Recent progress defining the activation of inflammasomes
Keywords: ASC, caspase-1, cell death, NLRs, pathogens
Abstract
Inflammasomes are multimeric protein complexes that regulate inflammatory responses and pyroptotic cell death to exert host defense against microbes. Intracellular pattern-recognition receptors such as nucleotide-binding domain and leucine-rich repeat receptors (NLRs) and absent in melanoma 2 like receptors (ALRs) assemble the inflammasome complexes in response to pathogens and danger or altered-self signals in the cell. Inflammasome sensors, in association with an adaptor protein—apoptosis-associated speck-like protein containing a caspase-activation and -recruitment domain (ASC)—activate inflammatory caspase-1 to enable the release of inflammatory cytokines and induce cell death, conferring host defense against pathogens. Beyond infectious diseases, the importance of inflammasomes is implicated in a variety of clinical conditions such as auto-inflammatory diseases, neuro-degeneration and metabolic disorders and the development of cancers. Understanding inflammasome activation and its molecular regulation can unveil therapeutic targets for controlling inflammasome-mediated disorders. In this review, we describe recent advances in inflammasome biology and discuss its activation, structural insights into inflammasome assembly and mechanisms for the execution of pyroptosis.
Introduction
Innate immunity is the first line of defense to recognize pathogen-associated molecular patterns (PAMPs) and discriminate between self and non-self patterns. The innate immune system utilizes a fixed set of germline-encoded pattern-recognition receptors (PRRs) to engage PAMPs and eliminate the pathogens from the host system (1). The components of the bacterial cell wall, proteins of secretory systems and microbial nucleic acids are some of the conventional PAMPs recognized by the PRRs. Emerging evidence indicates that PRRs can also sense host-associated physiological aberrations called danger-associated molecular patterns (DAMPs) (2). The DAMPs can vary from host cell damage to the release of cellular contents such as ATP, perturbations in potassium and uric acid crystals.
The PRRs are evolutionarily conserved in most living organisms including plants. These PRRs are localized to various cellular compartments to engage microbial PAMPs arriving through various routes of entry. On the basis of their subcellular localization, PRRs are classified as membrane-anchoring PRRs and intracellular PRRs. Membrane-anchored PRRs include Toll-like receptors (TLRs) and C-type lectin receptors (CLRs) that are localized to the plasma membrane and endosomes to sense the PAMPs and DAMPs that are present in the extracellular compartment (3). Nucleotide-binding oligomerization domain (NOD) and leucine-rich repeat (LRR) receptors (NLRs), retinoic acid inducible gene I (RIG-I)-like receptors (RLRs) and absent in melanoma 2 (AIM2)-like receptors (ALRs) are intracellular PRRs localized within the cytoplasm (4).
NLRs and ALRs are distinct from the other PRRs in their domain architecture and function (5, 6). After the discovery of the first member of the NLR family—nucleotide-binding oligomerization domain-containing protein 1 (NOD1), also called NLR-family caspase-activation and -recruitment domain (CARD)-containing 1 (NLRC1)—multiple NLRs have been identified in humans and mice (2, 6). Upon activation, some of the NLRs assemble macro-molecular protein complexes called inflammasomes in the cytosol (7). This inflammasome assembly consists of NLR or ALR and a bipartite protein called apoptosis-associated speck-like protein containing a caspase-activation and -recruitment domain (ASC) to initiate caspase-1 activation. The NLR-nucleated assembly of the ASC specks in the cytosol is considered as a hallmark for inflammasome assembly (4). The activation of caspase-1 results in the proteolytic processing of pro-inflammatory cytokines IL-1β and IL-18 and execution of a type of inflammatory cell death called pyroptosis (8, 9).
Inflammasomes are widely recognized for their role in pathogen recognition and host defense. However, constitutive or deregulated activation of inflammasomes is associated with multiple autoimmune and auto-inflammatory disorders and with the development of cancers, neuro-degeneration and metabolic diseases. Therefore, the activation of inflammasome and its functions need to be tightly regulated in order to avoid unintended host tissue damage while inducing the pathogen-killing inflammatory responses. Thus, a better understanding of inflammasome biology enables therapeutic modulation of its activity in multiple disorders. In this review, we report recent advances pertaining to inflammasome activation, its structural assembly and functional effector mechanisms.
Inflammasomes
The ability of NLRs to assemble into inflammasomes was first ascribed to NLR-family pyrin domain (PYD)-containing 1 (NLRP1) (10, 11). On the basis of the current literature, five PRRs are confirmed to assemble the inflammasome complex after sensing their respective stimuli: NLRP1, NLRP3, NLRC4, AIM2 and pyrin (9, 12). These inflammasomes are considered as canonical inflammasomes as they convert pro-caspase-1 into the catalytically active capsase-1. The canonical inflammasome activation is complemented by a non-canonical pathway, which promotes activation of capsase-11 (in mice) and caspase-4 and caspase-5 (in humans) (13). These caspases in turn activate NLRP3 inflammasomes or caspase-1 (14). Caspase-1 converts substrates such as pro-IL-1β, pro-IL-18 and gasdermin-D (a pyroptosis inducer) into their active forms upon inflammasome activation (9). In addition to these inflammasome-forming receptors, other innate receptors of NLR and ALR families such as NLRP2, NLRP6, NLRP7, NLRP12, IFI16 (interferon-γ-inducible protein 16) and RIG-I are also reported to activate caspase-1 (15–20). The activation mechanism of each inflammasome is described below.
The NLRP1 inflammasome
NLRP1 was the first NLR reported to form an inflammasome complex (7). Whereas humans have only one NLRP1 protein, the mouse NLRP1 gene is highly polymorphic and encodes multiple paralogs of the NLRP1: NLRP1a, 1b and 1c. Human NLRP1 contains an N-terminal PYD, a NOD, LRRs, the function to find domain (FIIND) and a CARD at the C-terminus, but mouse NLRP1 molecules lack a PYD (Fig. 1A).
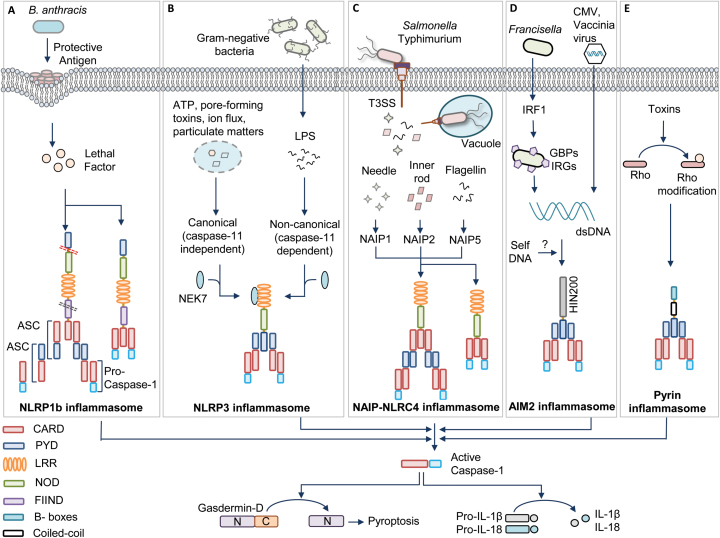
Fig. 1.
Inflammasome activation mechanisms. (A) Bacillus anthracis toxin containing protective antigen and lethal factor activate the human (contains pyrin domain) NLRP1 inflammasome by inducing cleavage at the N-terminal linker region (red dotted lane). Auto-proteolysis at the FIIND domain (black dotted lane) is also required for NLRP1 activation. The NLRP1 inflammasome activates caspase-1 via ASC-dependent recruitment to the inflammasome complex or by direct association with caspase-1 through CARD–CARD interactions. (B) Various pathogen-derived ligands (PAMPs) and physiological aberrations (DAMPs) activate NLRP3. Assembly of the NLRP3 inflammasome is categorized into canonical (caspase-11 independent) and non-canonical (caspase-11 dependent) inflammasome activation. NEK7 is an upstream activator of NLRP3 inflammasome assembly. (C) Pathogenic bacteria such as Salmonella enterica subspecies Typhimurium operate a T3SS to release effector proteins into the cytosol. These pathogen-associated proteins are recognized by a family of NAIPs and they further recruit NLRC4 to assemble the inflammasome complex. NLRC4 enables ASC-dependent or direct CARD–CARD interaction-dependent casapse-1 activation. (D) The DNA viruses such as cytomegalovirus (CMV) and vaccinia virus and intracellular bacteria such as Francisella release DNA during infection for activating the AIM2 inflammasome. IRF1 induces the expression of GBPs and IRGs to liberate DNA for AIM2 recognition. (E) Pyrin detects the modifications of Rho induced by Rho-inactivating toxins. All these inflammasomes recruit an adaptor protein called ASC, which contains PYD and CARD. ASC further brings caspase-1 to the inflammasome complex by CARD–CARD interactions for its activation. Activated caspase-1 drives the cleavage of pro-inflammatory cytokines pro-IL-1β and pro-IL-18 and also the processing of gasdermin-D protein for executing pyroptosis.
The NLRP1b allele of mice is polymorphic and exists as five different variants. Two of these variants are susceptible for Anthrax lethal toxin from Bacillus anthracis. The Anthrax toxin consists of a pore-forming protective antigen and a lethal factor that enters through the pore created by protective antigen (Fig. 1A) (21). The lethal factor activates NLRP1b by cleaving at its N-terminal domain. New studies suggest that the human NLRP1 and mouse NLRP1a and NLRP1b are activated by proteolysis within an N-terminal linker region (22, 23). This proteolysis processing is reported as a general mechanism of NLRP1 activation (22).
Evolutionary analysis of NLRP1 in primates indicates that the N-terminal linker region has evolved under positive selection, suggesting pathogen-elicited selective pressure at this region. However, lethal factor cleaves both lethal toxin-sensitive and lethal toxin-resistant forms of NLRP1, indicating the requirement for an additional activation step after the cleavage (23, 24). In addition, further studies reported that FIIND in the human NLRP1 and the lethal toxin-resistant mouse NLRP1b undergoes auto-proteolytic processing that leads to the assembly of the inflammasome (25).
A very recent study suggests that gain-of-function mutations in the human NLRP1 gene enhance its self-oligomerization and lead to spontaneous inflammasome activation in primary keratinocytes (26). These gain-of-function mutations interfere with the auto-inhibitory function of the PYD and disrupt interactions of PYD and LRRs of NLRP1 to relieve it from its inactive conformation (26). All these studies indicate that the spatial separation of PYD and LRRs and cleavage of NLRP1 at its N-terminus might relieve its auto-inhibition and perhaps cleavage at FIIND may separate the N-terminus of the NLRP1 from ASC complexes once the ASC speck formation is initiated.
The NLRP3 inflammasome
NLRP3 (also known as cryopyrin and NALP3) is the best-studied inflammasome. Initial observations suggested that NLRP3 triggers activation of caspase-1 under in vitro conditions (27). Further, three independent studies demonstrated NLRP3-dependent caspase-1 activation under physiological conditions and its role in host defense and immune responses (28–30). Studies from our laboratory and others demonstrated a critical role for the NLRP3 inflammasome in host responses against influenza and fungal infections (31–35). The NLRP3 is activated in response to a diverse array of PAMPs derived from various microbes and DAMPs such as nucleic acids, uric acid crystals, asbestos, aluminium hydroxide and silica (4, 8, 36).
The activation of NLRP3 inflammasomes in macrophages requires two stimuli. The first signal, called priming, is provided by an inflammatory stimulus such as TLRs and TNF-α receptor (TNFR) that leads to NF-κB-mediated NLRP3 expression and post-translational modifications of NLRP3 (37). In addition, this NF-κB signaling induces the expression of pro-IL-1β and pro-IL18 cytokines, which are the important effectors of inflammasome activation. The second signal, called the activation signal, provided by PAMPs or DAMPs initiates inflammasome assembly to promote caspase-1-mediated IL-1β and IL-18 release and pyroptosis. However, human monocytes require only a priming signal for IL-1β secretion and perhaps this priming signal is sufficient to mediate the activation of caspase-1 (38). The sensing of a variety of stimuli by NLRP3 with no evidence of direct interaction of these stimuli with NLRP3 strongly indicates that NLRP3 may be activated by a common cellular mechanism, which converges the signals from different stimuli (Fig. 1B).
Different mechanisms have been proposed for NLRP3 activation, including potassium efflux, calcium influx, reactive oxygen species (ROS), oxidized mitochondrial DNA, translocation of cardiolipin from the inner mitochondrial membrane, phagosome destabilization, perturbation in cell volume and pore-formation mechanisms driven by the host or bacteria (4, 39). Several studies suggest that potassium efflux is associated with many of these stimuli and low intracellular potassium is sufficient to activate the NLRP3 inflammasome assembly. This indicates that potassium efflux can be a downstream intracellular convergence point to activate NLRP3 (40). However, it is yet to be determined whether NLRP3 can directly sense a decrease in the levels of potassium or NLRP3 requires additional cellular proteins to sense the low levels of potassium. Several potassium-independent mechanisms of NLRP3 activation were also reported recently (41, 42). Imiquimod and CL097, small molecular ligands of TLR7, inhibit quinone oxidoreductase NQO2 and mitochondrial complex I. This results in the induction of ROS production and thiol oxidation, which triggers NIMA-related kinase 7 (NEK7)-dependent NLRP3 activation (41). In addition, a drop in NADH levels and mitochondrial ROS production leads to disruption of glycolytic flux in the cell that activates NLRP3 inflammasome and pyroptosis (42). Recent studies from our laboratory suggest that Z-DNA-binding protein 1/DNA-dependent activator of interferon regulatory factors (ZBP1/DAI), the apoptosis adaptor protein Fas-associated death domain (FADD), caspase-8 and Toll or interleukin-1 receptor containing adaptor inducing interferon-β (TRIF) act as apical mediators of canonical and non-canonical NLRP3 activation (43–46). In addition, recent reports identified an unexpected role for NEK7, an important protein in the formation of mitotic spindles and separation of centrosomes, for the activation of NLRP3 in response to both canonical and non-canonical stimuli (Fig. 1B) (47–49). NEK7 is required downstream of potassium efflux, physically interacts with NLRP3 and is required for NLRP3 oligomerization. Surprisingly, the kinase activity of NEK7 is not important for NLRP3 activation, but its catalytic domain is vital for NLRP3 interaction. In addition, mitotic cells show reduced inflammasome activation compared with the cells in interphase (49). This suggests that NEK7 enables a regulatory switch to balance cell division and inflammasome activity.
Current studies suggest that post-translational modifications of NLRP3, such as ubiquitination and phosphorylation, play an important role in NLRP3 activation. NLRP3 is reportedly ubiquitinated at its NOD and LRRs before its activation (50). Removal of ubiquitin chains by a deubiquitinase, BRCA1/BRCA2-containing complex subunit 3 (BRCC3), is required for the activation of the NLRP3 inflammasome (50). The phosphorylation of NLRP3 is also reported to regulate its function. Protein tyrosine phosphatase, non-receptor type 22 (PTPN22) dephosphorylates NLRP3 leading to efficient activation of NLRP3 inflammasome and IL-1β release (51). These studies suggest that the priming of NLRP3 expression is immediately followed by its ubiquitination and/or phosphorylation to regulate aberrant activation of NLRP3. More studies are needed to further understand the intricate mechanisms of NLRP3 activation.
The NLRC4 inflammasome
Initial reports on NLRC4 indicated its similarity to apoptotic protease-activating factor (APAF1). Mouse NLRC4 is activated by bacterial flagellin to assemble the inflammasome complex (52–54). Further studies showed that NLRC4 is also activated by the inner rod and needle proteins of the type III secretion system (T3SS) of bacteria (Fig. 1C) (55, 56). Similar to NLRP3, the bacterial ligands do not directly interact with NLRC4.
It was later identified that proteins belonging to the NLR-family apoptosis inhibitory proteins (NAIPs) are the sensors acting upstream of NLRC4 inflammasome assembly. NAIPs directly bind to the pathogen ligands such as flagellin (NAIP5 and NAIP6) or rod and needle proteins of the T3SS (NAIP1 and NAIP2) and recruit NLRC4 to assemble the inflammasome complex (57). Interestingly, the ligand sensing by NAIPs is conferred by the NOD, but not the LRR domain, which was considered to be essential for ligand binding (58). Humans have only one NAIP protein, and it senses the needle protein of the bacterial T3SS but does not respond to the rod proteins or flagellin (56, 59). In contrast, a recent report showed that human NAIP has two isoforms and one of the isoforms expressed in human monocyte-derived macrophages responds to Salmonella flagellin (60).
The CARD domain of NLRC4 can directly interact with the CARD domain of caspase-1 in the absence of ASC (Fig. 1C). This direct interaction of NLRC4 with caspase-1 partly explains its ability to activate pyroptosis independently of ASC (61). NLRC4 also recruits caspase-8 into its inflammasome complex, which is speculated to induce cell death functions of NLRC4 (62). The presence of ASC in the NLRC4 complex enhances NLRC4-mediated IL1-β and IL-18 release by aggregating large ASC specks. It is reported that the activating mutation in the nucleotide-binding domain (NBD), a subdomain of NOD, of the NLRC4 leads to spontaneous inflammasome activation that causes recurrent macrophage-activation syndrome (63).
The phosphorylation of the highly conserved S533, mediated by the kinase PCKδ, is reported to be required for NLRC4 inflammasome assembly during Salmonella typhimurium infection (64). However, another report argues that PKCδ is dispensable for NLRC4 activation (65). A recent study found that the phosphorylation of NLRC4 at S533 occurs upstream of flagellin detection by NAIP5 (66). This suggests that the NLRC4 activation requires a two-step mechanism where phosphorylation of S533 primes the NLRC4 for its subsequent activation by NAIP5.
The AIM2 inflammasome
AIM2 is a cytosolic receptor for double-stranded DNA (dsDNA) and assembles an inflammasome complex to activate caspase-1 (67–70). The discovery of AIM2 has been made based on the fact that the DNA from host and microbes can trigger ASC-dependent caspase-1 activation, which is independent of NLR signaling (71). AIM2 consists of a PYD and a hematopoietic interferon-inducible nuclear protein with a 200-amino acid repeat (HIN200) domain. The positively charged surface of the HIN domain binds to the DNA and the PYD recruits ASC to assemble the inflammasome complex (67, 68, 70).
Various DNA viruses including vaccinia virus and cytomegalovirus and intracellular bacterial infections such as Listeria monocytogenes, Francisella tularensis, Streptococcus pneumonia and Mycobacterium tuberculosis activate the AIM2 inflammasome to orchestrate host defense (Fig. 1D) (72–75). In the case of intracellular bacterial infections, type I interferon signaling contributes to the activation of the AIM2 inflammasome (72, 76). Molecular mechanistic studies from our laboratory and others show that Francisella infection induces type I interferon-dependent interferon regulatory factor-1 (IRF1) expression that further induces the expression of guanylate-binding proteins (GBPs) and immunity-related GTPases (IRGs) for promoting the lysis of the bacteria to liberate DNA ligands (Fig. 1D) (77–79).
Apart from responding to infectious agents, AIM2 is also associated with several other human diseases. Increased expression of AIM2 is associated with auto-inflammatory diseases such as systemic lupus erythematosus, psoriasis and abdominal aortic aneurysm (80–82). Recognition of self-DNA in the cytosol of keratinocytes by AIM2 drives IL-1β release and auto-inflammation in psoriasis (81). However, a reduction in AIM2 expression is associated with prostate and colorectal cancer (83, 84). Further studies from our laboratory and others indicated that AIM2-deficient mice are highly susceptible to colorectal cancer and this is because of the role of AIM2 in restraining stem cell proliferation (83). Further, a new study showed that PKM2, a pyruvate kinase that catalyses a final rate limiting step of glycolysis, promotes AIM2 and NLRP3 inflammasome activation in sepsis (85). In addition, a recent study shows that AIM2 mediates caspase-1-dependent cell death in response to radiation-induced DNA damage and the AIM2-deficient mice are protected from radiation-induced gastrointestinal syndrome and hematopoietic failure (86). Another study showed that an inhibitor of HIV aspartyl protease, Nelfinavir, disrupts nuclear envelope integrity. This leads to the release of DNA into the cytosol, which activates AIM2 inflammasomes (87). These additional roles of AIM2 suggest the possibility that AIM2 could sense self-DNA to regulate cellular functions.
The pyrin inflammasome
Pyrin is encoded by the gene MEFV, mutations in which are associated with an auto-inflammatory disease called familial Mediterranean fever (FMF) (88, 89). The mouse pyrin molecule consists of a PYD, two B-boxes and a coiled-coil domain, and the human pyrin molecule consists of an extra C-terminal domain called the B30.2 domain. Some of the mutations in the B30.2 domain are associated with FMF.
In initial studies, pyrin was recognized as a negative regulator of inflammasomes and secretion of IL-1β (90, 91). However, Chae et al. showed that FMF is caused by gain-of-function mutations in the pyrin-encoding gene Mefv (88, 89). Supporting this evidence, Gavrilin et al. (92) reported that type VI secretion system (T6SS) effectors of Burkholderia cenocepacia activates pyrin inflammasome. In addition, recent studies demonstrate that the pyrin inflammasome is triggered by Rho-modifying toxins that are produced by bacteria, such as Clostridium difficile (TcdB), Histophilus somni (IbpA), Vibrio parahemolyticus (VopS), Clostridium botulinum (C3) and Burkholderia cenocepacia (93, 94). These toxins induce covalent modifications in the Rho switch I region such as glycosylation, adenylation, ADP ribosylation and deamination (Fig. 1E) (93, 94). Pyrin senses these modifications instead of directly interacting with Rho. Rho modifications are essential for other cellular functions and perhaps pyrin is repressed under these homeostatic conditions.
Multiple pathways that inhibit the pyrin inflammasome have been identified including pyrin phosphorylation, cytoskeletal alterations and the mevalonate pathway. Two recent studies indicate that Rho activates the serine threonine kinases protein kinase N1 (PKN1) and PKN2 and they in turn phosphorylate pyrin (95, 96). Further, 14-3-3 proteins bind to the phosphorylated form of pyrin to regulate its activation. A mutation at phosphorylation site S242 of pyrin, which attenuates 14-3-3 binding, is reported to cause pyrin-associated auto-inflammation with neutrophilic dermatosis (95). A direct interaction of pyrin with actin has also been reported, suggesting a possibility for the sensing of Rho-mediated downstream events by pyrin (97). A recent study demonstrated that a mutation of WD-repeat-containing protein 1 (Wdr1), which encodes a factor that regulates actin depolymerization, causes pyrin-mediated auto-inflammation and thrombocytopenia (98). Another study identified negative regulation of pyrin inflammasome through protein geranylgeranylation, which is a protein post-translational modification catalyzed by geranylgeranyl transferase I (GGTase I). Perturbations in the mevalonate pathway that generate substrates for protein geranylgeranylation or absence of GGtase I lead to constitutive activation of pyrin inflammasome (99).
Pyrin is also categorized as a member of the tripartite motif (TRIM) family and these proteins actively participate in autophagy. Recent evidence indicates that the B30.2 domain is involved in recognizing autophagy-targeted cargo and participates in the autophagic degradation of NLRP1, NLRP3 and caspase-1 (100). However, the understanding of precise regulatory mechanisms of the pyrin inflammasome needs further studies.
Structural insights into inflammasome activation and assembly
Research done over the last decade in the inflammasome field described the signals that trigger inflammasome assembly. However, the structural mechanism of inflammasome assembly is poorly understood, and studies are emerging to describe the structure of the inflammasome.
It was proposed that inflammasomes attain a wheel-shaped structure similar to the apoptosome complex because of their functional similarity (39). Recent studies used negative-stain cryo-electron microscopy (cryo-EM) of purified NLRC4 in association with NAIP2–flagellin or NAIP5–PrgJ (a rod protein) to describe inflammasome assembly (101–104). These studies show that the NLRC4 inflammasome attains a wheel-like or disc-like structure with 10–12 protomers unlike the apoptosome, which is a heptameric wheel structure.
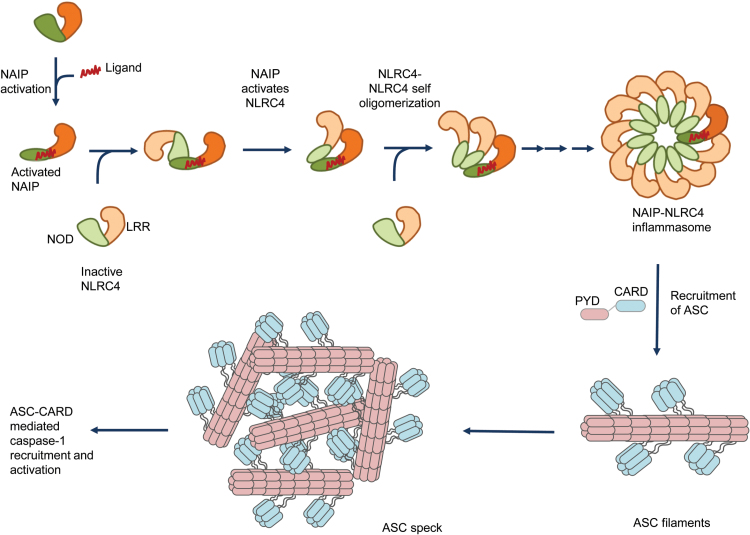
Typically, the binding of NAIPs to their respective ligand initiates recruitment of the NLRC4 protomers to NAIPs and NAIP nucleates NLRC4 oligomerization (Fig. 2). Assembly of wheel-shaped inflammasomes requires NAIP–NLRC4 and NLRC4–NLRC4 interactions, and these interactions are predominantly mediated by the NOD of these proteins (Fig. 2). On the basis of these studies, activated NAIP binds to an NLRC4 protomer and induces conformational changes in NLRC4 that make its oligomerization surface accessible for the subsequent NLRC4 protomer interaction to propagate the self-oligomerization process. These structures of the NLRC4 inflammasome lack CARDs, which precludes the understanding of their spatial orientation in the inflammasome complex.
Fig. 2.
Assembly of the NLRC4 inflammasome complex and ASC specks. Bacterial ligands such as flagellin, rod and needle proteins of the T3SS bind to NAIPs, which relieves the auto-inhibitory conformation and enables the accessibility of the oligomerization/interacting surface. The interacting surface of NAIP acts as a scaffold and recruits inactive NLRC4. The binding of NLRC4 monomer to the NAIP leads to the fully activated conformation of the NLRC4. The fully activated NLRC4 in turn exposes its interaction surface to further recruit another NLRC4 protomer to progress the self-oligomerization process. This chain of co-operative recruitment assembles a wheel- or disc-shaped inflammasome complex, which has inner ring of NODs and an outer ring of LRR domains. The available structures for the NLRC4 inflammasome lack CARD domains, which preclude the prediction of its orientation in the inflammasome complex. The activated inflammasome recruits ASC, an adaptor protein. ASC further undergoes self-polymerization to form filamentous structures. These ASC filaments aggregate to form macro-molecular complexes known as ASC specks. The exposed CARD domains of ASC in turn recruit caspase-1 that also forms filamentous structures (not shown). The recruitment of caspase-1 to the ASC filaments leads to caspase-1 activation.
These proposed structures provide the basis for understanding the process of inflammasome assembly. The importance of the NOD in this assembly is highlighted by the fact that the NOD is conserved amongst NLRs and mutations within the NOD are highly associated with auto-inflammatory disorders. Future studies should investigate (i) why NAIPs do not undergo self-oligomerization, (ii) how the CARD is arranged in the inflammasome wheel-like structure and (iii) whether all inflammasome-forming NLRs attain a similar structure.
Interestingly, previous literature suggests that NLRC4 physically interacts with NLRP3 and both are present in the same inflammasome complex during Salmonella infection (105, 106). Our recent study also indicates that the AIM2 and NLRP3 activation during Aspergillus infection leads to the assembly of a single inflammasome platform (107). The interaction of different NLRs indicates the possibility that multiple NLRs might be recruited to a single inflammasome complex during infections. AIM2 does not contain a NOD that induces self-oligomerization but it still assembles into an inflammasome complex. Therefore, it was proposed that DNA bound to the HIN domain of AIM2 acts as a scaffold to assemble the AIM2 inflammasome (108). A recent study suggests that the HIN domain of AIM2 forms filament-like structure with dsDNA (109).
Inflammasome assembly recruits ASC to the complex through PYD–PYD interaction, and it further recruits caspase-1 through CARD–CARD interactions (Fig. 2). The recruitment of ASC to the inflammasome complex induces macro-molecular aggregate structures of 1–2 μM in size named ASC puncta or specks (4, 110, 111). ASC specks are formed after receptor activation and are released into the extracellular space to further enhance inflammatory responses.
Structural studies by cryo-EM and solid-state nuclear magnetic resonance spectroscopy indicate that ASC forms filament-like structures and through the PYD it can further form long helically shaped filaments (111). ASC specks promote recruitment of caspase-1 to the filaments through CARD-mediated interactions (112). A recent study suggests that mutations in the CARD of ASC led to the formation of ASC filaments but not ASC specks (113). Further, ASC speck formation is required for efficient processing of IL-1β but dispensable for the activation of gasdermin-D and pyroptosis induction (113).
In summary, ligands bound to the NAIP trigger assembly of the NLRC4 inflammasome that recruits the ASC and caspase-1 to generate the macro-molecular ASC specks, which mediate robust cellular responses.
Recent insights into the mechanisms of pyroptotic cell death
Canonical inflammasome-induced caspase-1 activation or the non-canonical inflammasome-induced caspase-4, caspase-5 (humans) and caspase-11 (mice) triggers an inflammatory form of cell death called pyroptosis (114). Pyroptosis is a lytic form of programmed cell death characterized by cell swelling and release of cellular contents through lysis. Pyroptosis is initiated in response to sensing of pathogens or host-derived perturbations.
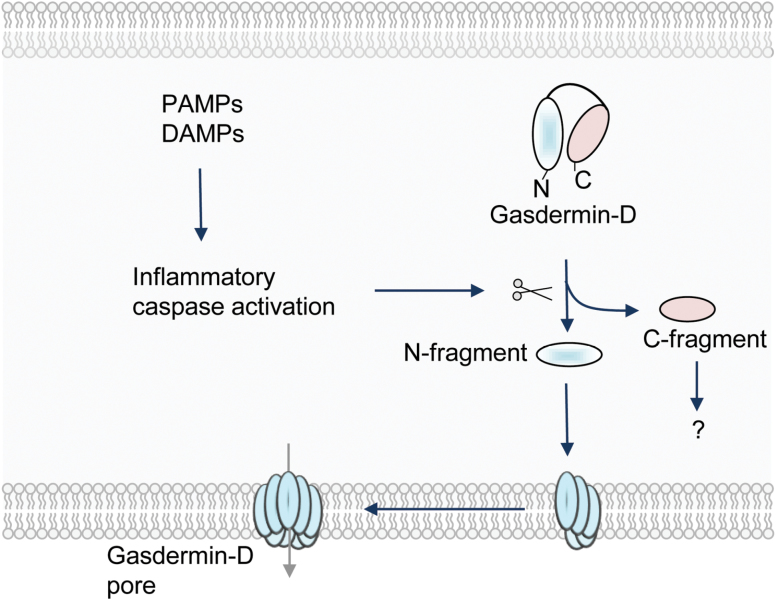
The molecular mechanisms that mediate pyroptosis were poorly understood for a long time. Recent studies show that the proteins from the gasdermin family execute inflammatory caspase-induced pyroptosis (13, 115). The gasdermin gene family is highly conserved in vertebrates and consists of four paralogous genes in humans (GSDMA, GSDMB, GSDMC and GSDMD), whereas mice lack Gsdmb (116). All gasdermin family members share a similar N-terminal domain. Gasdermin-D is a substrate for inflammatory caspases and is cleaved between the N-terminal and C-terminal domains (Fig. 3). The N-terminal domain of gasdermin-D is sufficient to induce pyroptotic cell death, and the C-terminal domain blocks the cell death function through auto-inhibition (Fig. 3) (115, 117).
Fig. 3.
Induction of pyroptosis mediated by gasdermin-D. Pathogen-derived PAMPs or physiological aberrations (DAMPs) activate inflammatory caspases. These caspases cleave gasdermin-D to separate its N- and C-terminal domains. The N-terminal domain of gasdermin-D is targeted to the membrane and assembles large, permeable pore complexes in the plasma membrane of the cell to induce pyroptosis.
Biochemical and structural studies have further unraveled the mechanism of gasdermin-mediated cell death. These studies show that gasdermin family members (GSDMD, GADMA3 and GSDMA) bind membrane lipids and exhibit membrane-disrupting cytotoxicity (118–120). Dye-loaded liposome-based assays indicated that the N-terminal domain of gasdermin-D targets liposome membranes to form pores sized ~10–20 nm, whereas the C-terminal domain or full-length gasdermin-D remains soluble (Fig. 3). These studies confirmed that the pyroptotic death is executed through the recruitment of the N-terminal domain of gasdermin-D to the plasma membrane to assemble large pores that impair cell membrane integrity. Ding et al. (119) further show that the pores formed by most gasdermin proteins contain 16 symmetric protomers.
In addition, the crystal structure of GSDMA3 showed the auto-inhibitory conformation of gasdermin domains and explains their inhibition in the absence of caspase-dependent cleavage. The understanding of gasdermin function and its mechanism of action will open up new therapeutic targets for controlling inflammatory diseases.
Conclusions
Inflammasome biology is a rapidly expanding field and the past 2–3 years have provided a tremendous advancement in understanding of inflammasome activation and function. Most importantly, studies detailing the structural components of inflammasome assembly, the mechanism of pyroptosis induction and genetic mutations in the NLRs associated with disease progression have expanded the existing knowledge about inflammasomes. These exciting findings have also raised many new questions in the field. However, the functions and specific ligands for many NLRs still remain unknown. The interplay between NLR activation and cellular homeostasis and its impact on physiological functions is still under investigation. Furthermore, ambiguities about receptors that activate inflammasome-complex formation still exist in the field. Therefore, in the coming years, it will be crucial to understand receptor activation mechanisms and the receptor structural requirements for ligand recognition to design novel immunotherapies.
Funding
US National Institutes of Health (AI101935, AI124346, AR056296 and CA163507 to T.-D.K.); the American Lebanese Syrian Associated Charities (to T.-D.K.).
Acknowledgements
We apologize to all the investigators whose research could not be appropriately cited because of the journal’s space limitations. S.K. and T.-D.K. contributed to the writing of this review.
Conflict of interest statement: The authors declared no conflict of interest.
References
- 1. Takeuchi O., Akira S. 2010. Pattern recognition receptors and inflammation. Cell 140:805. [DOI] [PubMed] [Google Scholar]
- 2. Schroder K., Tschopp J. 2010. The inflammasomes. Cell 140:821. [DOI] [PubMed] [Google Scholar]
- 3. Brubaker S. W., Bonham K. S., Zanoni I., et al. 2015. Innate immune pattern recognition: a cell biological perspective. Annu. Rev. Immunol. 33:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Man S. M., Kanneganti T. D. 2015. Regulation of inflammasome activation. Immunol. Rev. 265:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cridland J. A., Curley E. Z., Wykes M. N., et al. 2012. The mammalian PYHIN gene family: phylogeny, evolution and expression. BMC Evol. Biol. 12:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Harton J. A., Linhoff M. W., Zhang J., et al. 2002. Cutting edge: CATERPILLER: a large family of mammalian genes containing CARD, pyrin, nucleotide-binding, and leucine-rich repeat domains. J. Immunol. 169:4088. [DOI] [PubMed] [Google Scholar]
- 7. Martinon F., Burns K., Tschopp J. 2002. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell 10:417. [DOI] [PubMed] [Google Scholar]
- 8. Kanneganti T. D., Lamkanfi M., Núñez G. 2007. Intracellular NOD-like receptors in host defense and disease. Immunity 27:549. [DOI] [PubMed] [Google Scholar]
- 9. Man S. M., Kanneganti T. D. 2016. Converging roles of caspases in inflammasome activation, cell death and innate immunity. Nat. Rev. Immunol. 16:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bertin J., Nir W. J., Fischer C. M., et al. 1999. Human CARD4 protein is a novel CED-4/Apaf-1 cell death family member that activates NF-kappaB. J. Biol. Chem. 274:12955. [DOI] [PubMed] [Google Scholar]
- 11. Inohara N., Koseki T., del Peso L., et al. 1999. Nod1, an Apaf-1-like activator of caspase-9 and nuclear factor-kappaB. J. Biol. Chem. 274:14560. [DOI] [PubMed] [Google Scholar]
- 12. Sharma D., Kanneganti T. D. 2016. The cell biology of inflammasomes: mechanisms of inflammasome activation and regulation. J. Cell Biol. 213:617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kayagaki N., Warming S., Lamkanfi M., et al. 2011. Non-canonical inflammasome activation targets caspase-11. Nature 479:117. [DOI] [PubMed] [Google Scholar]
- 14. Wang S., Miura M., Jung Y. K., et al. 1998. Murine caspase-11, an ICE-interacting protease, is essential for the activation of ICE. Cell 92:501. [DOI] [PubMed] [Google Scholar]
- 15. Elinav E., Strowig T., Kau A. L., et al. 2011. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 145:745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kerur N., Veettil M. V., Sharma-Walia N., et al. 2011. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell Host Microbe 9:363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Khare S., Dorfleutner A., Bryan N. B., et al. 2012. An NLRP7-containing inflammasome mediates recognition of microbial lipopeptides in human macrophages. Immunity 36:464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Minkiewicz J., de Rivero Vaccari J. P., Keane R. W. 2013. Human astrocytes express a novel NLRP2 inflammasome. Glia 61:1113. [DOI] [PubMed] [Google Scholar]
- 19. Poeck H., Bscheider M., Gross O., et al. 2010. Recognition of RNA virus by RIG-I results in activation of CARD9 and inflammasome signaling for interleukin 1 beta production. Nat. Immunol. 11:63. [DOI] [PubMed] [Google Scholar]
- 20. Vladimer G. I., Weng D., Paquette S. W., et al. 2012. The NLRP12 inflammasome recognizes Yersinia pestis. Immunity 37:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Boyden E. D., Dietrich W. F. 2006. Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat. Genet. 38:240. [DOI] [PubMed] [Google Scholar]
- 22. Chavarría-Smith J., Mitchell P. S., Ho A. M., et al. 2016. Functional and evolutionary analyses identify proteolysis as a general mechanism for NLRP1 inflammasome activation. PLoS Pathog. 12:e1006052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chavarría-Smith J., Vance R. E. 2013. Direct proteolytic cleavage of NLRP1B is necessary and sufficient for inflammasome activation by anthrax lethal factor. PLoS Pathog. 9:e1003452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hellmich K. A., Levinsohn J. L., Fattah R., et al. 2012. Anthrax lethal factor cleaves mouse nlrp1b in both toxin-sensitive and toxin-resistant macrophages. PLoS One 7:e49741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Frew B. C., Joag V. R., Mogridge J. 2012. Proteolytic processing of Nlrp1b is required for inflammasome activity. PLoS Pathog. 8:e1002659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhong F. L., Mamaï O., Sborgi L., et al. 2016. Germline NLRP1 mutations cause skin inflammatory and cancer susceptibility syndromes via inflammasome activation. Cell 167:187. [DOI] [PubMed] [Google Scholar]
- 27. Martinon F., Agostini L., Meylan E., et al. 2004. Identification of bacterial muramyl dipeptide as activator of the NALP3/cryopyrin inflammasome. Curr. Biol. 14:1929. [DOI] [PubMed] [Google Scholar]
- 28. Kanneganti T. D., Ozören N., Body-Malapel M., et al. 2006. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature 440:233. [DOI] [PubMed] [Google Scholar]
- 29. Mariathasan S., Weiss D. S., Newton K., et al. 2006. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature 440:228. [DOI] [PubMed] [Google Scholar]
- 30. Martinon F., Pétrilli V., Mayor A., et al. 2006. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature 440:237. [DOI] [PubMed] [Google Scholar]
- 31. Allen I. C., Scull M. A., Moore C. B., et al. 2009. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity 30:556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Thomas P. G., Dash P., Aldridge J. R., Jr, et al. 2009. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity 30:566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gross O., Poeck H., Bscheider M., et al. 2009. Syk kinase signalling couples to the Nlrp3 inflammasome for anti-fungal host defence. Nature 459:433. [DOI] [PubMed] [Google Scholar]
- 34. Kanneganti T. D., Body-Malapel M., Amer A., et al. 2006. Critical role for Cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA. J. Biol. Chem. 281:36560. [DOI] [PubMed] [Google Scholar]
- 35. Lamkanfi M., Malireddi R. K., Kanneganti T. D. 2009. Fungal zymosan and mannan activate the cryopyrin inflammasome. J. Biol. Chem. 284:20574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lamkanfi M., Kanneganti T. D. 2010. Nlrp3: an immune sensor of cellular stress and infection. Int. J. Biochem. Cell Biol. 42:792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bauernfeind F. G., Horvath G., Stutz A., et al. 2009. Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J. Immunol. 183:787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Netea M. G., Nold-Petry C. A., Nold M. F., et al. 2009. Differential requirement for the activation of the inflammasome for processing and release of IL-1beta in monocytes and macrophages. Blood 113:2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Broz P., Dixit V. M. 2016. Inflammasomes: mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 16:407. [DOI] [PubMed] [Google Scholar]
- 40. Muñoz-Planillo R., Kuffa P., Martínez-Colón G, et al. 2013. K+ efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity 38:1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Groß C. J., Mishra R., Schneider K. S., et al. 2016. K(+) efflux-independent NLRP3 inflammasome activation by small molecules targeting mitochondria. Immunity 45:761. [DOI] [PubMed] [Google Scholar]
- 42. Sanman L. E., Qian Y., Eisele N. A., et al. 2016. Disruption of glycolytic flux is a signal for inflammasome signaling and pyroptotic cell death. Elife 5:e13663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gurung P., Anand P. K., Malireddi R. K., et al. 2014. FADD and caspase-8 mediate priming and activation of the canonical and noncanonical Nlrp3 inflammasomes. J. Immunol. 192:1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gurung P. Burton A., and Kanneganti T. D. 2016. NLRP3 inflammasome plays a redundant role with caspase 8 to promote IL-1beta-mediated osteomyelitis. Proc. Natl Acad. Sci. USA 113:4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gurung P., Malireddi R. K., Anand P. K., et al. 2012. Toll or interleukin-1 receptor (TIR) domain-containing adaptor inducing interferon-β (TRIF)-mediated caspase-11 protease production integrates Toll-like receptor 4 (TLR4) protein- and Nlrp3 inflammasome-mediated host defense against enteropathogens. J. Biol. Chem. 287:34474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kuriakose T., Man S. M., Malireddi R. K., et al. 2016. ZBP1/DAI is an innate sensor of influenza virus triggering the NLRP3 inflammasome and programmed cell death pathways. Sci. Immunol. 1:aag2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. He Y., Zeng M. Y., Yang D., et al. 2016. NEK7 is an essential mediator of NLRP3 activation downstream of potassium efflux. Nature 530:354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schmid-Burgk J. L., Chauhan D., Schmidt T., et al. 2016. A genome-wide CRISPR (clustered regularly interspaced short palindromic repeats) screen identifies NEK7 as an essential component of NLRP3 inflammasome activation. J. Biol. Chem. 291:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shi H., Wang Y., Li X., et al. 2016. NLRP3 activation and mitosis are mutually exclusive events coordinated by NEK7, a new inflammasome component. Nat. Immunol. 17:250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Py B. F., Kim M. S., Vakifahmetoglu-Norberg H., et al. 2013. Deubiquitination of NLRP3 by BRCC3 critically regulates inflammasome activity. Mol. Cell 49:331. [DOI] [PubMed] [Google Scholar]
- 51. Spalinger M. R., Kasper S., Gottier C., et al. 2016. NLRP3 tyrosine phosphorylation is controlled by protein tyrosine phosphatase PTPN22. J. Clin. Invest. 126:1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Poyet J. L., Srinivasula S. M., Tnani M., et al. 2001. Identification of Ipaf, a human caspase-1-activating protein related to Apaf-1. J. Biol. Chem. 276:28309. [DOI] [PubMed] [Google Scholar]
- 53. Franchi L., Amer A., Body-Malapel M., et al. 2006. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat. Immunol. 7:576. [DOI] [PubMed] [Google Scholar]
- 54. Miao E. A., Alpuche-Aranda C. M., Dors M., et al. 2006. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat. Immunol. 7:569. [DOI] [PubMed] [Google Scholar]
- 55. Miao E. A., Mao D. P., Yudkovsky N., et al. 2010. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc. Natl Acad. Sci. USA 107:3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhao Y., Yang J., Shi J., et al. 2011. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature 477:596. [DOI] [PubMed] [Google Scholar]
- 57. Kofoed E. M., Vance R. E. 2011. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature 477:592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tenthorey J. L., Kofoed E. M., Daugherty M. D., et al. 2014. Molecular basis for specific recognition of bacterial ligands by NAIP/NLRC4 inflammasomes. Mol. Cell 54:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yang J., Zhao Y., Shi J., et al. 2013. Human NAIP and mouse NAIP1 recognize bacterial type III secretion needle protein for inflammasome activation. Proc. Natl Acad. Sci. USA 110:14408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kortmann J., Brubaker S. W., Monack D. M. 2015. Cutting edge: inflammasome activation in primary human macrophages is dependent on flagellin. J. Immunol. 195:815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mariathasan S., Newton K., Monack D. M., et al. 2004. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature 430:213. [DOI] [PubMed] [Google Scholar]
- 62. Man S. M., Tourlomousis P., Hopkins L., et al. 2013. Salmonella infection induces recruitment of Caspase-8 to the inflammasome to modulate IL-1β production. J. Immunol. 191:5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Canna S. W., de Jesus A. A., Gouni S., et al. 2014. An activating NLRC4 inflammasome mutation causes autoinflammation with recurrent macrophage activation syndrome. Nat. Genet. 46:1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Qu Y., Misaghi S., Izrael-Tomasevic A., et al. 2012. Phosphorylation of NLRC4 is critical for inflammasome activation. Nature 490:539. [DOI] [PubMed] [Google Scholar]
- 65. Suzuki S., Franchi L., He Y., et al. 2014. Shigella type III secretion protein MxiI is recognized by Naip2 to induce Nlrc4 inflammasome activation independently of Pkcδ. PLoS Pathog. 10:e1003926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Matusiak M., Van Opdenbosch N., Vande Walle L., et al. 2015. Flagellin-induced NLRC4 phosphorylation primes the inflammasome for activation by NAIP5. Proc. Natl Acad. Sci. USA 112:1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Fernandes-Alnemri T., Yu J. W., Datta P., et al. 2009. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature 458:509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hornung V., Ablasser A., Charrel-Dennis M., et al. 2009. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 458:514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bürckstümmer T., Baumann C., Blüml S., et al. 2009. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat. Immunol. 10:266. [DOI] [PubMed] [Google Scholar]
- 70. Roberts T. L., Idris A., Dunn J. A., et al. 2009. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science 323:1057. [DOI] [PubMed] [Google Scholar]
- 71. Muruve D. A., Pétrilli V., Zaiss A. K., et al. 2008. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature 452:103. [DOI] [PubMed] [Google Scholar]
- 72. Fernandes-Alnemri T., Yu J. W., Juliana C., et al. 2010. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat. Immunol. 11:385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rathinam V. A., Jiang Z., Waggoner S. N., et al. 2010. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat. Immunol. 11:395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Jones J. W., Kayagaki N., Broz P., et al. 2010. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc. Natl Acad. Sci. USA 107:9771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sauer J. D., Witte C. E., Zemansky J., et al. 2010. Listeria monocytogenes triggers AIM2-mediated pyroptosis upon infrequent bacteriolysis in the macrophage cytosol. Cell Host Microbe 7:412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Henry T., Brotcke A., Weiss D. S., et al. 2007. Type I interferon signaling is required for activation of the inflammasome during Francisella infection. J. Exp. Med. 204:987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Man S. M., Karki R., Malireddi R. K., et al. 2015. The transcription factor IRF1 and guanylate-binding proteins target activation of the AIM2 inflammasome by Francisella infection. Nat. Immunol. 16:467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Man S. M., Karki R., Sasai M., et al. 2016. IRGB10 liberates bacterial ligands for sensing by the AIM2 and Caspase-11-NLRP3 inflammasomes. Cell 167:382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Meunier E., Wallet P., Dreier R. F., et al. 2015. Guanylate-binding proteins promote activation of the AIM2 inflammasome during infection with Francisella novicida. Nat. Immunol. 16:476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Dihlmann S., Erhart P., Mehrabi A., et al. 2014. Increased expression and activation of absent in melanoma 2 inflammasome components in lymphocytic infiltrates of abdominal aortic aneurysms. Mol. Med. 20:230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Dombrowski Y., Peric M., Koglin S., et al. 2011. Cytosolic DNA triggers inflammasome activation in keratinocytes in psoriatic lesions. Sci. Transl. Med. 3:82ra38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Javierre B. M., Fernandez A. F., Richter J., et al. 2010. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 20:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Man S. M., Zhu Q., Zhu L., et al. 2015. Critical role for the DNA sensor AIM2 in stem cell proliferation and cancer. Cell 162:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wilson J. E., Petrucelli A. S., Chen L., et al. 2015. Inflammasome-independent role of AIM2 in suppressing colon tumorigenesis via DNA-PK and Akt. Nat. Med. 21:906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Xie M., Yu Y., Kang R., et al. 2016. PKM2-dependent glycolysis promotes NLRP3 and AIM2 inflammasome activation. Nat. Commun. 7:13280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Hu B., Jin C., Li H. B., et al. 2016. The DNA-sensing AIM2 inflammasome controls radiation-induced cell death and tissue injury. Science 354:765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Di Micco A., Frera G., Lugrin J., et al. 2016. AIM2 inflammasome is activated by pharmacological disruption of nuclear envelope integrity. Proc. Natl Acad. Sci. USA 113:E4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Chae J. J., Cho Y. H., Lee G. S., et al. 2011. Gain-of-function Pyrin mutations induce NLRP3 protein-independent interleukin-1β activation and severe autoinflammation in mice. Immunity 34:755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. French FMF Consortium. 1997. A candidate gene for familial Mediterranean fever. Nat. Genet. 17:25. [DOI] [PubMed] [Google Scholar]
- 90. Chae J. J., Wood G., Masters S. L., et al. 2006. The B30.2 domain of pyrin, the familial Mediterranean fever protein, interacts directly with caspase-1 to modulate IL-1beta production. Proc. Natl Acad. Sci. USA 103:9982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Hesker P. R., Nguyen M., Kovarova M., et al. 2012. Genetic loss of murine pyrin, the Familial Mediterranean fever protein, increases interleukin-1β levels. PLoS One 7:e51105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Gavrilin M. A., Abdelaziz D. H., Mostafa M., et al. 2012. Activation of the pyrin inflammasome by intracellular Burkholderia cenocepacia. J. Immunol. 188:3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Aubert D. F., Xu H., Yang J., et al. 2016. A Burkholderia type vi effector deamidates Rho GTPases to activate the Pyrin inflammasome and trigger inflammation. Cell Host Microbe 19:664. [DOI] [PubMed] [Google Scholar]
- 94. Xu H., Yang J., Gao W., et al. 2014. Innate immune sensing of bacterial modifications of Rho GTPases by the Pyrin inflammasome. Nature 513:237. [DOI] [PubMed] [Google Scholar]
- 95. Masters S. L., Lagou V., Jéru I., et al. 2016. Familial autoinflammation with neutrophilic dermatosis reveals a regulatory mechanism of pyrin activation. Sci. Transl. Med. 8:332ra45. [DOI] [PubMed] [Google Scholar]
- 96. Park Y. H., Wood G., Kastner D. L., et al. 2016. Pyrin inflammasome activation and RhoA signaling in the autoinflammatory diseases FMF and HIDS. Nat. Immunol. 17:914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Waite A. L., Schaner P., Hu C., et al. 2009. Pyrin and ASC co-localize to cellular sites that are rich in polymerizing actin. Exp. Biol. Med. 234:40. [DOI] [PubMed] [Google Scholar]
- 98. Kim M. L., Chae J. J., Park Y. H., et al. 2015. Aberrant actin depolymerization triggers the pyrin inflammasome and autoinflammatory disease that is dependent on IL-18, not IL-1β. J. Exp. Med. 212:927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Akula M. K., Shi M., Jiang Z., et al. 2016. Control of the innate immune response by the mevalonate pathway. Nat. Immunol. 17:922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Kimura T., Jain A., Choi S. W., et al. 2015. TRIM-mediated precision autophagy targets cytoplasmic regulators of innate immunity. J. Cell Biol. 210:973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Diebolder C. A., Halff E. F., Koster A. J., et al. 2015. Cryoelectron tomography of the NAIP5/NLRC4 inflammasome: implications for NLR activation. Structure 23:2349. [DOI] [PubMed] [Google Scholar]
- 102. Halff E. F., Diebolder C. A., Versteeg M., et al. 2012. Formation and structure of a NAIP5-NLRC4 inflammasome induced by direct interactions with conserved N- and C-terminal regions of flagellin. J. Biol. Chem. 287:38460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Hu Z., Zhou Q., Zhang C., et al. 2015. Structural and biochemical basis for induced self-propagation of NLRC4. Science 350:399. [DOI] [PubMed] [Google Scholar]
- 104. Zhang L., Chen S., Ruan J., et al. 2015. Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization. Science 350:404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Man S. M., Hopkins L. J., Nugent E., et al. 2014. Inflammasome activation causes dual recruitment of NLRC4 and NLRP3 to the same macromolecular complex. Proc. Natl Acad. Sci. USA 111:7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Qu Y., Misaghi S., Newton K., et al. 2016. NLRP3 recruitment by NLRC4 during Salmonella infection. J. Exp. Med. 213:877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Karki R., Man S. M., Malireddi R. K., et al. 2015. Concerted activation of the AIM2 and NLRP3 inflammasomes orchestrates host protection against Aspergillus infection. Cell Host Microbe 17:357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Jin T., Perry A., Jiang J., et al. 2012. Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity 36:561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Lu A., Li Y., Yin Q., et al. 2015. Plasticity in PYD assembly revealed by cryo-EM structure of the PYD filament of AIM2. Cell Discov. 1:15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Baroja-Mazo A., Martín-Sánchez F., Gomez A. I., et al. 2014. The NLRP3 inflammasome is released as a particulate danger signal that amplifies the inflammatory response. Nat. Immunol. 15:738. [DOI] [PubMed] [Google Scholar]
- 111. Franklin B. S., Bossaller L., De Nardo D., et al. 2014. The adaptor ASC has extracellular and ‘prionoid’ activities that propagate inflammation. Nat. Immunol. 15:727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Lu A., Magupalli V. G., Ruan J., et al. 2014. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell 156:1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Dick M. S., Sborgi L., Rühl S., et al. 2016. ASC filament formation serves as a signal amplification mechanism for inflammasomes. Nat. Commun. 7:11929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Fink S. L., Cookson B. T. 2005. Apoptosis, pyroptosis, and necrosis: mechanistic description of dead and dying eukaryotic cells. Infect. Immun. 73:1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Shi J., Zhao Y., Wang K., et al. 2015. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526:660. [DOI] [PubMed] [Google Scholar]
- 116. Tanaka S., Mizushina Y., Kato Y., et al. 2013. Functional conservation of Gsdma cluster genes specifically duplicated in the mouse genome. G3 (Bethesda) 3:1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Kayagaki N., Stowe I. B., Lee B. L., et al. 2015. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 526:666. [DOI] [PubMed] [Google Scholar]
- 118. Aglietti R. A., Estevez A., Gupta A., et al. 2016. GsdmD p30 elicited by caspase-11 during pyroptosis forms pores in membranes. Proc. Natl Acad. Sci. USA 113:7858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Ding J., Wang K., Liu W., et al. 2016. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature 535:111. [DOI] [PubMed] [Google Scholar]
- 120. Sborgi L., Rühl S., Mulvihill E., et al. 2016. GSDMD membrane pore formation constitutes the mechanism of pyroptotic cell death. EMBO J. 35:1766. [DOI] [PMC free article] [PubMed] [Google Scholar]