A transcriptional serenAID
Keywords: activation-induced cytidine deaminase, AID, noncoding RNA targeting
Abstract
During an immune response, activated B cells may undergo class switch recombination (CSR), a molecular rearrangement that allows B cells to switch from expressing IgM and IgD to a secondary antibody heavy chain isotype such as IgG, IgA or IgE. Secondary antibody isotypes provide the adaptive immune system with distinct effector functions to optimally combat various pathogens. CSR occurs between repetitive DNA elements within the immunoglobulin heavy chain (Igh) locus, termed switch (S) regions and requires the DNA-modifying enzyme activation-induced cytidine deaminase (AID). AID-mediated DNA deamination within S regions initiates the formation of DNA double-strand breaks, which serve as biochemical beacons for downstream DNA repair pathways that coordinate the ligation of DNA breaks. Myriad factors contribute to optimal AID targeting; however, many of these factors also localize to genomic regions outside of the Igh locus. Thus, a current challenge is to explain the specific targeting of AID to the Igh locus. Recent studies have implicated noncoding RNAs in CSR, suggesting a provocative mechanism that incorporates Igh-specific factors to enable precise AID targeting. Here, we chronologically recount the rich history of noncoding RNAs functioning in CSR to provide a comprehensive context for recent and future discoveries. We present a model for the RNA-guided targeting of AID that attempts to integrate historical and recent findings, and highlight potential caveats. Lastly, we discuss testable hypotheses ripe for current experimentation, and explore promising ideas for future investigations.
Introduction
Following initial exposure to an immunogen, the adaptive immune system provides a mechanism to activate antigen-specific B cells in order to produce antibodies and facilitate an immune response. Antibodies are secreted forms of the B cell antigen-receptor (BCR) and are composed of immunoglobulin (Ig) heavy (IgH) and light (IgL) chains; the N-terminal region of the IgH and IgL chains comprise the antigen-recognition domain, whereas the C-terminal domain of IgH mediates downstream signaling and effector functions following antigen binding (Fig. 1).
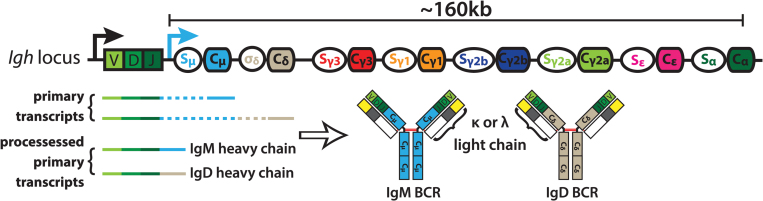
Fig. 1.
Organization of the mouse immunoglobulin heavy chain locus. Mature B cells express both IgM and IgD after exiting the bone marrow and migrating to secondary lymphoid tissues. Individual constant regions (CH) span ~160 kb of chromosome 12, and consist of an intronic switch (S) region (indicated by unfilled ovals) located upstream of its matching CH (indicated by filled rectangles). Cδ is a notable exception; it is preceded by a non-canonical S region called σδ. B cells co-express of IgM and IgD B cell antigen-receptors (BCRs) by virtue of the alternative splicing of primary transcripts, and assembly with either kappa (κ) or lambda (λ) light chains. Solid arrows indicate transcription start sites, dashed lines within transcripts indicate introns, solid lines within transcripts indicate exons, red lines indicate disulfide bonds.
A hallmark of the adaptive immune system is the ability to recognize a vast array of antigens, a property mediated by the diverse repertoire of B cell Ig sequences. This diversity is created in part through V(D)J recombination, a process that assembles the N-terminal variable (V), diversity (D) and joining (J) segments in different combinations. Following the completion of V(D)J recombination, mature B cells exit the bone marrow and migrate to secondary lymphoid organs where they may encounter antigens, become activated and undergo class switch recombination (CSR).
CSR is a molecular rearrangement that deletes and recombines portions of the immunoglobulin heavy chain (Igh) locus, exchanging the default Cμ constant region (CH) gene segment for an alternate set of CH genes (Cγ, Cε, Cα). CSR occurs between repetitive DNA elements, termed switch (S) regions, which precede each CH gene segment, and requires the DNA-modifying enzyme activation-induced cytidine deaminase (AID) (1–3). AID deamination within S regions initiates the formation of DNA double-strand breaks (DSBs) (4, 5), which are then repaired primarily by non-homologous end-joining [reviewed in (6)] (Fig. 2A). This acts to simultaneously excise the intervening CH segments and juxtapose the upstream V(D)J region with a new downstream CH. The excised fragment can ligate to form a closed circle that will undergo transcription and processing, and whose presence is a molecular proxy for CSR (Fig. 2B).
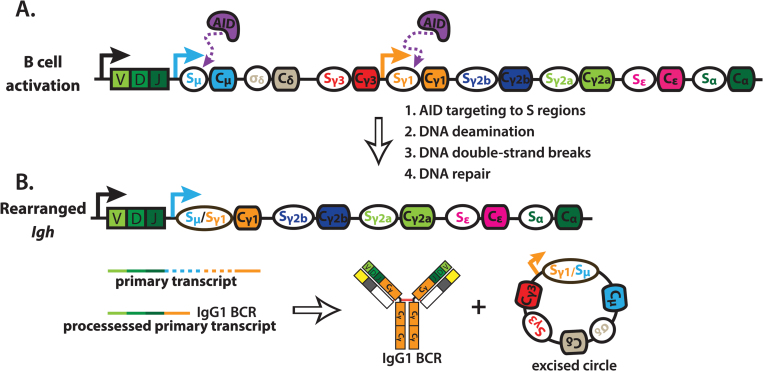
Fig. 2.
Class switch recombination. (A) B cell activation can induce expression of activation-induced cytidine deaminase (AID) and transcription of downstream CH regions (orange arrow) driven by cytokine-responsive elements. This recruits AID to the downstream S region, as well as Sμ, which is constitutively transcribed. AID-mediated DNA deamination within S regions initiates the formation of DNA double-strand breaks (DSBs), and DNA repair occurs primarily, but not exclusively, through non-homologous end joining. (B) Repair of DNA DSBs results in a deletional-recombination event that juxtaposes the upstream V(D)J segment with a new downstream CH (indicated by Cγ1). Additionally, the excised region can ligate to form a closed circle. The class switched B cell now expresses a new B cell antigen-receptor (BCR), represented as IgG1. Solid arrows indicate transcription start sites, dashed lines within transcripts indicate introns, solid lines within transcripts indicate exons, red lines indicate disulfide bonds. AID, activation-induced cytidine deaminase.
CSR thus enables a B cell to switch from expressing default IgM and IgD to a secondary heavy chain isotype, providing the adaptive immune system with distinct effector functions and properties to optimally combat a vast array of pathogens it may encounter.
Understanding the biochemical properties and molecular mechanisms that govern AID targeting has been paramount since its discovery, partly because of the implications that aberrant AID targeting has in the genesis and treatment of B cell malignancies (7). Nearly two decades of research have revealed myriad factors that contribute to AID targeting, including protein–protein interactions, protein–nucleic acid interactions and megabase-scale chromatin organization and looping [CSR and AID targeting reviewed extensively in (6, 8, 9)]. While these factors undoubtedly contribute to efficient AID targeting and CSR, many do not localize specifically to the Igh locus. A current challenge is to decipher how this complex milieu of molecular interactions supports efficient AID targeting to the Igh locus, while minimizing deleterious mis-targeting driven by the same processes.
The CSR field arose with a cluster of discoveries that identified noncoding RNAs emanating from the Igh locus. Recent studies have highlighted new roles for these RNAs in targeting AID (10–12), and suggest a provocative mechanism that contributes to Igh-specific targeting (10) analogous to CRISPR-Cas9 gene editing. Amidst a growing paradigm of noncoding RNAs targeting DNA- or chromatin-modifying factors (13), recounting the bountiful history of CSR and noncoding RNAs serves to highlight the significance of recent findings while providing an opportunity to spark new ideas.
Here, we aim to provide a comprehensive, chronological review of noncoding RNAs and transcription in AID targeting, focusing largely on CSR. We begin with the discovery of Igh noncoding transcripts and the mystery of their role in CSR. Next, we examine the biochemical properties of these transcripts, with a focus on their ability to form R-loops, or RNA/DNA hybrids and secondary structures like G-quadruplexes. We then examine transcription-related factors, as well as the RNA molecules themselves, demonstrated to contribute to AID targeting. We conclude with a model for RNA-guided AID targeting, and highlight the important questions, caveats and hypotheses this model evokes.
Prelude to transcription
Over 40 years ago, Klinman and colleagues (14) used the splenic focus system to show that a single B cell clone could produce antibodies of multiple isotypes, but with an identical variable region. Building upon previous findings that a single B cell could have both surface IgM and cytoplasmic IgG (15), secrete both IgM and IgG (16) and single-cell-derived splenic foci could produce monoclonal antibodies containing both IgM and IgG1 (17), the biological phenomenon of CSR was laid bare for the emergent field to unravel. Merely 3 years later, Honjo and Kataoka (18) utilized myeloma tumor lines expressing various IgH isotypes to describe the linear organization of the Igh locus and presciently postulate the ‘allelic deletion’ model for CSR.
Spurred by these groundbreaking discoveries, the developing CSR field progressed rapidly through the turn of the century, evolving in response to major discoveries and intertwining with various fields such as splicing, DNA damage and repair and chromatin biology. The discovery of AID and its essential role in antibody diversification (1–3) kindled the ensuing debate over AID’s nucleic acid substrate, inspiring the DNA deamination model (5) and precipitating the identification of downstream players in the CSR pathway such as the DNA glycosylase UNG (19). In turn, this discovery fueled a race to uncover single-stranded DNA (ssDNA) as the target of AID’s enzymatic activity, as well as the dependence on transcription for deamination (20–24). However, nearly two decades before AID and UNG, there was the peculiar observation that noncoding RNA transcripts originated from within the Igh locus, and the notion they may be playing a role in CSR.
Uncovering germline transcripts, starting with T cells
Early studies on B cell tumor lines first characterized the expression of two differentially spliced Cμ mRNAs representing either membrane (μm) or secreted forms (μs) (25–27). However, studies searching for immunoglobulin expression in T cells (pre-dating the discovery of the T cell antigen-receptor) first observed Cμ-containing transcripts differing in size to μm and μs (28, 29). Northern blot analyses using probes spanning Cμ revealed these alternative transcripts underwent splicing, and while similar to μm and μs at their 3′ ends, their 5′ ends originated within the JH–Cμ intron and did not include a variable region (30) (Fig. 3).
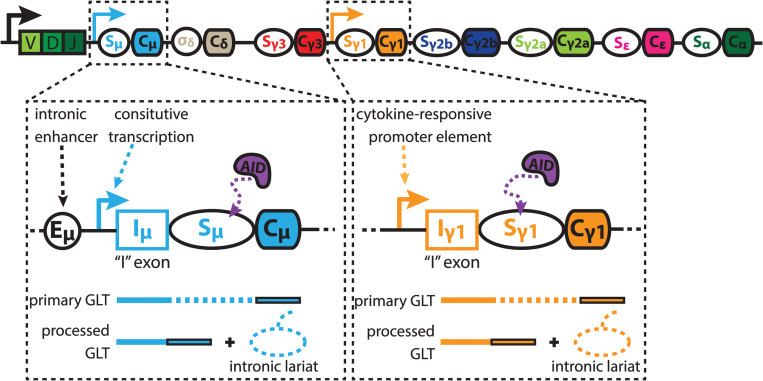
Fig. 3.
Genesis and processing of germline transcripts. Each constant region (CH) contains an individual transcriptional unit that produces noncoding germline transcripts (GLTs). Within each unit, transcription initiates upstream of the switch (S) regions, and is promoted by mitogen and/or cytokine-responsive elements (right box), with the exception of Cμ GLTs which are driven by the μ enhancer (Eμ) and are constitutively expressed (left box). Transcription proceeds through the intervening, or I-exon, intronic S regions and CH exons to produce a primary GLT. Splicing of the I-exon to the 5′ exon of the downstream CH gene creates a processed GLT, and an intronic S region lariat that can undergo debranching to become a linear S region transcript. Solid arrows indicate transcription start sites, dashed lines within transcripts indicate introns, solid lines within transcripts indicate exons. AID, activation-induced cytidine deaminase.
Additional studies using transformed B cell lines also detected Cμ containing RNAs lacking variable regions, designating them as either ‘sterile’ or ‘Iμ’ transcripts, hereafter termed germline transcripts (GLTs) (31, 32). Cμ GLTs were found to originate at heterogeneous sites downstream of the Cμ enhancer (Eμ) and promoter region, and contained a cryptic splice site ~700 bp downstream of initiation. This created an exon, now referred to as the intervening or ‘I-exon’, that was spliced to the 5′ Cμ constant region (32) (Fig. 3).
Experimentation on transformed B cell lines continued to fuel CSR discoveries; the isolation of pure IgM+ cells from the I.29 mouse B cell lymphoma, which contained a mixture of cells expressing membrane-bound IgM and IgA of the same idiotype (33, 34), was a key breakthrough. IgM+ cells could be induced to undergo CSR in vitro to IgA, IgE or IgG2b by treatment with LPS in combination with a monoclonal antibody raised against the I.29 Ig idiotype (35). This crucially allowed a controlled comparison of GLTs and genomic rearrangements at the Igh locus before, during and after CSR. Amongst the first clues that GLTs functioned during CSR was the detection of RNAs transcribed from an unrearranged Cα region in IgM+ I.29 cells undergoing IgA CSR (36). These observations extended to other isotypes as well; in Abelson murine leukemia virus (A-MuLV)-transformed pre-B cell lines, expression of Cγ2b GLTs preempted IgG2b CSR (37).
A flurry of studies followed examining the influence of various mitogens and cytokines on GLTs. LPS was shown to induce Cγ2b and Cγ3 GLTs prior to IgG2b and IgG3 CSR, respectively, whereas IL-4 had an inhibitory effect on these GLTs (38, 39). Alternatively, IL-4 induced Cγ1 and Cε GLTs prior to the appearance of membrane-bound IgG1 and IgE mRNAs, respectively (39–44). IFNγ induced Cγ2b GLTs, but repressed Cγ1 and Cε GLTs (39, 45, 46), and TGFβ increased Cα GLTs and IgA CSR, but repressed Cε GLTs (45, 47). The observed cytokine-induced transcription of GLTs was rapid; Cγ1 and Cε transcripts were detected only 4 hours after cytokine stimulation (42, 43).
These studies established an overwhelmingly clear consensus within the field: mitogens and cytokines directed CSR to particular isotypes by activating germline transcription upstream of the desired CH region, and in some cases abrogating transcription in undesired CH regions. This prompted the idea that activation of chromatin regions could provide access not only to RNA polymerases, but also to a yet unidentified recombinase (it would be more than a decade before AID was discovered). Notably, the concept of transcription priming recombination was heavily influenced by findings of unrearranged variable regions undergoing transcription prior to rearrangement (48, 49), the observation that transcription could enhance variable region recombination (50) and the positive correlation between transcription and recombination in Saccharomyces cerevisiae (51). Indeed, the magnitude of Cα GLTs was shown to correlate directly with I.29 clones that had increased IgA CSR (40).
Molecular cloning reveals conserved GLT structure
Simultaneous to the mitogen and cytokine studies, parallel efforts were honing in on defining the molecular structure and function of the various GLTs. The Cγ2b GLT was first cloned from A-MuLV-transformed cells, demonstrating the transcripts initiated 5′ of Cγ2b, in accord with previous findings of Cμ GLTs (32, 52). The cloning of GLTs from Cα (47, 53), Cε (44, 54) and Cγ3 (55) was rapidly published within the next year. Additionally, the discovery of regulatory DNA elements abutting transcription initiation sites in CH loci revealed sequences that conferred LPS and IL-4 responsive transcription (56).
Strikingly, these studies uncovered conserved features within all GLTs: initiation of transcription upstream of the S region, the presence of an I-exon 5′ to the S region that was unlikely to code for a protein, and splicing of this exon to the 5′ exon of the downstream CH gene (Fig. 3). Stavnezer and colleagues succinctly characterized the implications of these findings:
‘The fact that the germ line transcripts that have been characterized have similar structures argues that their transcription does have a function and is not simply an accidental by-product of the accessibility of the CH gene.’ (53)
Molecular peculiarities of S regions
Due to the observation that breakpoints in CSR occurred within the intronic S regions of CH genes (57), a leading hypothesis was that S region sequences possessed unique characteristics that targeted CSR. Mouse S regions are composed of 1–10 kilobases (kb) of tandem repeats that are G-rich on the non-template strand DNA (57, 58). Both the sequence and number of tandem repeats vary between different mouse CH S regions. Sμ, Sα and Sε are composed of pentameric repeats, whereas Sγ1, Sγ2b and Sγ3 are composed of longer 49 bp repeats (59–64) (reviewed in (57)). A notable exception is the non-canonical switch-like region located upstream of Cδ, referred to as σδ. On rare occasions, recombination between Sμ and σδ results in IgD CSR, producing a B cell solely expressing IgD (65–68). The intronic S regions are excised from GLTs during splicing, connecting the I-exon to a downstream CH exon (Fig. 3).
Among the seminal studies of the molecular properties of S sequences, Wells and colleagues cloned various G-rich Sα repeats into a plasmid in a search of peculiar DNA structures (69). By assaying the sensitivity of Sα repeats to S1 and P1 nucleases, as well as a battery of chemical probes including dimethyl sulfate, bromoacetaldehyde, osmium tetroxide and diethyl pyrocarbonate, Wells and colleagues concluded that Sα repeats adopted a non-B-DNA structure, suggesting a potential intramolecular triple-strand (69). Aiming to identify proteins that specifically bound to S sequences, Sen and Gilbert (70) fortuitously observed that purine-rich S sequence ssDNA fragments migrated unexpectedly during electrophoretic mobility shift assays. Intriguingly, the complementary pyrimidine-rich strand did not exhibit a similar migration. Dimethyl sulfate treatment revealed stretches of Gs that were completely protected from methylation, suggestive of Hoogsteen bonding (71), and leading to a paradigmatic model of a parallel, four stranded structure, termed ‘G4-DNA’ (70).
By the early 1990s, the potential role of GLTs in CSR had become a major focus of the field, leading S region structure studies to broaden their purview to include transcription-induced structures. In a landmark study, Reaban and Griffin (72) observed that transcription of a supercoiled plasmid containing 2.3 kb of Sα sequence curiously led to a loss of supercoiling. Intriguingly, supercoiling relaxation only occurred when Sα was transcribed in the physiological orientation. Treatment of transcribed plasmids with various nucleases revealed sensitivity to RNase H and mung bean nuclease, suggesting the presence of an RNA–DNA hybrid and ssDNA (collectively referred to as an R-loop). Treatment with RNase H abolished sensitivity to mung bean nuclease, suggesting that formation of the RNA–DNA hybrid was necessary for the presence of ssDNA. Addition of Sα transcripts without polymerase did not lead to altered mobility, suggesting that the observed R-loop was transcription-dependent (72). In the context of the previously hypothesized S region DNA intramolecular-triple strand (69, 70), Reaban and Griffin postulated a model whereby transcription leads to formation of an R-loop (Fig. 4A), which then helps to stabilize the DNA triple-strand (72). Further work from Griffin and colleagues found that the Sα R-loops could extend up to 140 nucleotides, formed independently of supercoiling, and were remarkably stable, withstanding temperatures up to 95°C (73).
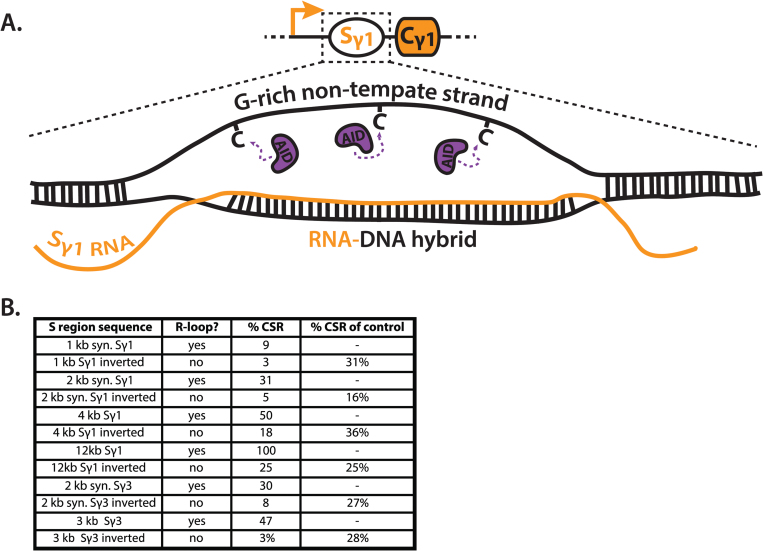
Fig. 4.
R-loops and AID targeting. (A) Transcription through S regions leads to the formation R-loops, consisting of an RNA–DNA hybrid and G-rich non-template single-stranded DNA (ssDNA). R-loops can extend over 1000 bp, as well as several hundred base pairs upstream of S regions. Current models hypothesize the availability of ssDNA is a key factor in targeting activation-induced cytidine deaminase (AID) to R-loops. (B) Table quantifying the contribution of R-loops to class switch recombination. Data were extracted from three studies that compared the ability of various S regions to potentiate IgG1 class switch recombination (CSR) at the endogenous mouse locus, with that of their inverted sequences (74–76). Because the inverted sequences do not support R-loop formation but are identical in size and sequence, an analysis of these comparisons can indicate the contribution of R-loops to CSR. This analysis was posited by Lieber and colleagues (77), and extended to include additional studies. ‘R-loop?’ indicates if the sequence has been shown to form an R-loop. ‘%CSR’ indicates the relative IgG1 CSR frequency, which is defined by the ratio of allele-specific hybridomas produced in various S region replacements, compared with that of the wild-type Sγ1 sequence. ‘%CSR of control’ indicates the frequency of CSR supported by inverted sequences relative to their non-inverted controls. kb, kilobase; syn, synthetic; AID, activation-induced cytidine deaminase.
Building upon the Sα studies (72, 73), transcription of Sμ, Sγ2b and Sγ3 sequences also formed stable R-loops in vitro, and only when transcribed in the physiological orientation (78, 79). Similar to Sα (72), these R-loops were also remarkably stable, persisting after phenol–chloroform extraction, protease treatment and temperatures up to 65°C (78). Most importantly, all S regions examined formed stable R-loops, suggesting a conserved mechanism during CSR and leading Reaban and Griffin to conclude:
‘The formation of such RNA–DNA hybrids in transcriptionally active switch regions may be important in switch recombination.’ (72)
Extrachromosomal recombination assays support a role for S region transcription
Early studies demonstrated that retroviral integration of S sequences could mediate recombination in pre-B cell lines (80), paving the road for the extrachromosomal recombination assay. In this assay, DNA substrates containing S region sequences flanking a lethal marker gene are transfected into B cells, which are then stimulated with cytokines to undergo CSR. Recombination between extrachromosomal S regions deletes the intervening lethal gene, allowing for recovery and selection of substrates in Escherichia coli. This provided a valuable tool to quickly assay the ability of various S region sequences, promoters, transcriptional orientations and cell lines to promote CSR.
In primary B cells, the addition of a promoter upstream of S regions led to a 10-fold increase in extrachromosomal recombination, and was dependent on a G-rich non-template strand (81). Extrachromosomal recombination occurred at significantly higher rates when S region transcription occurred in the physiological orientation (82), although some recombination was observed to occur between Sμ in the forward direction and Sα in reverse (83). Additionally, extrachromosomal substrates underwent more efficient recombination in B cell lines versus T cell or non-lymphoid lines, supporting the idea of a B cell-specific recombination factor (AID was still 5 years from discovery) (82, 84).
Modifying CH loci to better understand GLTs
Multiple lines of evidence accumulated implicating a role for GLTs in CSR. The conserved structure of GLTs, as well as the ability of all S regions to form R-loops, strongly implied a conserved function for transcription through CH loci. To formally address this, the CSR field heavily relied upon the genetic modification of CH loci within mouse embryonic stem (ES) cells. Modified ES cells were then injected into blastocysts, leading to the development of chimeric mice with altered CH loci. Initially, large sections of CH regions were removed to determine the role of GLTs in CSR.
Deletion of 1.7 kb located 5′ of Sγ1 removed promoter elements, the Iγ1-exon and splice donor site, leading to a complete loss of IgG1 CSR. Intriguingly, in mice heterozygous for the targeted allele, wild-type alleles could undergo normal CSR, suggesting GLTs were unable to act in trans on the targeted allele (85). Similarly, replacement of the promoter and Iγ2b-exon with a bacterial neomycin resistance gene in the opposite transcriptional orientation completely blocked Cγ2b GLTs and IgG2b CSR in homozygous mice (86). Lastly, deletion of a region including JH segments as well as Eμ led to the absence of Cμ GLTs and severely disrupted IgG1 CSR (87). While the Cμ locus was impaired in this study, the downstream CH genes remained intact. Intriguingly, the Sγ1 region was still rearranged upon stimulation to induce IgG1 CSR, resulting from internal Sγ1 deletions (87).
Collectively, these studies demonstrated the importance of transcription at both donor and acceptor CH regions to mediate efficient CSR, and that regulation of transcription at donor and acceptor CH regions occurred independently.
Building on these results, subsequent studies aimed to precisely replace different elements within CH genes while maintaining transcription. This would specify if GLTs per se, versus transcription in general, was required for CSR. Alt and colleagues replaced the LPS-IL4 responsive element and Iε-exon with either the Igh enhancer and V gene promoter, or an LPS inducible Eμ enhancer and V gene promoter. The former manipulation lead to constitutive transcription from the Cε locus and the ability to undergo some degree of CSR to IgE in the absence of IL-4, as measured by PCR-based assay (88). This suggested that transcription through the Cε locus, as opposed to other IL-4 responsive elements or the Iε exon itself, was sufficient to permit detectable levels of IgE CSR. The latter manipulation lead to LPS-responsive transcription and 10–100-fold lower CSR as measured by flow cytometry and IgE secretion (89). Together, these studies suggested that transcription per se was sufficient to support detectable levels of CSR; however, an intact I-exon and/or promoter regions were necessary for optimal CSR.
A crucial study focusing on the Cγ1 locus suggested a potential role for the splicing of GLTs in promoting CSR. A 5′ region containing the IL-4 responsive promoter and the Iγ1-exon was replaced with a human metallothionein (hMT) IIA promoter and IgG1 CSR was measured in mice with or without the presence of a 114-bp fragment of the Iγ1-exon containing the splice donor site. Mice with the splice donor site underwent comparable IgG1 CSR to wild-type controls, whereas those lacking the splice donor site did not. Control experiments demonstrated that mice with and without the splice donor site had equal rates of Sγ1 transcription as measured by nuclear run-on assay. However, the quality of Cγ1 GLTs, assayed by northern blot analysis with hMT probes, was severely compromised in those lacking the splice donor site (90).
These results built upon previous findings from Alt and colleagues (88, 89), suggesting the I-exon was not required for CSR. Additionally, they demonstrated that stable Sγ1 GLTs were sufficient to confer optimal CSR, and suggested a critical role for splicing. Radbruch and colleagues concluded:
‘The most intriguing speculation is that switch transcripts are part of the switch recombinase, providing the specificity to target distinct switch regions.’ (90)
A study manipulating the Cα locus provided a wrinkle to the evolving role of GLTs in CSR. Davis and colleagues replaced a 1.4-kb region of the Cα locus with a human hypoxanthine phosphoribosyltransferase (HPRT) minigene, deleting the entire Iα exon and splice donor site, while leaving the Cα promoter intact (91). Surprisingly, this did not affect IgA CSR as measured by surface IgA staining and IgA secretion. These data initially appeared to conflict with Radbruch and colleagues’ previous findings (90) because of the absence of the Iα splice donor site. However, Davis and colleagues noted that the HPRT minigene contained an intron and was transcribed in the physiological direction, leading to the detection of HPRT transcripts. This suggested that transcription through the Sα sequences and subsequent splicing may occur; however, an assay was not performed to confirm the presence of Sα containing GLTs (91).
Radbruch and colleagues (90) noted an important caveat within their initial study; the 114-bp fragment of the Iγ1-exon may have contained a critical recombination control element in addition to the splice donor site. To address this caveat and the recent data from Davis and colleagues (91), Radbruch and colleagues performed an elegantly controlled study. They replaced the endogenous Cγ1 promoter, Iγ1-exon and splice donor site with heterologous components: a hMT promoter, a bacterial sequence derived I-exon and an adenoviral splice donor site (92). While absence of the splice donor site completely disrupted CSR, addition of the wild-type or heterologous splice donor site was sufficient to restore IgG1 CSR to appreciable levels (92). This unequivocally demonstrated that the Iγ1-exon or surrounding sequences did not contain a recombination element, in agreement with Davis and colleagues’ conclusions on the Iα-exon (91).
It is important to reiterate that mice lacking the Iγ1 splice donor site were deemed transcriptionally active via nuclear run-on assays, however, no stable GLTs were detected by northern blot analysis of total RNA (90). This suggests the absence of a splice donor site prevented the stable transcription of the downstream Sγ1 region and Cγ1 exons. Thus, it was impossible to distinguish between the requirements of splicing per se, from that of stable Sγ1 GLTs. Nonetheless, in light of recent findings on the role of S region transcripts (10) (discussed in detail later), Radbruch and colleague’s conclusions would hit their mark:
‘Either the processing machinery or the processed transcripts are involved in class switch recombination.’(92)
In corroboration, Alt and colleagues (93) performed an analogous study to Davis and colleagues (91), replacing the Iγ2b-exon and splice donor site with a bacterial neomycin resistance gene transcribed in the physiological orientation. They found that IgG2b CSR was unperturbed in ES cell-derived chimeric mice, and crucially demonstrated the presence of Sγ2b-containing transcripts. This led to the notion that transcription through S regions may be sufficient to enable CSR, although Alt and colleagues (93) noted that the presence of cryptic splice sites within the neomycin resistance gene might have also contributed. In a parallel study of the Cμ locus, insertion of a neomycin resistance gene similarly rescued a CSR defect caused by deletion of Eμ and other Cμ control elements (94).
Over a span of 5 years, these studies collectively modified the Cμ (87, 94), Cγ1 (85, 90, 92), Cγ2b (86, 93), Cε (88, 89) and Cα (91) transcriptional control elements in order to better understand the role of GLTs. These pivotal studies strongly suggested that transcription of CH loci, and more precisely the generation of stable S transcripts, was necessary for efficient CSR. Importantly, the dependency on transcriptional orientation was in accord with results from R-loop and extrachromosomal recombination studies, and overwhelmingly pointed to a precise role for the G-rich S regions transcripts in targeting CSR.
An activation-induced interlude
The identification of AID by cDNA subtractive hybridization in the CH12 B cell line was a momentous discovery (1). AID was shown to be required for both CSR and somatic hypermutation (SHM) in a mouse knock-out model (2), and AID deficiency was linked to hyper-IgM syndrome and a lack of SHM in humans (3). Within the span of 3 months, five independent groups demonstrated that ssDNA was the molecular target of AID’s deaminase activity (20–24). Transcription was shown to induce AID deamination activity on the non-template strand of dsDNA (20, 23, 24), and the specific deamination of S regions was dependent upon transcription in the physiological orientation (20). The role of GLTs and R-loops in CSR were now reassessed from a new molecular perspective: how do these molecules facilitate AID targeting to S regions?
R-loops in an AID world
S region R-loops were hypothesized as vital molecular structures responsible for exposing ssDNA targeted by AID during CSR (Fig. 4A). R-loops had been detected in vitro using a wide variety of techniques including atomic force microscopy (95); however, it was unknown if they occurred in vivo and if they were necessary for CSR. To identify R-loops at endogenous S regions, Lieber and colleagues developed a technique that coupled sodium bisulfite treatment of nucleic acids with DNA sequencing (96). Sodium bisulfite selectively deaminates single-stranded cytidines, converting them into uridines, which can be detected and quantified using PCR amplification and DNA sequencing. In a sodium bisulfite experiment, sequenced clones containing long stretches of C to U conversion are indicative of R-loop formation throughout a locus.
Lieber and colleagues applied this technique to detect R-loops formed during in vitro transcription of various S region templates. They found that transcription of Sγ3 repeats led to long stretches (>100 bp) of conversion along the G-rich non-template strand, but not the C-rich strand (96). This was dependent upon the transcriptional orientation, as previously observed for S region R-loop formation (72,73,78,79). Lieber and colleagues adapted their protocol to boost sensitivity, and remarkably detected long stretches of conversion extending upwards of 1 kb in length at the endogenous Sγ3 and Sγ2b regions in primary B cells stimulated with LPS (96).
The remarkable length and stability of R-loops is understandable considering the known thermodynamic properties of nucleic acids: purine-rich RNA can form a more stable duplex with pyrimidine-rich DNA than purine-rich DNA forms with pyrimidine-rich DNA (97, 98). Likewise, the dependence upon transcription in the physiological orientation is consistent with the observation that purine-rich RNA can form a more stable duplex with pyrimidine-rich DNA, than pyrimidine-rich RNA does with purine-rich DNA (97, 98).
The majority of Sγ3 and Sγ2b R-loops were found to occur within S regions, and some were detected spanning the entire 2 kb Sγ3 region (99). R-loops could extend up to 600 bp downstream of S regions; however, the declining G-richness of the non-template strand outside of S regions was hypothesized to play a major role in demarcating R-loop boundaries (99). In accord, reduction of G density from 46 to 29% on the non-template strand was found to decrease both R-loop length and CSR in CH12 cell lines (100). Additionally, the pattern of AID deamination in ung–/–msh–/– mice (which lack uracil-DNA glycosylase and a mismatch-recognition protein) revealed long stretches of mutations within Sμ that correlated with the locations of R-loops (101, 102), strongly supporting a role for R-loops targeting AID.
The importance of S regions in CSR
Previous genetic manipulations of CH regions clearly suggested that transcription of S regions was a critical determinant for efficient CSR. The formation of R-loops within S regions now provided a mechanistic outcome for the requirement of transcription. However, the precise requirement for endogenous S regions had not been firmly established. Alt and colleagues addressed this gap in knowledge by deleting, inverting or replacing 12 kb of the Sγ1 region, including 8 kb of conserved Sγ1 repeats (74). Importantly, transcription and splicing of GLTs from the modified Sγ1 loci were unaffected during these perturbations. Analysis of allele-specific ELISAs and hybridomas revealed that deletion of the Sγ1 region resulted in a near complete loss of IgG1 CSR. Inversion of Sγ1 decreased IgG1 CSR to approximately 25% of wild-type levels (74), in agreement with R-loop formation only occurring during transcription in the physiological orientation (72, 73, 78, 79, 96).
Intriguingly, replacement of the Sγ1 region with 1 kb of random, non-repetitive G-rich sequence reduced CSR to 7% of wild-type levels, in spite of being approximately 10% the length of wild-type Sγ1. The inversion of this sequences to produce a C-rich transcript was unable to support detectable levels of CSR (74). These results were in accord with the in vitro formation of R-loops during transcription of the G-rich, but not C-rich RNA sequence (74). This suggested specific S region motifs were not required, as a random sequence could support appreciable levels of CSR. It also supported the concept that the most important consequence of S region transcription was the formation of R-loops and subsequent exposure of ssDNA.
Studies replacing the endogenous Sγ1 region with 1, 2 or 4 kb of Sγ1 repeats demonstrated a direct correlation between S region length and CSR efficiency (75). Additionally, replacement of Sγ1 with 2 kb of Sγ3, or 2 kb of Sγ1 as a size-matched control, led to equal IgG1 CSR, suggesting that the various S regions were interchangeable (76).
I can do anything better than μ?
S region transcription and R-loop formation had convincingly been demonstrated to play a vital role in CSR. However, a handful of studies modifying Sμ revealed important details. Deletion of all Sμ tandem repeats (ΔSμTR) (~3 kb) resulted in a 50% decrease in IgG1 CSR, suggesting the core repeats were not necessary, but increased CSR efficiency (103). In support of this, most ΔSμTR-derived IgG1 hybridomas had only rearranged the productive Igh allele, whereas wild-type-derived hybridomas frequently rearranged both alleles (94, 103–105). Because the majority of breakpoints in Sγ1, Sγ2b, Sγ3, Sε and Sα occur within the tandem repeat regions, this result initially appeared surprising. However, Sμ is unique in that approximately 40% of breakpoints fall outside of the tandem repeat region (57). Supporting this, deletion of all remaining Sμ pentameric motifs in addition to the tandem repeat region (ΔIμ–Cμ) resulted in a more severe reduction in CSR (106).
The CSR phenotype of ΔIμ–Cμ and ΔSμTR B cells correlated with their abilities to form R-loops. Because R-loops were found to initiate hundreds of base pairs upstream of the Sμ tandem repeats, the presence of R-loops in ΔSμTR B cells was no surprise (101). Predictably, R-loops could not be detected in ΔIμ–Cμ B cells. However, in a minority of ΔIμ–Cμ cells that were able to undergo successful CSR, breakpoints were found within the Iμ-exon. This demonstrated that CSR in the absence of R-loops and the Sμ region was possible, albeit at drastically lower levels (106).
What’s an R-loop worth to CSR? Ask a frog
Because of its AT-rich non-template strand (unlike G-rich sequences in mammals), the Xenopus laevis Sμ (XSμ) region offered a unique perspective on the precise contributions of R-loops to CSR. Predictably, a 4-kb XSμ region did not support transcription-dependent R-loop formation, although replacement of endogenous mouse Sγ1 with 4 kb XSμ supported 25% of wild-type IgG1 CSR in a hybridoma assay (107).
Insertion of a random 4-kb sequence resulted in a further 10-fold decrease in CSR, suggesting the presence of sequences within XSμ that supported AID targeting independent of R-loop formation (107). Breakpoint analysis within XSμ identified an enrichment for CSR junctions at or adjacent to ‘AGCT’ sequences, a variation of the recently characterized ‘DGYW’ AID hotspot motif (107, 108). The addition of replication protein A (RPA; this binds ssDNA and stabilizes its structure) was necessary to mediate AID deamination of XSμ sequences in vitro, supporting a model where RPA helps target AID to hotspot motifs in the absence of R-loops to mediate CSR (107). This model was consistent with the observation that SHM substrates unable to form R-loops required RPA for AID-mediated deamination (109).
AID’s function in SHM (occurring in fish, amphibians, birds, mammals) evolved approximately 100 million years before its function in CSR (occurring in amphibians, birds, mammals) (110, 111). Lieber and colleagues discussed the concept that G stretches within S regions evolved in mammals with CSR to more efficiently produce R-loops, and subsequently increase ssDNA exposure for AID deamination (77). To quantify the relative contribution of R-loops, Lieber and colleagues (77) cleverly compared CSR frequencies between experiments where Sγ1 has been inverted (74) or replaced with XSμ (107). In both cases, sequences supporting R-loops endowed 4-fold higher CSR than sequences that did not. If this analysis is extended to include two additional studies and limited only to various size-matched, inverted S regions (75, 76), the average benefit of forming RNA–DNA hybrids is a 3.9 ± 1.2-fold increase in CSR (Fig. 4B).
These XSμ studies demonstrated that while S region R-loops may have evolved to support more efficient CSR, a high-density of DGYW motifs throughout S regions can support AID deamination through an RPA-dependent, R-loop-independent mechanism. Importantly, both of these mechanisms are transcription dependent.
Deamination of template-strand DNA, in spite of RNA–DNA hybrids
R-loops provide a clear mechanism to account for AID targeting of the non-template strand. However, the formation of a thermodynamically stable RNA–DNA hybrid protects the template-strand DNA from deamination. Because AID deaminates both strands of DNA equally during CSR and SHM (102, 112), it remained a mystery how template-strand DNA strand was efficiently targeted. The RNA exosome was discovered to play an important role in targeting AID to both DNA strands, shedding light on this conundrum.
The RNA exosome is a conserved RNA degradation/processing complex that plays a vital role in RNA metabolism and transcriptional regulation (113). AID was found to interact with RNA exosome components in extracts from both CH12 and primary B cells stimulated to undergo CSR (11). Chromatin immunoprecipitation (ChIP) experiments revealed that the core RNA exosome component Exosc3 localized to both Sμ and Sγ1 in primary B cells stimulated to undergo IgG1 CSR. Strikingly, the addition of purified core RNA exosome components dramatically increased AID deamination of non-template strand DNA in vitro (11).
In a follow-up study, conditional deletion of Exosc3 in B cells decreased both SHM and CSR (114). Intriguingly, this deletion also revealed myriad transcription start site-associated antisense transcripts that correlated with AID-dependent translocation hotspots. CHiP experiments revealed that AID localized to these regions, and was dependent on the presence of Exosc3. Lastly, these regions also formed RNA–DNA hybrids, which were increased upon deletion of Exosc3 (114). Of note, the catalytic subunit of the RNA exosome was not required to target deamination of template-strand DNA in vitro (11). This demonstrated that exonucleolytic removal of the RNA transcript is not required for AID to access the template-strand DNA, suggesting a yet unidentified mechanism. Together, these studies demonstrate a role for the RNA exosome in targeting AID to loci with RNA–DNA hybrids, as well as mediating its deamination of the template-strand DNA.
In addition to the RNA exosome, RNase H was postulated to mediate template-strand deamination after a particularly curious finding. Surprisingly, treatment of S region R-loops with RNase H increased the amount of sodium bisulfite conversion along the template strand (96). This suggested that digestion of the hybridized RNA molecule could lead to an abrupt collapse of the R-loop and misalignment of S region repeats, precipitating the exposure of ssDNA to AID (96). While it is currently unknown if this mechanism occurs in vivo, a recent study overexpressing RNase H in transgenic mice found that CSR was unaffected, but template-strand deamination was marginally increased (115). More work is required to determine if endogenous levels of RNase H can support this mechanism.
Targeting AID with transcriptional machinery
Following the discovery of AID, numerous transcription-centric factors have been proposed to contribute to its targeting. An early study suggested an interaction with the RNA polymerase complex based on reciprocal co-immunoprecipitation experiments with AID and RNA polymerase II (Pol II) (116).
More recently, a study identified the RNA Pol II pausing factor Spt5 in an shRNA screen for factors required in CSR (117). In support of an interaction between AID and RNA Pol II, Spt5 and AID co-immunoprecipitated in both fibroblasts and CH12 cell lines. The genomic localization of Spt5, defined by CHiP-seq experiments, correlated with regions that had higher rates of mutation in a mouse model overexpressing AID, suggesting a strong correlation between the localization of Spt5 and AID (117). AID was also shown to interact with components of the splicing machinery; purification of AID complexes identified an interaction with the splicing factor PTBP2 (118). PTBP2 knockdown experiments revealed this interaction was important for efficient CSR, recalling previous findings on the requirement of processed GLTs (90, 92).
S region transcripts target AID to the Igh locus
Nearly four decades after the discovery of GLTs, a recent study provided evidence for an Igh-specific factor contributing to AID targeting: the S region transcripts themselves. Sμ and Sα RNAs transcribed in vitro were found to interact with AID in CH12 extracts, as did aptamer-tagged Sα RNAs expressed in vivo (10). The AID–RNA interaction was specific for transcripts generated in the physiological direction; reverse Sμ and Sα sequences did not interact with AID. Intriguingly, Sμ and Sα RNAs were found to fold into G-quadruplex structures that were necessary for their interaction with AID, as disruption of G-quadruplex structure via G to C mutations abolished direct AID binding (10). Notably, these RNA G-quadruplex structures echoed Sen and Gilbert’s discovery of ‘G4-DNA’ structures within S region DNA sequences (70). To determine the relevance of the AID–RNA interaction, a single point mutation in AID (G133V) was identified that completely disrupted direct RNA binding to Sμ and Sα. Importantly, this mutation did not interfere with ssDNA binding or AID deamination of ssDNA in vitro. Expression of AID-G133V was unable to complement IgG1 CSR in AID-deficient primary B cells, and localization of AID G133V to Sμ and Sγ1 was completely abolished (10).
These results suggested that direct AID–RNA binding is necessary for targeting AID to S regions, and predicts that depletion of S region transcripts would phenocopy the observed AID-G133V-dependent defect in CSR. To deplete S region transcripts, knock down of the lariat-debranching enzyme DBR1 was hypothesized to interfere with the processing of GLTs and the formation of soluble S region transcripts. Indeed, DBR1 knockdown in CH12 cells resulted in decreased AID localization to S regions as well as decreased IgA CSR. Remarkably, exogenous expression of forward Sα, but not reverse, rescued AID localization to Sα. Additionally, simultaneous exogenous expression of forward Sμ and forward Sα, but not their reverse counterparts, rescued IgA CSR (10).
A model for RNA-dependent AID targeting
In light of these findings, Radbruch and colleagues conclusions 20 years prior proved resoundingly prescient:
‘An attractive idea is that the spliced switch transcripts or the spliced intronic switch regions would interact with the recombination machinery. Either in trans, as a component of the switch recombinase, or in cis, rehybridizing with the DNA template.’ (92)
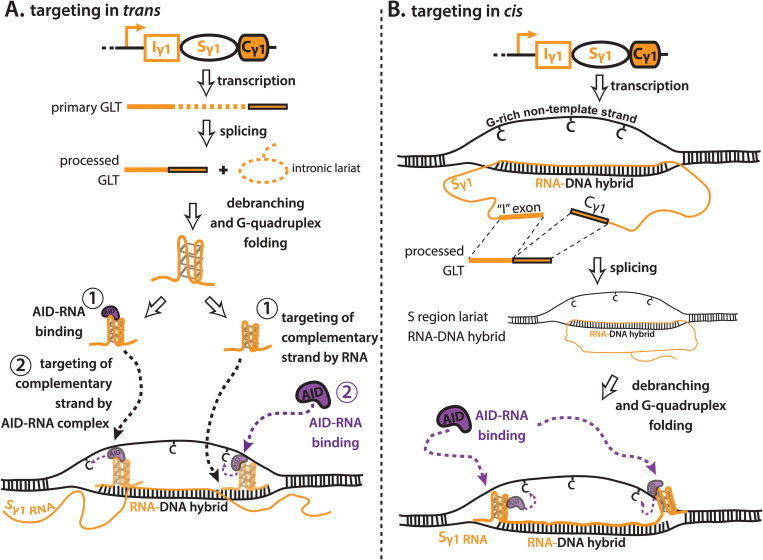
These data are consistent with a working model whereby processed S region transcripts serve to guide AID to the S regions from which they were transcribed (Fig. 5A and B). Splicing of GLTs liberates intronic S region lariats, which are debranched and can fold into G-quadruplex structures, enabling a direct interaction with AID. AID–RNA complexes are then targeted to S region DNA via the base complementarity between the S region transcripts and their DNA templates. In this sense, the proposed mechanism for RNA-dependent AID targeting is analogous to the mechanism for guide RNAs targeting Cas9 (119). This working model elicits many questions, predictions and caveats, which are discussed below.
Fig. 5.
Model for RNA-dependent targeting of AID during class switch recombination. (A) Targeting of AID–RNA complexes in trans. Germline transcripts (GLTs) are transcribed from downstream CH regions upon B cell activation, and undergo splicing to form a processed GLT and an intronic switch (S) region lariat. The S region lariat is debranched to form a linear S region transcript, which can fold into a G-quadruplex secondary RNA structure endowed by the G-richness of S region non-template strand DNA. It is currently unknown if debranching is required for G-quadruplex folding. G-quadruplexes can interact with soluble AID, and AID–RNA complexes are guided to the S region by the strand complementarity between the G-quadruplex and the S region from which it was transcribed (left path). Alternatively, the G-quadruplex can target to the S region independently of AID binding, and then recruit AID via AID–RNA binding (right path). For trans targeting, G-quadruplexes must compete with existing RNA–DNA hybrids for access to template-strand DNA. (B) Targeting of AID–RNA complexes in cis. GLTs are produced from downstream CH regions upon B cell activation; however, they remain attached to their template strand DNA via formation of an R-loop. The GLT undergoes splicing to liberate a processed GLT, while the S region lariat remains annealed as part of the R-loop. Debranching and folding into G-quadruplex secondary RNA structures mediates the recruitment of AID via AID–RNA binding. For cis targeting, G-quadruplexes are not required to compete with RNA–DNA hybrids for access to template-strand DNA; AID, activation-induced cytidine deaminase.
Cis vs trans targeting of AID–RNA complexes
S region transcripts can potentially target AID either in cis or in trans. Exogenous expression of S region transcripts was sufficient to rescue CSR in DBR1 knockdown cells (10), suggesting a trans mechanism is sufficient to support AID–RNA complex targeting to S regions (Fig. 5A). An important consideration for a trans model is that S region RNA–DNA hybrids are remarkably stable; trans targeting of AID–RNA complexes will have to compete with RNA–DNA hybrids in order to anneal to the template-strand DNA. Alternatively, the RNA exosome, Rnase H or R-loop collapse may expose single-strand template DNA that can be accessed by AID–RNA complexes in trans. Because template and non-template strands are deaminated equally during CSR (102), the template strand must be accessible at some stage of CSR. This particular stage may provide a window for base complementarity to occur between the RNA in the AID–RNA complex, and the template strand DNA.
Another possibility is that AID–RNA complexes are targeted in cis (Fig. 5B). For this to occur, a region of the unprocessed GLT (presumably part of the S region because of its ability form R-loops) may remain attached to the template-strand DNA as a component of an RNA–DNA hybrid. The remaining regions of the GLT are free to undergo co-transcriptional splicing and debranching, liberating a soluble processed GLT and allowing the S region RNA that is not annealed to fold into a G-quadruplex structure and recruit AID.
Additional roles for the AID–RNA interaction?
As a corollary of having reduced G-density, the AT-rich non-template strand of Xenopus S regions is unlikely to support G-quadruplex structures (although this has not been verified experimentally). If this is true, Xenopus S regions cannot utilize a mechanism for AID–RNA complex targeting that relies upon S region RNA binding to AID in a G-quadruplex-dependent manner.
Replacement of the endogenous mouse Sγ1 region with 4 kb of XSμ was sufficient to support 25% of wild-type CSR levels (107), suggesting that AID was still capable of targeting S regions in the absence of G-quadruplex-containing S transcripts. This suggests that AID deficient in binding G-quadruplex RNA, i.e AID-G133V, should be sufficient to undergo some degree of targeting to S regions and subsequent CSR. Intriguingly, AID-G133V was utterly unable to localize to S regions or rescue CSR in complementation experiments performed in AID-deficient primary B cells. One interpretation of these data is that AID-G133V is unable to utilize an R-loop-independent, RPA-dependent mechanism for targeting S regions. This hypothesis can be tested by determining if AID-G133V is still capable of binding RPA as previously described (109).
A more interesting interpretation of these data is that AID’s interaction with RNA confers an allosteric activation of enzymatic activity, facilitates AID oligomerization or initiates the formation of a multimeric complex containing AID and other components. In either of these scenarios, RNA binding would confer maximal enzymatic activity and efficient CSR. In the context of AID-G133V, the absence of G-quadruplex binding not only results in defective AID targeting, but also renders AID relatively inert due to its inability to undergo allosteric activation, oligomerize or form a multimeric complex. In the case of XSμ replacing mouse Sγ1, AID can still be effectively targeted through non-R-loop RPA-dependent mechanisms and potentially interact with soluble G-quadruplexes and RNAs from different loci in order achieve maximal enzymatic activity.
A thorough biochemical analysis of AID–RNA binding interactions, as well as the effect of RNA binding on AID enzymatic activity, will shed light on the potential for these mechanisms. Importantly, mutations that disrupt either G-quadruplex binding or render AID catalytically inactive will allow for the biochemical dissection of these two separate functions, and provide critical controls. Intriguingly, the working model for RNA-dependent targeting of AID predicts that AID can be targeted to loci outside the Igh locus by exogenously expressing a fusion transcript that contains a G-quadruplex motif for binding AID, and a targeting motif that is complementary to the loci of interest, analogous to CRISPR-Cas9 gene editing. This hypothesis can readily be tested in human or mouse cells lines using fluorescence microscopy.
G-quadruplexes and AID targeting in B cell malignancies
The discovery of S region RNAs interacting with AID was provocative due to the potential Igh locus-specific component endowed by base complementarity targeting. Ironically, while an Igh RNA guide may endow specificity for the Igh locus, the G-quadruplex binding property of AID may permit other loci to spuriously recruit AID by forming G-quadruplex structures. Intriguingly, in vitro transcription of the c-MYC and BCL6 genes, which are both common IGH translocation partners in B cell lymphomas, has been reported to promote R-loop formation, as well as G-quadruplex structures formed within the ssDNA portion of the R-loop, called ‘G-loops’ (120, 121). Using electron microscopy, AID was observed to bind to G-loop structures in vitro, suggesting that the G-quadruplex binding property of AID may contribute to aberrant targeting and malignant IGH translocations (121).
It is possible AID-targeting to c-MYC and BCL6 could be driven by a combination of DNA G-quadruplex and RNA G-quadruplex interactions. Likewise, it is possible that DNA G-quadruplex structures may play a role in targeting AID to the Igh locus. However, in sodium bisulfite experiments analyzing R-loops at S regions, no G-quadruplexes were detected along the exposed G-rich non-template ssDNA. In contrast, a loxP site was readily detected as a short stretch of unmodified bases, by virtue of its stem loop secondary structure protecting paired bases from modification (101). While further experimentation is needed to confirm these results, current evidence suggests mouse S region G-quadruplexes may be limited to RNA structures. Regardless, AID-G133V will be an invaluable tool to assess the contribution of G-quadruplexes to aberrant AID targeting.
Conclusions
Recounting 40 years of CSR discoveries highlights the complexity of AID targeting. Myriad factors play a role in targeting AID to the Igh locus, including R-loops, splicing, the transcriptional machinery, RPA, the RNA exosome and noncoding S region transcripts. Perplexingly, none of these factors can account for Igh locus-specific localization, but rather localize throughout the genome. A major challenge remains to decipher the remarkable specificity of AID targeting to the Igh locus, and how mistakes might lead to B cell malignancies, which given the high wire nature of CSR and SHM, are much less frequent than would seem likely. Clearly, evolution has taken great interest in minimizing the risk of CSR and SHM, which emphasizes their benefit to survival in a world filled with pathogens and other dangers.
Funding
This work was supported by grants from the National Institutes of Health (1RO1AI072194 and 1RO1AI124186) to J.C. W.T.Y was supported by a T32 training fellowship (4T32CA009149-40).
Acknowledgements
We thank members of the Chaudhuri laboratory for helpful discussions and J. Yewdell for thoughtful comments.
Conflicts of Interest statement: The authors declared no conflicts of interest.
References
- 1. Muramatsu M., Sankaranand V. S., Anant S., et al. 1999. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem. 274:18470. [DOI] [PubMed] [Google Scholar]
- 2. Muramatsu M. Kinoshita K. Fagarasan S. Yamada S. Shinkai Y. and Honjo T. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102:553. [DOI] [PubMed] [Google Scholar]
- 3. Revy P., Muto T., Levy Y., et al. 2000. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2). Cell 102:565. [DOI] [PubMed] [Google Scholar]
- 4. Rada C. Di Noia J. M. and Neuberger M. S. 2004. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol. Cell 16:163. [DOI] [PubMed] [Google Scholar]
- 5. Petersen-Mahrt S. K. Harris R. S. and Neuberger M. S. 2002. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature 418:99. [DOI] [PubMed] [Google Scholar]
- 6. Matthews A. J. Zheng S. DiMenna L. J. and Chaudhuri J. 2014. Regulation of immunoglobulin class-switch recombination: choreography of noncoding transcription, targeted DNA deamination, and long-range DNA repair. Adv. Immunol. 122:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lieber M. R. 2016. Mechanisms of human lymphoid chromosomal translocations. Nat. Rev. Cancer 16:387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pavri R. and Nussenzweig M. C. 2011. AID targeting in antibody diversity. Adv. Immunol. 110:1. [DOI] [PubMed] [Google Scholar]
- 9. Casellas R. Basu U. Yewdell W. T. Chaudhuri J. Robbiani D. F. and Di Noia J. M. 2016. Mutations, kataegis and translocations in B cells: understanding AID promiscuous activity. Nat. Rev. Immunol. 16:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zheng S. Vuong B. Q. Vaidyanathan B. Lin J. Y. Huang F. T. and Chaudhuri J. 2015. Non-coding RNA generated following lariat debranching mediates targeting of AID to DNA. Cell 161:762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Basu U., Meng F. L., Keim C., et al. 2011. The RNA exosome targets the AID cytidine deaminase to both strands of transcribed duplex DNA substrates. Cell 144:353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Laffleur B., Basu U. and Lim J. 2017. RNA exosome and non-coding RNA-coupled mechanisms in AID-mediated genomic alterations. J. Mol. Biol., in press. doi:10.1016/j.jmb.2016.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rinn J. L. and Chang H. Y. 2012. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gearhart P. J., Sigal N. H. and Klinman N. R. 1975. Production of antibodies of identical idiotype but diverse immunoglobulin classes by cells derived from a single stimulated B cell. Proc. Natl Acad. Sci. USA 72:1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pernis B. Forni L. and Amante L. 1971. Immunoglobulins as cell receptors. Ann. N. Y. Acad. Sci. 190:420. [DOI] [PubMed] [Google Scholar]
- 16. Nossal G. J. Warner N. L. and Lewis H. 1971. Incidence of cells simultaneously secreting IgM and IgG antibody to sheep erythrocytes. Cell. Immunol. 2:41. [DOI] [PubMed] [Google Scholar]
- 17. Press J. L. and Klinman N. R. 1973. Monoclonal production of both IgM and IgG1 antihapten antibody. J. Exp. Med. 138:300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Honjo T. and Kataoka T. 1978. Organization of immunoglobulin heavy chain genes and allelic deletion model. Proc. Natl Acad. Sci. USA 75:2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rada C. Williams G. T. Nilsen H. Barnes D. E. Lindahl T. and Neuberger M. S. 2002. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr. Biol. 12:1748. [DOI] [PubMed] [Google Scholar]
- 20. Chaudhuri J. Tian M. Khuong C. Chua K. Pinaud E. and Alt F. W. 2003. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature 422:726. [DOI] [PubMed] [Google Scholar]
- 21. Bransteitter R., Pham P., Scharff M. D. and Goodman M. F. 2003. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc. Natl Acad. Sci. USA 100:4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dickerson S. K. Market E. Besmer E. and Papavasiliou F. N. 2003. AID mediates hypermutation by deaminating single stranded DNA. J. Exp. Med. 197:1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ramiro A. R. Stavropoulos P. Jankovic M. and Nussenzweig M. C. 2003. Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nat. Immunol. 4:452. [DOI] [PubMed] [Google Scholar]
- 24. Sohail A. Klapacz J. Samaranayake M. Ullah A. and Bhagwat A. S. 2003. Human activation-induced cytidine deaminase causes transcription-dependent, strand-biased C to U deaminations. Nucleic Acids Res. 31:2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Alt F. W., Bothwell A. L., Knapp M., et al. 1980. Synthesis of secreted and membrane-bound immunoglobulin mu heavy chains is directed by mRNAs that differ at their 3’ ends. Cell 20:293. [DOI] [PubMed] [Google Scholar]
- 26. Rogers J., Early P., Carter C., et al. 1980. Two mRNAs with different 3’ ends encode membrane-bound and secreted forms of immunoglobulin mu chain. Cell 20:303. [DOI] [PubMed] [Google Scholar]
- 27. Early P., Rogers J., Davis M., et al. 1980. Two mRNAs can be produced from a single immunoglobulin mu gene by alternative RNA processing pathways. Cell 20:313. [DOI] [PubMed] [Google Scholar]
- 28. Kemp D. J., Harris A. W., Cory S. and Adams J. M. 1980. Expression of the immunoglobulin C mu gene in mouse T and B lymphoid and myeloid cell lines. Proc. Natl Acad. Sci. USA 77:2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kemp D. J. Wilson A. Harris A. W. and Shortman K. 1980. The immunoglobulin mu constant region gene is expressed in mouse thymocytes. Nature 286:168. [DOI] [PubMed] [Google Scholar]
- 30. Kemp D. J., Harris A. W. and Adams J. M. 1980. Transcripts of the immunoglobulin C mu gene vary in structure and splicing during lymphoid development. Proc. Natl Acad. Sci. USA 77:7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Alt F. W. Rosenberg N. Enea V. Siden E. and Baltimore D. 1982. Multiple immunoglobulin heavy-chain gene transcripts in Abelson murine leukemia virus-transformed lymphoid cell lines. Mol. Cell. Biol. 2:386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lennon G. G. and Perry R. P. 1985. C mu-containing transcripts initiate heterogeneously within the IgH enhancer region and contain a novel 5’-nontranslatable exon. Nature 318:475. [DOI] [PubMed] [Google Scholar]
- 33. Sitia R. Rubartelli A. and Hammerling U. 1981. Expression of 2 immunoglobulin isotypes, IgM and IgA, with identical idiotype in the B cell lymphoma I.29. J. Immunol. 127:1388. [PubMed] [Google Scholar]
- 34. Stavnezer J. Marcu K. B. Sirlin S. Alhadeff B. and Hammerling U. 1982. Rearrangements and deletions of immunoglobulin heavy chain genes in the double-producing B cell lymphoma I.29. Mol. Cell. Biol. 2:1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stavnezer J. Sirlin S. and Abbott J. 1985. Induction of immunoglobulin isotype switching in cultured I.29 B lymphoma cells. Characterization of the accompanying rearrangements of heavy chain genes. J. Exp. Med. 161:577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stavnezer-Nordgren J. and Sirlin S. 1986. Specificity of immunoglobulin heavy chain switch correlates with activity of germline heavy chain genes prior to switching. EMBO J. 5:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yancopoulos G. D. DePinho R. A. Zimmerman K. A. Lutzker S. G. Rosenberg N. and Alt F. W. 1986. Secondary genomic rearrangement events in pre-B cells: VHDJH replacement by a LINE-1 sequence and directed class switching. EMBO J. 5:3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lutzker S. Rothman P. Pollock R. Coffman R. and Alt F. W. 1988. Mitogen- and IL-4-regulated expression of germ-line Ig gamma 2b transcripts: evidence for directed heavy chain class switching. Cell 53:177. [DOI] [PubMed] [Google Scholar]
- 39. Severinson E. Fernandez C. and Stavnezer J. 1990. Induction of germ-line immunoglobulin heavy chain transcripts by mitogens and interleukins prior to switch recombination. Eur. J. Immunol. 20:1079. [DOI] [PubMed] [Google Scholar]
- 40. Stavnezer J., Radcliffe G., Lin Y. C., Nietupski J., Berggren L., Sitia R. and Severinson E. 1988. Immunoglobulin heavy-chain switching may be directed by prior induction of transcripts from constant-region genes. Proc. Natl Acad. Sci. USA 85:7704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rothman P. Lutzker S. Cook W. Coffman R. and Alt F. W. 1988. Mitogen plus interleukin 4 induction of C epsilon transcripts in B lymphoid cells. J. Exp. Med. 168:2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Berton M. T., Uhr J. W. and Vitetta E. S. 1989. Synthesis of germ-line gamma 1 immunoglobulin heavy-chain transcripts in resting B cells: induction by interleukin 4 and inhibition by interferon gamma. Proc. Natl Acad. Sci. USA 86:2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Esser C. and Radbruch A. 1989. Rapid induction of transcription of unrearranged S gamma 1 switch regions in activated murine B cells by interleukin 4. EMBO J. 8:483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gauchat J. F. Lebman D. A. Coffman R. L. Gascan H. and de Vries J. E. 1990. Structure and expression of germline epsilon transcripts in human B cells induced by interleukin 4 to switch to IgE production. J. Exp. Med. 172:463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shockett P. and Stavnezer J. 1991. Effect of cytokines on switching to IgA and alpha germline transcripts in the B lymphoma I.29 mu. Transforming growth factor-beta activates transcription of the unrearranged C alpha gene. J. Immunol. 147:4374. [PubMed] [Google Scholar]
- 46. Collins J. T. and Dunnick W. A. 1993. Germline transcripts of the murine immunoglobulin gamma 2a gene: structure and induction by IFN-gamma. Int. Immunol. 5:885. [DOI] [PubMed] [Google Scholar]
- 47. Lebman D. A., Nomura D. Y., Coffman R. L. and Lee F. D. 1990. Molecular characterization of germ-line immunoglobulin A transcripts produced during transforming growth factor type beta-induced isotype switching. Proc. Natl Acad. Sci. USA 87:3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Picard D. and Schaffner W. 1984. Unrearranged immunoglobulin lambda variable region is transcribed in kappa-producing myelomas. EMBO J. 3:3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yancopoulos G. D. and Alt F. W. 1985. Developmentally controlled and tissue-specific expression of unrearranged VH gene segments. Cell 40:271. [DOI] [PubMed] [Google Scholar]
- 50. Blackwell T. K., Moore M. W., Yancopoulos G. D., et al. 1986. Recombination between immunoglobulin variable region gene segments is enhanced by transcription. Nature 324:585. [DOI] [PubMed] [Google Scholar]
- 51. Thomas B. J. and Rothstein R. 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56:619. [DOI] [PubMed] [Google Scholar]
- 52. Lutzker S. and Alt F. W. 1988. Structure and expression of germ line immunoglobulin gamma 2b transcripts. Mol. Cell. Biol. 8:1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Radcliffe G. Lin Y. C. Julius M. Marcu K. B. and Stavnezer J. 1990. Structure of germ line immunoglobulin alpha heavy-chain RNA and its location on polysomes. Mol. Cell. Biol. 10:382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rothman P., Chen Y. Y., Lutzker S., et al. 1990. Structure and expression of germ line immunoglobulin heavy-chain epsilon transcripts: interleukin-4 plus lipopolysaccharide-directed switching to C epsilon. Mol. Cell. Biol. 10:1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rothman P. Lutzker S. Gorham B. Stewart V. Coffman R. and Alt F. W. 1990. Structure and expression of germline immunoglobulin gamma 3 heavy chain gene transcripts: implications for mitogen and lymphokine directed class-switching. Int. Immunol. 2:621. [DOI] [PubMed] [Google Scholar]
- 56. Rothman P. Li S. C. Gorham B. Glimcher L. Alt F. and Boothby M. 1991. Identification of a conserved lipopolysaccharide-plus-interleukin-4-responsive element located at the promoter of germ line epsilon transcripts. Mol. Cell. Biol. 11:5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dunnick W. Hertz G. Z. Scappino L. and Gritzmacher C. 1993. DNA sequences at immunoglobulin switch region recombination sites. Nucleic Acids Res. 21:365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shimizu A. and Honjo T. 1984. Immunoglobulin class switching. Cell 36:801. [DOI] [PubMed] [Google Scholar]
- 59. Sakano H. Maki R. Kurosawa Y. Roeder W. and Tonegawa S. 1980. Two types of somatic recombination are necessary for the generation of complete immunoglobulin heavy-chain genes. Nature 286:676. [DOI] [PubMed] [Google Scholar]
- 60. Dunnick W. Rabbitts T. H. and Milstein C. 1980. An immunoglobulin deletion mutant with implications for the heavy-chain switch and RNA splicing. Nature 286:669. [DOI] [PubMed] [Google Scholar]
- 61. Davis M. M. Kim S. K. and Hood L. E. 1980. DNA sequences mediating class switching in alpha-immunoglobulins. Science 209:1360. [DOI] [PubMed] [Google Scholar]
- 62. Kataoka T. Miyata T. and Honjo T. 1981. Repetitive sequences in class-switch recombination regions of immunoglobulin heavy chain genes. Cell 23:357. [DOI] [PubMed] [Google Scholar]
- 63. Nikaido T. Yamawaki-Kataoka Y. and Honjo T. 1982. Nucleotide sequences of switch regions of immunoglobulin C epsilon and C gamma genes and their comparison. J. Biol. Chem. 257:7322. [PubMed] [Google Scholar]
- 64. Stanton L. W. and Marcu K. B. 1982. Nucleotide sequence and properties of the murine gamma 3 immunoglobulin heavy chain gene switch region: implications for successive C gamma gene switching. Nucleic Acids Res. 10:5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Arpin C., de Bouteiller O., Razanajaona D., et al. 1998. The normal counterpart of IgD myeloma cells in germinal center displays extensively mutated IgVH gene, Cmu-Cdelta switch, and lambda light chain expression. J. Exp. Med. 187:1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chen K., Xu W., Wilson M., et al. 2009. Immunoglobulin D enhances immune surveillance by activating antimicrobial, proinflammatory and B cell-stimulating programs in basophils. Nat. Immunol. 10:889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rouaud P., Saintamand A., Saad F., et al. 2014. Elucidation of the enigmatic IgD class-switch recombination via germline deletion of the IgH 3’ regulatory region. J. Exp. Med. 211:975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Choi J. H., Wang K. W., Zhang D. et al. 2017. IgD class switching is initiated by microbiota and limited to mucosa-associated lymphoid tissue in mice. Proc. Natl Acad. Sci. USA 114:E1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Collier D. A. Griffin J. A. and Wells R. D. 1988. Non-B right-handed DNA conformations of homopurine.homopyrimidine sequences in the murine immunoglobulin C alpha switch region. J. Biol. Chem. 263:7397. [PubMed] [Google Scholar]
- 70. Sen D. and Gilbert W. 1988. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature 334:364. [DOI] [PubMed] [Google Scholar]
- 71. Hoogsteen K. 1959. The structure of crystals containing a hydrogen-bonded complex of 1-methylthymine and 9-methyladenine. Acta Cryst. 12:822. [Google Scholar]
- 72. Reaban M. E. and Griffin J. A. 1990. Induction of RNA-stabilized DNA conformers by transcription of an immunoglobulin switch region. Nature 348:342. [DOI] [PubMed] [Google Scholar]
- 73. Reaban M. E. Lebowitz J. and Griffin J. A. 1994. Transcription induces the formation of a stable RNA.DNA hybrid in the immunoglobulin alpha switch region. J. Biol. Chem. 269:21850. [PubMed] [Google Scholar]
- 74. Shinkura R. Tian M. Smith M. Chua K. Fujiwara Y. and Alt F. W. 2003. The influence of transcriptional orientation on endogenous switch region function. Nat. Immunol. 4:435. [DOI] [PubMed] [Google Scholar]
- 75. Zarrin A. A., Tian M., Wang J., Borjeson T. and Alt F. W. 2005. Influence of switch region length on immunoglobulin class switch recombination. Proc. Natl Acad. Sci. USA 102:2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zarrin A. A. Goff P. H. Senger K. and Alt F. W. 2008. Sgamma3 switch sequences function in place of endogenous Sgamma1 to mediate antibody class switching. J. Exp. Med. 205:1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Roy D. Yu K. and Lieber M. R. 2008. Mechanism of R-loop formation at immunoglobulin class switch sequences. Mol. Cell. Biol. 28:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Daniels G. A. and Lieber M. R. 1995. RNA:DNA complex formation upon transcription of immunoglobulin switch regions: implications for the mechanism and regulation of class switch recombination. Nucleic Acids Res. 23:5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Tian M. and Alt F. W. 2000. Transcription-induced cleavage of immunoglobulin switch regions by nucleotide excision repair nucleases in vitro. J. Biol. Chem. 275:24163. [DOI] [PubMed] [Google Scholar]
- 80. Ott D. E. Alt F. W. and Marcu K. B. 1987. Immunoglobulin heavy chain switch region recombination within a retroviral vector in murine pre-B cells. EMBO J. 6:577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Leung H. and Maizels N. 1992. Transcriptional regulatory elements stimulate recombination in extrachromosomal substrates carrying immunoglobulin switch-region sequences. Proc. Natl Acad. Sci. USA 89:4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Daniels G. A. and Lieber M. R. 1995. Strand specificity in the transcriptional targeting of recombination at immunoglobulin switch sequences. Proc. Natl Acad. Sci. USA 92:5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kinoshita K. Tashiro J. Tomita S. Lee C. G. and Honjo T. 1998. Target specificity of immunoglobulin class switch recombination is not determined by nucleotide sequences of S regions. Immunity 9:849. [DOI] [PubMed] [Google Scholar]
- 84. Leung H. and Maizels N. 1994. Regulation and targeting of recombination in extrachromosomal substrates carrying immunoglobulin switch region sequences. Mol. Cell. Biol. 14:1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Jung S. Rajewsky K. and Radbruch A. 1993. Shutdown of class switch recombination by deletion of a switch region control element. Science 259:984. [DOI] [PubMed] [Google Scholar]
- 86. Zhang J. Bottaro A. Li S. Stewart V. and Alt F. W. 1993. A selective defect in IgG2b switching as a result of targeted mutation of the I gamma 2b promoter and exon. EMBO J. 12:3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Gu H. Zou Y. R. and Rajewsky K. 1993. Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre-loxP-mediated gene targeting. Cell 73:1155. [DOI] [PubMed] [Google Scholar]
- 88. Xu L., Gorham B., Li S. C., Bottaro A., Alt F. W. and Rothman P. 1993. Replacement of germ-line epsilon promoter by gene targeting alters control of immunoglobulin heavy chain class switching. Proc. Natl Acad. Sci. USA 90:3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Bottaro A. Lansford R. Xu L. Zhang J. Rothman P. and Alt F. W. 1994. S region transcription per se promotes basal IgE class switch recombination but additional factors regulate the efficiency of the process. EMBO J. 13:665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Lorenz M. Jung S. and Radbruch A. 1995. Switch transcripts in immunoglobulin class switching. Science 267:1825. [DOI] [PubMed] [Google Scholar]
- 91. Harriman G. R. Bradley A. Das S. Rogers-Fani P. and Davis A. C. 1996. IgA class switch in I alpha exon-deficient mice. Role of germline transcription in class switch recombination. J. Clin. Invest. 97:477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Hein K. Lorenz M. G. Siebenkotten G. Petry K. Christine R. and Radbruch A. 1998. Processing of switch transcripts is required for targeting of antibody class switch recombination. J. Exp. Med. 188:2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Seidl K. J. Bottaro A. Vo A. Zhang J. Davidson L. and Alt F. W. 1998. An expressed neo® cassette provides required functions of the 1gamma2b exon for class switching. Int. Immunol. 10:1683. [DOI] [PubMed] [Google Scholar]
- 94. Bottaro A. Young F. Chen J. Serwe M. Sablitzky F. and Alt F. W. 1998. Deletion of the IgH intronic enhancer and associated matrix-attachment regions decreases, but does not abolish, class switching at the mu locus. Int. Immunol. 10:799. [DOI] [PubMed] [Google Scholar]
- 95. Mizuta R., Iwai K., Shigeno M., et al. 2003. Molecular visualization of immunoglobulin switch region RNA/DNA complex by atomic force microscope. J. Biol. Chem. 278:4431. [DOI] [PubMed] [Google Scholar]
- 96. Yu K. Chedin F. Hsieh C. L. Wilson T. E. and Lieber M. R. 2003. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 4:442. [DOI] [PubMed] [Google Scholar]
- 97. Roberts R. W. and Crothers D. M. 1992. Stability and properties of double and triple helices: dramatic effects of RNA or DNA backbone composition. Science 258:1463. [DOI] [PubMed] [Google Scholar]
- 98. Ratmeyer L. Vinayak R. Zhong Y. Y. Zon G. and Wilson W. D. 1994. Sequence specific thermodynamic and structural properties for DNA.RNA duplexes. Biochemistry 33:5298. [DOI] [PubMed] [Google Scholar]
- 99. Huang F. T., Yu K., Hsieh C. L. and Lieber M. R. 2006. Downstream boundary of chromosomal R-loops at murine switch regions: implications for the mechanism of class switch recombination. Proc. Natl Acad. Sci. USA 103:5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Zhang Z. Z. Pannunzio N. R. Hsieh C. L. Yu K. and Lieber M. R. 2014. The role of G-density in switch region repeats for immunoglobulin class switch recombination. Nucleic Acids Res. 42:13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Huang F. T., Yu K., Balter B. B., et al. 2007. Sequence dependence of chromosomal R-loops at the immunoglobulin heavy-chain Smu class switch region. Mol. Cell. Biol. 27:5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Xue K. Rada C. and Neuberger M. S. 2006. The in vivo pattern of AID targeting to immunoglobulin switch regions deduced from mutation spectra in msh2-/- ung-/- mice. J. Exp. Med. 203:2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Luby T. M. Schrader C. E. Stavnezer J. and Selsing E. 2001. The mu switch region tandem repeats are important, but not required, for antibody class switch recombination. J. Exp. Med. 193:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Hummel M. Berry J. K. and Dunnick W. 1987. Switch region content of hybridomas: the two spleen cell Igh loci tend to rearrange to the same isotype. J. Immunol. 138:3539. [PubMed] [Google Scholar]
- 105. Winter E. Krawinkel U. and Radbruch A. 1987. Directed Ig class switch recombination in activated murine B cells. EMBO J. 6:1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Khamlichi A. A. Glaudet F. Oruc Z. Denis V. Le Bert M. and Cogné M. 2004. Immunoglobulin class-switch recombination in mice devoid of any S mu tandem repeat. Blood 103:3828. [DOI] [PubMed] [Google Scholar]
- 107. Zarrin A. A., Alt F. W., Chaudhuri J., et al. 2004. An evolutionarily conserved target motif for immunoglobulin class-switch recombination. Nat. Immunol. 5:1275. [DOI] [PubMed] [Google Scholar]
- 108. Rogozin I. B. and Diaz M. 2004. Cutting edge: DGYW/WRCH is a better predictor of mutability at G:C bases in Ig hypermutation than the widely accepted RGYW/WRCY motif and probably reflects a two-step activation-induced cytidine deaminase-triggered process. J. Immunol. 172:3382. [DOI] [PubMed] [Google Scholar]
- 109. Chaudhuri J. Khuong C. and Alt F. W. 2004. Replication protein A interacts with AID to promote deamination of somatic hypermutation targets. Nature 430:992. [DOI] [PubMed] [Google Scholar]
- 110. Flajnik M. F. 2002. Comparative analyses of immunoglobulin genes: surprises and portents. Nat. Rev. Immunol. 2:688. [DOI] [PubMed] [Google Scholar]
- 111. Du Pasquier L. 2001. The immune system of invertebrates and vertebrates. Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 129:1. [DOI] [PubMed] [Google Scholar]
- 112. Milstein C., Neuberger M. S. and Staden R. 1998. Both DNA strands of antibody genes are hypermutation targets. Proc. Natl Acad. Sci. USA 95:8791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Kilchert C. Wittmann S. and Vasiljeva L. 2016. The regulation and functions of the nuclear RNA exosome complex. Nat. Rev. Mol. Cell Biol. 17:227. [DOI] [PubMed] [Google Scholar]
- 114. Pefanis E., Wang J., Rothschild G., et al. 2014. Noncoding RNA transcription targets AID to divergently transcribed loci in B cells. Nature 514:389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Maul R. W., Chon H., Sakhuja K., Cerritelli S. M., Gugliotti L. A., Gearhart P. J. and Crouch R. J. 2017. R-Loop depletion by over-expressed RNase H1 in mouse B cells increases activation-induced deaminase access to the transcribed strand without altering frequency of isotype switching. J. Mol. Biol., in press. doi:10.1016/j.jmb.2016.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Nambu Y., Sugai M., Gonda H., et al. 2003. Transcription-coupled events associating with immunoglobulin switch region chromatin. Science 302:2137. [DOI] [PubMed] [Google Scholar]
- 117. Pavri R., Gazumyan A., Jankovic M., et al. 2010. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell 143:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Nowak U. Matthews A. J. Zheng S. and Chaudhuri J. 2011. The splicing regulator PTBP2 interacts with the cytidine deaminase AID and promotes binding of AID to switch-region DNA. Nat. Immunol. 12:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Doudna J. A. and Charpentier E. 2014. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346:1258096. [DOI] [PubMed] [Google Scholar]
- 120. Duquette M. L. Pham P. Goodman M. F. and Maizels N. 2005. AID binds to transcription-induced structures in c-MYC that map to regions associated with translocation and hypermutation. Oncogene 24:5791. [DOI] [PubMed] [Google Scholar]
- 121. Duquette M. L. Huber M. D. and Maizels N. 2007. G-rich proto-oncogenes are targeted for genomic instability in B-cell lymphomas. Cancer Res. 67:2586. [DOI] [PubMed] [Google Scholar]