exposure time is critical in determining the efficacy of these products. Clinicians using these topical antimicrobial solutions as a sole therapy under the assumption of killing or eradicating biofilm should consider adopting multifaceted strategies that include sharp debridement as the gold standard.
Supplementary Material
Acknowledgements
We acknowledge the support of South West Sydney LHD who presented the lead author with an early career research award, allowing the undertaking of this project as part of a PhD thesis.
Funding
This research was funded by two separate industry innovation research grants provided by Mundipharma Australia and Eloquest Healthcare Inc. USA, which contributed to the analysis of tissue using DNA sequencing and microscopy techniques.
Transparency declarations
M. M. and G. S. have consulted for Smith & Nephew to undertake work not associated with this study but pertaining to producing educational material on biofilms in chronic wounds. All other authors have none to declare.
Supplementary data
Supplementary data Parts S1–S6 are available as Supplementary data at JAC Online.
References
- 1. Guest JF, Ayoub N, McIlwraith T. et al. Health economic burden that wounds impose on the National Health Service in the UK. BMJ Open 2015; 5: e009283.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bjarnsholt T, Kirketerp-Møller K, Jensen PØ. et al. Why chronic wounds will not heal: a novel hypothesis. Wound Repair Regen 2008; 16: 2–10. [DOI] [PubMed] [Google Scholar]
- 3. Wolcott RD, Rees EJ, Koenig LD. et al. Analysis of the chronic wound microbiota of 2,963 patients by 16S rDNA pyrosequencing. Wound Repair Regen 2015; 24: 163–74. [DOI] [PubMed] [Google Scholar]
- 4. Stewart P, Costerton J.. Antibiotic resistance of bacteria in biofilms. Lancet 2001; 358: 135–8. [DOI] [PubMed] [Google Scholar]
- 5. Buckingham-Meyer K, Goeres DM, Hamilton MA.. Comparative evaluation of biofilm disinfectant efficacy tests. J Microbiol Methods 2007; 70: 236–44. [DOI] [PubMed] [Google Scholar]
- 6. Leid JG, Willson CJ, Shirtliff ME. et al. The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN-γ-mediated macrophage killing. J Immunol 2005; 175: 7512–8. [DOI] [PubMed] [Google Scholar]
- 7. Wolcott RD. A study of biofilm-based wound management in subjects with critical limb ischaemia. J Wound Care 2008; 17: 145–18. [DOI] [PubMed] [Google Scholar]
- 8. Malone M, Goeres DM, Gosbell I. et al. Approaches to biofilm-associated infections: the need for standardized and relevant biofilm methods for clinical applications. Expert Rev Anti Infect Ther 2017; 15: 147–56. [DOI] [PubMed] [Google Scholar]
- 9. Ortega-Peña S, Hidalgo-González C, Robson MC. et al. In vitro microbicidal, anti-biofilm and cytotoxic effects of different commercial antiseptics. Int Wound J 2016; 14: 470–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sauer K, Thatcher E, Northey R. et al. Neutral super-oxidised solutions are effective in killing P. aeruginosa biofilms. Biofouling 2009; 25: 45–54. [DOI] [PubMed] [Google Scholar]
- 11. Brackman G, De Meyer L, Nelis HJ. et al. Biofilm inhibitory and eradicating activity of wound care products against Staphylococcus aureus and Staphylococcus epidermidis biofilms in an in vitro chronic wound model. J Appl Microbiol 2013; 114: 1833–42. [DOI] [PubMed] [Google Scholar]
- 12. Castaneda P, McLaren A, Tavaziva G. et al. Biofilm antimicrobial susceptibility increases with antimicrobial exposure time. Clin Orthop Relat Res 2016; 474: 1659–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ngo QD, Vickery K, Deva AK.. The effect of topical negative pressure on wound biofilms using an in vitro wound model. Wound Repair Regen 2012; 20: 83–90. [DOI] [PubMed] [Google Scholar]
- 14. Yang Q, Phillips PL, Sampson EM. et al. Development of a novel ex vivo porcine skin explant model for the assessment of mature bacterial biofilms. Wound Repair Regen 2013; 21: 704–14. [DOI] [PubMed] [Google Scholar]
- 15. Malone MJK, Jensen SO, Gosbell IB. et al. Effect of cadexomer iodine on the microbial load and diversity of chronic non-healing diabetic foot ulcers complicated by biofilm in vivo. J Antimicrob Chemother 2017; 72: 2093–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wolcott RD, Rumbaugh KP, James G. et al. Biofilm maturity studies indicate sharp debridement opens a time-dependent therapeutic window. J Wound Care 2010; 19: 320–8. [DOI] [PubMed] [Google Scholar]
- 17. Han A, Zenilman JM, Melendez JH. et al. The importance of a multifaceted approach to characterizing the microbial flora of chronic wounds. Wound Repair Regen 2011; 19: 532–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hengzhuang W, Wu H, Ciofu O. et al. Pharmacokinetics/pharmacodynamics of colistin and imipenem on mucoid and nonmucoid Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 2011; 55: 4469–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sotto A, Lina G, Richard JL. et al. Virulence potential of Staphylococcus aureus strains isolated from diabetic foot ulcers: a new paradigm. Diabetes Care 2008; 31: 2318–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jones CG. Chlorhexidine: is it still the gold standard? Periodontol 2000 1997; 15: 55–62. [DOI] [PubMed] [Google Scholar]
- 21. Ruiz-Linares M, Ferrer-Luque CM, Arias-Moliz T. et al. Antimicrobial activity of alexidine, chlorhexidine and cetrimide against Streptococcus mutans biofilm. Ann Clin Microbiol Antimicrob 2014; 13: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ferran AA, Liu J, Toutain P-L. et al. Comparison of the in vitro activity of five antimicrobial drugs against Staphylococcus pseudintermedius and Staphylococcus aureus biofilms. Front Microbiol 2016; 7: 1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Arias-Moliz MT, Ferrer-Luque CM, González-Rodríguez MP. et al. Eradication of Enterococcus faecalis biofilms by cetrimide and chlorhexidine. J Endod 2010; 36: 87–90. [DOI] [PubMed] [Google Scholar]
- 24. Simões M, Pereira MO, Vieira MJ.. Effect of mechanical stress on biofilms challenged by different chemicals. Water Res 2005; 39: 5142–52. [DOI] [PubMed] [Google Scholar]
- 25. Kanagalingam J, Feliciano R, Hah JH. et al. Practical use of povidone-iodine antiseptic in the maintenance of oral health and in the prevention and treatment of common oropharyngeal infections. Int J Clin Pract 2015; 69: 1247–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hosaka Y, Saito A, Maeda R. et al. Antibacterial activity of povidone iodine against an artificial biofilm of Porphyromonas gingivalis and Fusobacterium nucleatum. Arch Oral Biol 2012; 57: 364–8. [DOI] [PubMed] [Google Scholar]
- 27. Oduwole KO, Glynn AA, Molony DC. et al. Anti-biofilm activity of sub-inhibitory povidone-iodine concentrations against Staphylococcus epidermidis and Staphylococcus aureus. J Orthop Res 2010; 28: 1252–6. [DOI] [PubMed] [Google Scholar]
- 28. Hoekstra MJ, Westgate SJ, Mueller S.. Povidone-iodine ointment demonstrates in vitro efficacy against biofilm formation. Int Wound J 2017; 14: 172–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mah T-F, Pitts B, Pellock B. et al. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 2003; 426: 306–10. [DOI] [PubMed] [Google Scholar]
- 30. Lebeaux D, Chauhan A, Rendueles O. et al. From in vitro to in vivo models of bacterial biofilm-related infections. Pathogens 2013; 2: 288–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Summerfield A, Meurens F, Ricklin ME.. The immunology of the porcine skin and its value as a model for human skin. Mol Immunol 2015; 66: 14–21. [DOI] [PubMed] [Google Scholar]
- 32. Gabrilska RA, Rumbaugh KP.. Biofilm models of polymicrobial infection. Future Microbiol 2015; 10: 1997–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fazli M, Bjarnsholt T, Kirketerp-Møller K. et al. Nonrandom distribution of Pseudomonas aeruginosa and Staphylococcus aureus in chronic wounds. J Clin Microbiol 2009; 47: 4084–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hibbing ME, Fuqua C, Parsek MR. et al. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol 2010; 8: 15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bowler PG, Armstrong DG.. Wound microbiology and associated approaches to wound management. Clin Microbiol Rev 2001; 14: 244–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dalton T, Dowd SE, Wolcott RD. et al. An in vivo polymicrobial biofilm wound infection model to study interspecies interactions. PLoS One 2011; 6: e27317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gerding DN. Foot infections in diabetic patients: the role of anaerobes. Clin Infect Dis 1995; 20 Suppl 2: S283–8. [DOI] [PubMed] [Google Scholar]
- 38. Wolcott R, Dowd S.. The role of biofilms: are we hitting the right target? Plast Reconstr Surg 2011; 127 Suppl 1: 28S–35S. [DOI] [PubMed] [Google Scholar]
- 39. Caporaso JG, Kuczynski J, Stombaugh J. et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010; 7: 335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Loesche M, Gardner SE, Kalan L. et al. Temporal stability in chronic wound microbiota is associated with poor healing. J Invest Dermatol 2017; 137: 237–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Malone M, Gosbell IB, Dickson HG. et al. Can molecular DNA-based techniques unravel the truth about diabetic foot infections? Diabetes Metab Res Rev 2017; 33: doi:10.1002/dmrr.2834. [DOI] [PubMed] [Google Scholar]
- 42. Flanagan JL, Brodie EL, Weng L. et al. Loss of bacterial diversity during antibiotic treatment of intubated patients colonized with Pseudomonas aeruginosa. J Clin Microbiol 2007; 45: 1954–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.