Abstract
Background
High throughput molecular screening techniques allow the identification of multiple molecular alterations, some of which are actionable and can be targeted by molecularly targeted agents (MTA). We aimed at evaluating the relevance of using this approach in the frame of Institut Curie Molecular Tumor Board (MTB) to guide patients with cancer to clinical trials with MTAs.
Patients and methods
We included all patients presented at Institut Curie MTB from 4 October 2014 to 31 October 2017. The following information was extracted from the chart: decision to perform tumour profiling, types of molecular analyses, samples used, molecular alterations identified and those which are actionable, and inclusion in a clinical trial with matched MTA.
Results
736 patients were presented at the MTB. Molecular analyses were performed in 442 patients (60%). Techniques used included next-generation sequencing, comparative genomic hybridisation array and/or other techniques including immunohistochemistry in 78%, 51% and 58% of patients, respectively. Analyses were performed on a fresh frozen biopsy in 91 patients (21%), on archival tissue (fixed or frozen) in 326 patients (74%) and on both archival and fresh frozen biopsy in 25 patients (6%). At least one molecular alteration was identified in 280 analysed patients (63%). An actionable molecular alteration was identified in 207 analysed patients (47%). Forty-five analysed patients (10%) were enrolled in a clinical trial with matched MTA and 29 additional patients were oriented and included in a clinical trial based on a molecular alteration identified prior to the MTB analysis. Median time between date of specimen reception and molecular results was 28 days (range: 5–168).
Conclusions
The implementation of an MTB at Institut Curie enabled the inclusion of 10% of patients into a clinical trial with matched therapy.
Keywords: molecular tumour board, tumour profiling, targeted therapy, clinical trials
Key questions.
What is already known about this subject?
Results of retrospective analyses of tumour molecular screening programmes and non-randomised clinical trials assessing the clinical utility of using high-throughput genomic technologies are encouraging yet it still remains to be demonstrated that using molecular profiling to guide therapy improves patient outcome in oncology.
What does this study add?
Our study reports the Institut Curie Molecular Tumor Board 3 years’ experience since its creation in October 2014 until the end of October 2017. We show the feasibility and efficacy of molecular testing to orient patients towards clinical trials in delays compatible with clinical practice.
How might this impact on clinical practice?
A minority of patients were treated with matched targeted therapy within clinical trials. Main challenges remain in patients’ selection, low rate of identified druggable molecular alterations, as well as the unavailability of a trial with a molecularly targeted agent matching the molecular alteration identified.
Introduction
Some genomic alterations that lead to cancer cells’ development are actionable: proteins resulting from these genomic alterations can be targeted by molecularly targeted agents (MTA).1 2 Recent technological advances such as next-generation sequencing (NGS) make possible the identification of actionable molecular alterations for matched MTA in time frames compatible with clinical practice.3 4 MTAs such as tyrosine kinase inhibitors and monoclonal antibodies are used to treat various types of cancer.5–9 MTAs can be highly effective in some tumour types, and recent studies have suggested that molecular alterations could be targeted across different tumour types.10 Results of retrospective analyses of tumour molecular screening programmes11 and non-randomised clinical trials12–14assessing the clinical utility of using high-throughput genomic technologies were encouraging although not confirmed in the SHIVA01 randomised trial that showed no statistically significant difference in progression free survival (PFS) between MTA and control arms.15 Nevertheless, patients who crossed over in SHIVA01 compared favourably with results16 obtained in von Hoff study and in MOSCATO01 with PFS on matched targeted therapy to PFS on last non-matched treatment ratio exceeding 1.3 in 37% of patients.12 13
The Institut Curie implemented a molecular screening programme for patients with recurrent and/or metastatic cancers. Its main objective is to guide patients to clinical trials based on tumours’ molecular alterations. Paediatrics experience of the molecular tumour board (MTB) being already published,17 we report here our single-centre experience focusing on adult patients.
Patients and methods
Institut Curie molecular screening programme started in October 2014. The objective of this programme is to detect actionable molecular alterations in order to guide patients to relevant clinical trials. Patients with recurrent and/or metastatic cancer treated at the Institut Curie or in partner hospitals were enrolled in the programme. The MTB meets on a weekly basis and regroups medical oncologists, radiologists, pathologists, geneticists and bioinformaticians. Patients are discussed at the MTB on medical oncologists’ requests. Informed consent with regard to the collection of tumour and blood samples and molecular analysis is obtained from patients within the institutional general consent.
For each patient presented at the MTB, eligibility for potential clinical trials is discussed in terms of performance status and biological functions. When a patient is considered eligible for a potential inclusion in a clinical trial, the MTB specifies the types of molecular analyses required, the need to perform an on-purpose biopsy and the possibility to perform one from accessibility point of view. If an on-purpose biopsy is preferred but not feasible, analyses are performed on the most recent available sample. Techniques used include NGS, comparative genomic hybridisation array (CGHa), immunohistochemistry (IHC) and Sanger sequencing (online supplementary material and methods). When necessary, tumour DNA extraction is performed after evaluation of tumour cells content to allow macrodissection of the sample in order to increase tumour cellularity. Samples containing ≥30% of tumour cells were considered suitable for DNA extraction for targeted NGS as well as CGHa. When tumour cell content is below 30% yet ≥10% only targeted NGS and/or IHC are performed.
esmoopen-2018-000339supp001.pdf (299.6KB, pdf)
Screening of mutations is performed by targeted NGS using several panels ranging from 23 to 80 genes of theranostic interest. Molecular alterations are selected as follows: (1) focal amplification and pathogenic mutation or variant of unknown signification validated by biologists for oncogenes; (2) homozygous deletion, heterozygous deletion associated to mutation/variant of unknown signification/loss of protein expression in IHC, mutation/variant of unknown signification associated to loss of protein expression in IHC or loss of protein expression in IHC for tumour suppressor genes.
Results of the molecular analyses are discussed during a subsequent MTB based on previously reported recommendations for the interpretation and prioritisation of molecular alterations.18 If one or several molecular alterations are considered actionable, a recommendation is given for inclusion in a clinical trial at Institut Curie or other cancer centres.
Clinical and demographic data are collected and include age, gender, type of specimen, tumour type, number of previous treatments received, techniques used for molecular analyses, molecular alterations identified and inclusion or not in a clinical trial.
Results
Patient characteristics
From 4 October 2014 to 31 October 2017, up to 736 patients were registered at the MTB (table 1).
Table 1.
Patient characteristics
| n (%) | Median (range) | |
| Gender | ||
| Female | 459 (62) | |
| Male | 277 (38) | |
| Age (years) | 60 (18–87) | |
| Number of previous lines of treatment | 2 (0–17) | |
| Chemotherapy | 1 (0–10) | |
| Targeted therapy | 0 (0–4) | |
| Hormonotherapy | 0 (0–6) | |
| Immunotherapy | 0 (0–3) | |
| Drug combinations | 0 (0–9) | |
| Tumour location | ||
| Breast | 130 (18) | |
| Lung | 92 (13) | |
| Ovary | 76 (10) | |
| Head and neck | 73 (10) | |
| Colorectal | 67 (9) | |
| Cervix | 41 (6) | |
| Prostate | 37 (5) | |
| Uterus | 32 (4) | |
| Pancreas | 30 (4) | |
| Brain | 22 (3) | |
| Bone | 17 (2) | |
| Soft tissue | 16 (2) | |
| Eye | 15 (2) | |
| Liver | 11 (2) | |
| Salivary gland | 11 (2) | |
| Urothelial | 11 (2) | |
| Stomach | 7 (1) | |
| Vulva | 7 (1) | |
| Skin | 6 (1) | |
| Appendix | 4 (1) | |
| Gall bladder | 3 (0.4) | |
| Pleura | 3 (0.4) | |
| Lacrimal gland | 2 (0.3) | |
| Oesophagus | 2 (0.3) | |
| Adrenal gland | 2 (0.3) | |
| Thymus | 2 (0.3) | |
| Blood | 1 (0.1) | |
| Kidney | 1 (0.1) | |
| Unknown | 14 (2) | |
| Total | 736 (100) | |
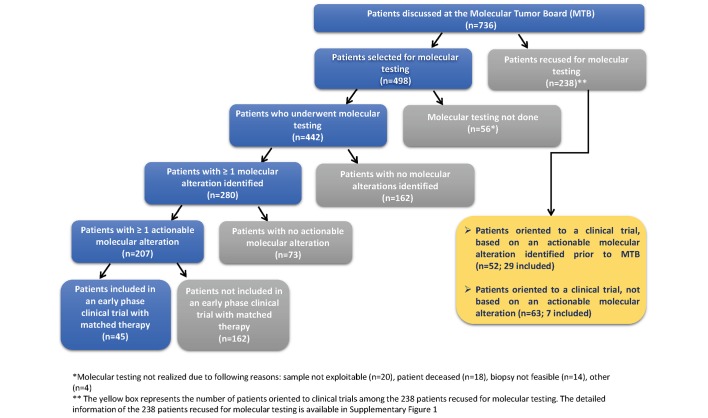
Patients were female in 62% of cases. Median age was 60 years (range 18–87). Patients had received a median of two previous lines of treatments mainly chemotherapies in the recurrent and/or metastatic setting. Most frequent tumours were breast, lung, ovarian, head and neck, and colorectal cancer (table 1). Of the 736 patients discussed, the MTB validated molecular screening for 498 patients (68%) (figure 1). For the remaining 238 patients (32%), molecular screening was not performed for: (1) patients oriented to a clinical trial based or not on an actionable molecular alteration identified prior to the MTB (n=115), and (2) patients not oriented to a clinical trial (n=123) for the following reasons: patients not eligible for a clinical protocol (n=38), biopsy required for analyses but not feasible (n=30), patients with stable disease (n=20), analysis already done or ongoing independently of the MTB (n=18), diagnosis not defined (n=5), tissue sample not available (n=4) and other reasons (n=8) (online supplementary figure 1).
Figure 1.
Flow chart of patients discussed in Institut Curie Molecular Tumor Board (MTB).
esmoopen-2018-000339supp002.pdf (908.8KB, pdf)
Molecular analyses
Molecular analyses were performed for 442 patients (60%) (figure 1). Analyses were performed on a fresh frozen biopsy in 91 patients (21%), on archival tissue (fixed or frozen) in 326 patients (74%) and on both archival and fresh frozen biopsy in 25 patients (6%) (online supplementary figure 2). When an on-purpose biopsy was requested, imaging-guided biopsies (for 88 patients) or resections (for 28 patients) were performed on liver (32%), skin (9%), lymph nodes (8%) peritoneum (7%), bone (6%), lung (4%), prostate (4%), breast (3%) and others localisations (27%). Molecular analyses were not realised in 56 patients due to the following main reasons: sample not exploitable (n=20), patient deceased (n=18), biopsy not feasible (n=14) and other reasons (n=4) (figure 1 and online supplementary figure 2). Among the 442 patients analysed, both NGS and CGHa were realised in 202 patients (46%), whereas NGS or CGHa alone was performed in 141 (32%) and 25 patients (5%), respectively. Seventy-four patients did not undergo NGS and CGH (17%) and were screened using other techniques including IHC, Sanger sequencing and/or microsatellites instability testing (online supplementary figure 3). Median time between the date of specimen reception and the molecular results was 28 days (range: 5–168). Median time between the date of the MTB (validation of indication of the molecular testing) and the molecular results confirmed in the second MTB was 35 days (range: 7–196).
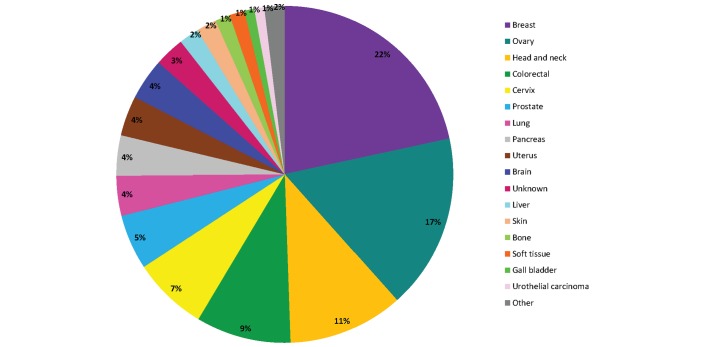
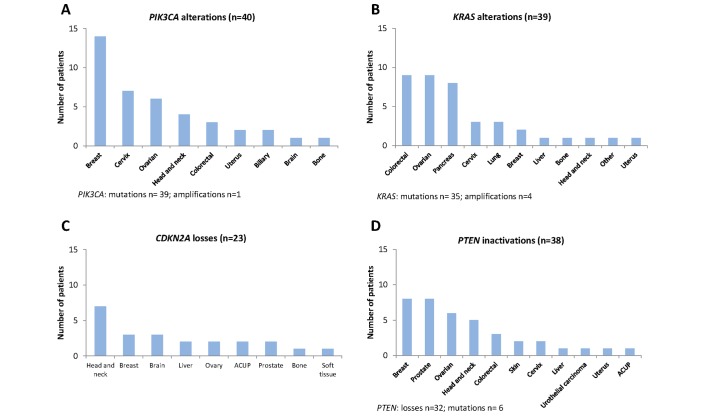
Of the 442 analysed samples, at least one molecular alteration was identified in 280 patients (63%), and 162 patients had no molecular alteration identified (37%). Of the 280 patients with at least one molecular alteration, 207 patients (74%) had at least one actionable molecular alteration identified (figure 1). Among the 207 patients with at least one actionable molecular alteration, most frequent tumour types included breast cancer (22%), ovarian cancer (17%), head and neck cancer (11%) and colorectal cancer (9%) (figure 2). Most frequent molecular alterations were present in the PIK3CA (19%), KRAS (19%), PTEN (18%) and CDKN2A (11%) genes (figures 3 and 4). The PI3K/AKT/MTOR pathway was the most frequently altered in the MTB population analysed (online supplementary figure 4).
Figure 2.
Tumour types of patients whose cancers harboured actionable molecular alterations in our series. ACUP, adenocarcinoma with unknown primary. Other: appendix (n=1), vagina (n=1), vulva (n=1), adrenal gland (n=1).
Figure 3.
Description of actionable molecular alterations identified. ACUP, adenocarcinoma with unknown primary.
Figure 4.
Frequency of molecular alterations per tumour type for KRAS mutations (A), PI3KCA mutations (B), CDKN2A losses (C) (loss=homozygous deletion), PTEN inactivations (D) (loss=homozygous deletion, or heterozygous deletion associated to mutation/variant of unknown significance/loss of expression in immunohistochemistry (IHC), or loss of expression in IHC without any other analysis). ACUP, adenocarcinoma with unknown primary.
Enrolment into clinical trials
Twenty-nine out of the 238 patients (12%) recused for molecular testing were oriented and enrolled in a clinical trial based on a molecular alteration identified prior to the MTB (figure 1). One hundred and sixty-two patients with an actionable molecular alteration were not enrolled in a clinical trial for the following reasons: patient’s death (n=34), no clinical trial available (n=30), patients lost to follow-up (n=25), exclusion criteria to enrolment in an early phase clinical trial (n=19), other approved treatment available (n=19), matched drug already received by the patient (n=10) and other reasons (n=25) (online supplementary figure 5).
Forty five out of the 442 patients who underwent molecular testing (10%) were included into a clinical trial (online supplementary table 1). The 45 patients enrolled in a clinical trial based on a molecular alteration identified in the frame of the MTB had received a median of two previous lines of treatments in the recurrent and/or metastatic setting and were predominantly females (67%). Median age was 58 years (range: 19–80). Most frequent tumour types included ovarian cancer (27%), breast cancer (13%), colorectal cancer (13%), cervical cancer (9%), prostate cancer (7%), uterine cancer (7%), pancreatic cancer (4%) and urothelial cancer (4%). Four patients out of the 37 evaluable patients for response (11%) experienced an objective response according to Response Evaluation Criteria In Solid Tumors (RECIST).
Discussion
Our study shows the feasibility of the implementation of molecular testing in the frame of a multidisciplinary MTB with an acceptable turnaround time. The median delay between the date MTB validating the molecular testing and the molecular results was 35 days (range: 7–196). These delays are in accordance with previous reports of molecular screening programmes with 32 days in the Princess Margaret IMPACT/COMPACT,19 and 26 days in the MD Anderson study from consent to genomic report.20 Median time between the date of specimen reception and the molecular results was 28 days (range: 5–168) comparable to the median time of 21 and 26 days (range: 14–27) from biopsy to molecular board reported in the MOSCATO01 study12 and SHIVA01 trials,4 respectively.
At least one molecular alteration was identified in 280 analysed patients (63%). An actionable molecular alteration was identified in 207 analysed patients (47%). Detection rate of actionable alterations was comparable to other studies ranging from 28% to 49% of tumours with actionable mutations.19–22 Differences can be discussed in terms of the number of genes screened within targeted NGS panels and additional techniques such as CGHa and IHC performed.
Ten per cent of analysed patients were enrolled in a clinical trial based on an actionable molecular alteration identified in the frame of the MTB. The rate of enrolment in clinical trials with MTAs based on tumour molecular profiles is consistent among the different screening programmes reported until the present ranging from 4% to 7% of patients screened in the different programmes.16 19 20 Higher rates were observed in recent clinical trials with 18% of patients enrolled in the MOSCATO01 study12 who were subsequently treated in MTA trials and 27% of patients screened in SHIVA01 who were treated in the frame of the trial15 and which could be explained with the availability of the MTAs in the frame of the trial, at least for SHIVA01.
One hundred and sixty-two patients with an actionable alteration were not enrolled in clinical trials mainly due to death or poor performance status. This point calls into question the fact that clinical trials are proposed to patients too late in the evolution of their disease; when their clinical condition is fragile and/or other organ deterioration impairs inclusion in clinical trials. In addition, many patients were not enrolled due to the unavailability of a trial with an MTA matching the molecular alteration identified.
It is important to point out that although the use of MTAs in oncology has shown improvement of patients’ survival in different cancer types when the corresponding actionable molecular alteration is present, it remains to be demonstrated that using molecular profiling in large prescreening series to guide therapy improves patient outcome in oncology.
Last, we did not find any actionable molecular alterations for 162 patients. Our molecular analyses were limited to panels ranging from 23 to 80 tested genes. We potentially would have found more alterations with a whole exome sequencing23 or with an RNA sequencing which detects viable fusion genes. NGS contributed to identify 90% of the nearly 10 000 gene fusions identified in cancer.24 25 The assessments of new biomarkers for which new clinical trials are available will be crucial to increase the 10% accrual following screening in the MTB. Homologous recombination-deficient patients may be eligible to Olaparib trials26 and POLE/POLD1 mutated, high mutational load patients or patients with microsatellite instability-high may be eligible to checkpoint inhibitor trials.27
Conclusion
The implementation of an MTB at Institut Curie enabled the inclusion of 10% of patients into a clinical trial with matched therapy. Although molecular screening is implemented and running in a routine basis in the frame of the MTB, main challenges remain in patients’ selection, low rate of identified druggable molecular alterations, as well as the unavailability of a trial with an MTA matching the molecular alteration identified. An optimal approach pending funding for molecular testing would be to detect molecular alterations in order to anticipate new lines of treatment in case of progression after approved strategies. On the opposite, some patients with actionable alterations were not enrolled for clinical deterioration or death which highlights the challenge to select patients for enrolment in clinical trials.
Footnotes
CB and CM contributed equally.
Funding: This work is supported by the SiRIC (Site de Recherche Intégré contre le Cancer INCa-DGOS-4654).
Competing interests: None declared.
Patient consent: Obtained.
Ethics approval: Institutional Review Board.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646–74. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 2. Tran B, Dancey JE, Kamel-Reid S, et al. . Cancer genomics: technology, discovery, and translation. J Clin Oncol 2012;30:647–60. 10.1200/JCO.2011.39.2316 [DOI] [PubMed] [Google Scholar]
- 3. Rodón J, Saura C, Dienstmann R, et al. . Molecular prescreening to select patient population in early clinical trials. Nat Rev Clin Oncol 2012;9:359–66. 10.1038/nrclinonc.2012.48 [DOI] [PubMed] [Google Scholar]
- 4. Le Tourneau C, Paoletti X, Servant N, et al. . Randomised proof-of-concept phase II trial comparing targeted therapy based on tumour molecular profiling vs conventional therapy in patients with refractory cancer: results of the feasibility part of the SHIVA trial. Br J Cancer 2014;111:17–24. 10.1038/bjc.2014.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gray SW, Hicks-Courant K, Cronin A, et al. . Physicians' attitudes about multiplex tumor genomic testing. J Clin Oncol 2014;32:1317–23. 10.1200/JCO.2013.52.4298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chapman PB, Hauschild A, Robert C, et al. . Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 2011;364:2507–16. 10.1056/NEJMoa1103782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Maemondo M, Inoue A, Kobayashi K, et al. . Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010;362:2380–8. 10.1056/NEJMoa0909530 [DOI] [PubMed] [Google Scholar]
- 8. Shaw AT, Kim DW, Nakagawa K, et al. . Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med 2013;368:2385–94. 10.1056/NEJMoa1214886 [DOI] [PubMed] [Google Scholar]
- 9. Hortobagyi GN. Trastuzumab in the treatment of breast cancer. N Engl J Med 2005;353:1734–6. 10.1056/NEJMe058196 [DOI] [PubMed] [Google Scholar]
- 10. Alexandrov LB, Nik-Zainal S, Wedge DC, et al. . Signatures of mutational processes in human cancer. Nature 2013;500:415–21. 10.1038/nature12477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tsimberidou AM, Wen S, Hong DS, et al. . Personalized medicine for patients with advanced cancer in the phase I program at MD Anderson: validation and landmark analyses. Clin Cancer Res 2014;20:4827–36. 10.1158/1078-0432.CCR-14-0603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Massard C, Michiels S, Ferté C, et al. . High-Throughput Genomics and Clinical Outcome in Hard-to-Treat Advanced Cancers: Results of the MOSCATO 01 Trial. Cancer Discov 2017;7:586–95. 10.1158/2159-8290.CD-16-1396 [DOI] [PubMed] [Google Scholar]
- 13. Von Hoff DD, Stephenson JJ, Rosen P, et al. . Pilot study using molecular profiling of patients' tumors to find potential targets and select treatments for their refractory cancers. J Clin Oncol 2010;28:4877–83. 10.1200/JCO.2009.26.5983 [DOI] [PubMed] [Google Scholar]
- 14. André F, Bachelot T, Commo F, et al. . Comparative genomic hybridisation array and DNA sequencing to direct treatment of metastatic breast cancer: a multicentre, prospective trial (SAFIR01/UNICANCER). Lancet Oncol 2014;15:267–74. 10.1016/S1470-2045(13)70611-9 [DOI] [PubMed] [Google Scholar]
- 15. Le Tourneau C, Delord JP, Gonçalves A, et al. . Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): a multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol 2015;16:1324–34. 10.1016/S1470-2045(15)00188-6 [DOI] [PubMed] [Google Scholar]
- 16. Belin L, Kamal M, Mauborgne C, et al. . Randomized phase II trial comparing molecularly targeted therapy based on tumor molecular profiling versus conventional therapy in patients with refractory cancer: cross-over analysis from the SHIVA trial. Ann Oncol 2017;28:590–6. 10.1093/annonc/mdw666 [DOI] [PubMed] [Google Scholar]
- 17. Pincez T, Clément N, Lapouble E, et al. . Feasibility and clinical integration of molecular profiling for target identification in pediatric solid tumors. Pediatr Blood Cancer 2017;64:e26365 10.1002/pbc.26365 [DOI] [PubMed] [Google Scholar]
- 18. Le Tourneau C, Kamal M, Tsimberidou AM, et al. . Treatment Algorithms Based on Tumor Molecular Profiling: The Essence of Precision Medicine Trials. J Natl Cancer Inst 2016;108:djv362 10.1093/jnci/djv362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stockley TL, Oza AM, Berman HK, et al. . Molecular profiling of advanced solid tumors and patient outcomes with genotype-matched clinical trials: the Princess Margaret IMPACT/COMPACT trial. Genome Med 2016;8:109 10.1186/s13073-016-0364-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Meric-Bernstam F, Brusco L, Shaw K, et al. . Feasibility of Large-Scale Genomic Testing to Facilitate Enrollment Onto Genomically Matched Clinical Trials. J Clin Oncol 2015;33:2753–62. 10.1200/JCO.2014.60.4165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Harris MH, DuBois SG, Glade Bender JL, et al. . Multicenter Feasibility Study of Tumor Molecular Profiling to Inform Therapeutic Decisions in Advanced Pediatric Solid Tumors: The Individualized Cancer Therapy (iCat) Study. JAMA Oncol 2016. 10.1001/jamaoncol.2015.5689 [DOI] [PubMed] [Google Scholar]
- 22. Hyman DM, Taylor BS, Baselga J, et al. . Implementing Genome-Driven Oncology. Cell 2017;168:584–99. 10.1016/j.cell.2016.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Postel-Vinay S, Boursin Y, Massard C, et al. . Seeking the driver in tumours with apparent normal molecular profile on comparative genomic hybridization and targeted gene panel sequencing: what is the added value of whole exome sequencing? Ann Oncol 2016;27:344–52. 10.1093/annonc/mdv570 [DOI] [PubMed] [Google Scholar]
- 24. Mertens F, Johansson B, Fioretos T, et al. . The emerging complexity of gene fusions in cancer. Nat Rev Cancer 2015;15:371–81. 10.1038/nrc3947 [DOI] [PubMed] [Google Scholar]
- 25. Reeser JW, Martin D, Miya J, et al. . Validation of a Targeted RNA Sequencing Assay for Kinase Fusion Detection in Solid Tumors. J Mol Diagn 2017;19:682–96. 10.1016/j.jmoldx.2017.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. National Library of Medicine. A Two-stage Simon Design Phase II Study for Non-BRCA MBC Patients With HRD Treated With Olaparib Single Agent (NOBROLA). https://clinicaltrials.gov/ct2/show/NCT03367689?term=HRD&rank=2
- 27. National Library of Medicine. TAPUR: Testing the Use of Food and Drug Administration (FDA) Approved Drugs That Target a Specific Abnormality in a Tumor Gene in People With Advanced Stage Cancer (TAPUR). https://clinicaltrials.gov/ct2/show/NCT02693535?term=mutational+load&rank=6
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
esmoopen-2018-000339supp001.pdf (299.6KB, pdf)
esmoopen-2018-000339supp002.pdf (908.8KB, pdf)