Abstract
Cancer stem cells (CSCs) represent a subset of cells within tumours that exhibit self-renewal and tumour seeding capacity. CSCs are typically refractory to conventional treatments and have been associated to metastasis and relapse. Salinomycin operates as a selective agent against CSCs through mechanisms that remain elusive. Here, we provide evidence that a synthetic derivative of salinomycin, which we named ironomycin (AM5), exhibits a more potent and selective activity against breast CSCs in vitro and in vivo, accumulates and sequesters iron in lysosomes. In response to the ensuing cytoplasmic depletion of iron, cells triggered the degradation of ferritin in lysosomes, leading to further iron loading in this organelle. Iron-mediated production of reactive oxygen species promoted lysosomal membrane permeabilization, activating a cell death pathway consistent with ferroptosis. These findings reveal the prevalence of iron homeostasis in breast CSCs, pointing towards iron and iron-mediated processes as potential targets against these cells.
During embryogenesis, epithelial cells undergo extensive epigenetic reprogramming, allowing them to transdifferentiate and acquire physical properties of mesenchymal cells1. In this manner, cells can detach from primary tissues and migrate to distant locations. This process, known as epithelial-to-mesenchymal transition (EMT), is accompanied by the reverse transition, namely MET, giving rise to specialized tissues in metazoan development. The paradigm of cancer stem cells (CSCs) defines the existence of subpopulations of cells able to harness similar mechanisms to disseminate, initiate and sustain tumour growth1–3. Downregulation of signalling and cell adhesion proteins such as E-cadherin, and activation of several other pathways confer these cells motility and invasion capacity. Although CSCs typically represent a small fraction of solid tumours, it has been shown in leukaemia and melanoma that the frequency of cells exhibiting similar properties can be higher4,5. CSCs have been shown to be refractory to conventional treatments and can cause relapse. Furthermore, CSCs are inherently difficult to isolate and to maintain in culture making it impractical to screen small molecules for effecacy against these cells. This obstacle has prompted the artificial induction of EMT to produce cells displaying properties reminiscent of those of CSCs suitable for high-throughput phenotypic screening6–8. High-throughput screening (HTS) coupled to the use of physiologically relevant models identified lovastatin as a potent drug against leukaemia stem cells through a mechanism involving the inhibition of hydroxymethylglutaryl-coenzyme A (HMG-CoA) reductase9. In contrast, salinomycin (Sal) has been identified by HTS as a selective agent against experimentally induced CSCs6, and its activity associated to the intrinsic amphiphilic behaviour of the natural product, facilitating the transport of polar alkali metals through lipophilic membranes10. Sal has been shown to inhibit Wingless-related integration site (Wnt) signalling11 and to perturb autophagic flux12 among other effects. Sal has also been shown to have therapeutic potential in humans13. However, explicit mechanisms underlying the pleiotropic nature of Sal and its ability to eradicate CSCs are unknown.
Results and discussion
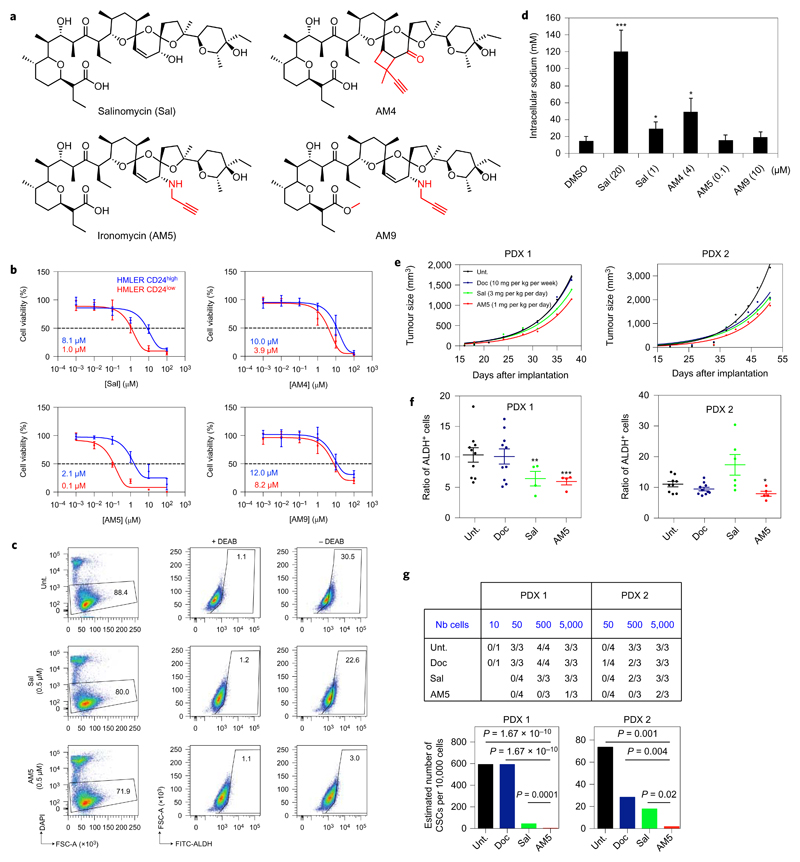
To identify mechanisms at work, we sought to generate a small library of structural variants with enhanced potency (Fig. 1a, Supplementary Fig. 1 and Supplementary Information). We reasoned that more effective derivatives would allow us to operate at lower doses, potentially dissociating the generic toxicity linked to ion transport from a selective effect against CSCs. Prior work in this area includes the development of remarkable synthetic strategies and the production of original collections of Sal derivatives14–19. In our hands, molecular editing of Sal using a chemoselective oxidation followed by a stereoselective reductive amination at C20 led to the alkyne derivative AM5 along with its methylated counterpart AM9. The C18=C19 double bond was functionalized through a [π2s + π2s] photocycloaddition yielding the apolar alkyne derivative AM4. Sal derivatives were first evaluated against transformed human mammary epithelial HMLER CD44high/CD24low cells (HMLER CD24low), a previously established model of human breast CSCs6,7, and a control isogenic cell line (HMLER CD24high). The zwitterionic derivative AM5 displayed a ~tenfold higher potency against HMLER CD24low cells compared to Sal with an IC50 value of ~100 nM whilst maintaining selectivity over control cells (Fig. 1b). The negatively charged AM4 retained some selectivity compared to Sal, whereas the methylated derivative AM9 was significantly less potent and less selective compared to AM5 indicating that the free carboxylate at C1 is functionally required. AM5 selectively targeted the aldehyde dehydrogenase positive (ALDH+) subpopulation of another model of CSCs, namely iCSCL-10A2 cells20, more effectively than Sal (Fig. 1c), and exhibited little toxicity against primary breast cells (Supplementary Fig. 2). These drugs abolished the capacity of HMLER CD24low cells to form colonies at low concentrations (Supplementary Fig. 3), and AM5 prevented these cells from developing tumourspheres in suspension, a well-established characteristic of CSCs, at doses as low as 30 nM (Supplementary Fig. 3). Conversely, AM9 was ineffective, validating the carboxylate as a required motif to alter CSC maintenance. These data illustrated the general susceptibility of AM5 to selectively target CSCs in vitro according to cell surface markers and ALDH activity. Then, each derivative was evaluated for its propensity to carry sodium through the plasma membrane of CSCs by monitoring changes in fluorescence of the sodium-binding benzofuran isophtalate (SBFI)21. While Sal induced a fast increase in intracellular sodium at a dose as high as twenty times the IC50 value, AM5 had no effect at doses effective against the proliferation of HMLER CD24low cells (Fig. 1d). This data challenged the idea that Sal selectively kills CSCs by directly altering membrane potentials6. AM5 prevented tumour growth in human breast cancer MCF-7 cells xenograft-bearing mice without generic toxicity, attested by a constant body weight throughout treatment and the integrity of peripheral tissues (Supplementary Fig. 4), whereas a fivefold higher concentration was lethal, suggesting a specific mechanism of action at low doses. Sal and AM5 reduced tumour growth in two early passage patient-derived xenografts (PDXs)22, where the clinically approved drug docetaxel (Doc) was less effective (Fig. 1e). Most importantly, this effect was associated with a reduced ratio of ALDH+ cells (Fig. 1f), and a decreased tumour-seeding capacity of tumour cells treated in vivo without detectable toxicity at effective doses, with AM5 being more potent than Sal and Doc (Fig. 1g and Supplementary Fig. 5). These data provided solid evidence that AM5 selectively targets CSCs in vivo.
Figure 1. Sal and AM5 alter the maintenance of CSCs independently of sodium transport.
a, Molecular structures of Sal and synthetic derivatives. b, Dose–response viability curves of HMLER CD24low and control cells treated for 72 h as indicated. Data points and error bars, mean values and s.d. of three biological replicates. c, In vitro effect of Sal and AM5 against ALDH+ iCSCL-10A2 cell subpopulation treated for 48 h measured by flow cytometry. DEAB, ALDH inhibitor. d, Quantification of sodium uptake by ratiometric fluorescence in HMLER CD24low cells treated as indicated. Bars and error bars, mean values and s.d. of three biological replicates. e, In vivo antitumour effect of Sal and AM5 against PDX in NOD/scid mice treated as indicated by means of intra-peritoneal injections (n ≥ 4 per condition per PDX). f, Quantification of the proportion of residual ALDH+ cells in PDX treated as in e measured by flow cytometry. Bars and error bars, mean values and s.d. g, Tumour-seeding capacity of cells treated in vivo as in e and estimated number of CSCs calculated by extreme limiting dilution analysis (ELDA) software. P values, χ2 pairwise test. In d,f *P < 0.05, **P < 0.01, ***P < 0.001, Student’s t-test.
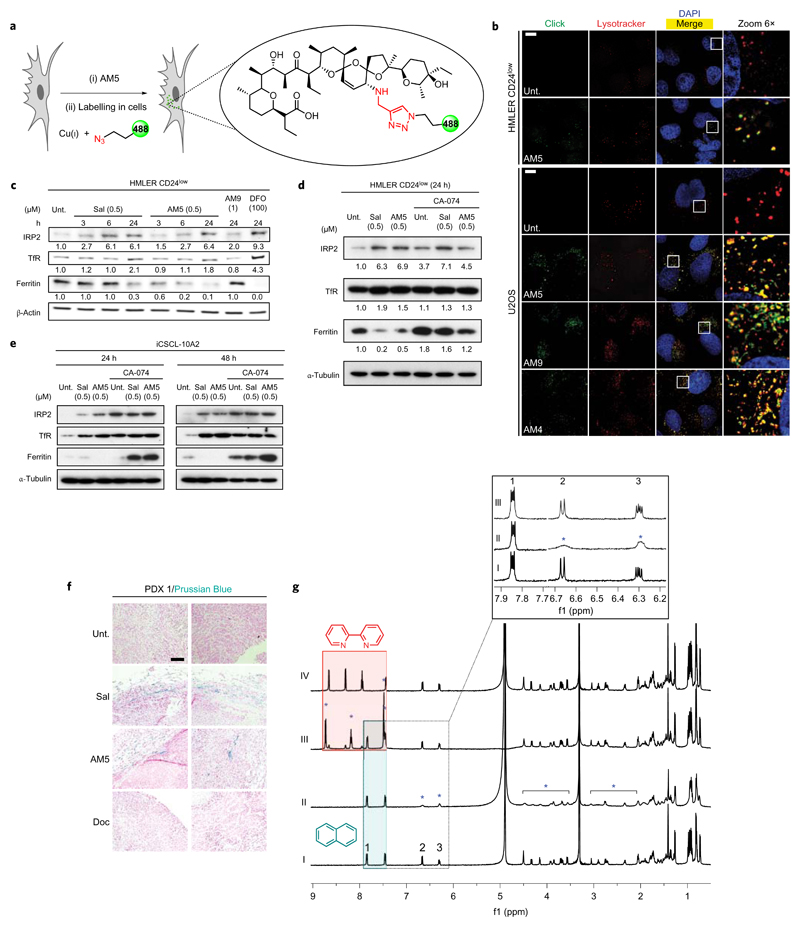
To detect derivatives of Sal, we functionalized alkynes in cells by means of in situ click chemistry, a strategy virtually applicable to any molecule (Fig. 2a)23–25. Sal surrogates co-localized with chemical and biochemical markers of lysosomes, including a lysotracker, the Ras-related protein Rab7 and the lysosomal-associated membrane protein 1 (Lamp1), in HMLER CD24low and human osteosarcoma U2OS cells (Fig. 2b and Supplementary Figs 6 and 7), demonstrating that these compounds physically accumulate in the lysosomal compartment irrespective of the overall charge and without altering the lysosomal pH according to acridine orange staining (Supplementary Fig. 8). In particular, the closely related derivative AM4, devoid of a protonable amine, also accumulated in lysosomes lending strong support to the notion that Sal targets this organelle. Lowering the temperature to block endocytic processes reduced the uptake of a Texas Red (TR)-dextran and the lysosomotropic small molecule artesumycin26, but had no effect on the cellular distribution of AM5 (Supplementary Fig. 9). Moreover, AM5 did not co-localize with the early endosome antigen 1 marker EEA1 (Supplementary Fig. 9). These data argued in favour of an endocytosis-independent entry mechanism in accord with the ability of Sal to freely diffuse across lipophilic membranes10. In comparison, AM5 did not target the ER, mitochondria or the Golgi apparatus (Supplementary Fig. 10). Because Sal can interact with alkali metals, and given that intracellular iron is tightly regulated and transits through lysosomal compartments, we explored the effect of Sal on iron homeostasis. Treatment of HMLER CD24low and iCSCL-10A2 cells with Sal or AM5 induced a response characteristic of cytoplasmic depletion of iron27, including increased levels of iron-responsive element-binding protein 2 (IRP2) and transferrin receptor (TfR) along with reduced levels of ferritin (Fig. 2c). A similar response was observed when cells were treated with the iron chelating agent deferoxamine (DFO). These results are consistent with the idea that these small molecules block the release of iron from lysosomes. Sal and AM5 also promoted a re-localization of ferritin to the lysosomal compartment, whose degradation was prevented by CA-074, an inhibitor of the lysosomal protease cathepsin B (Fig. 2d,e and Supplementary Fig. 11). In line with the lysosomal degradation of ferritin28 and further loading of iron in this organelle, iron(II)-mediated reduction of the fluorogenic probe RhoNox-1 (ref. 29) revealed that treatment with Sal or AM5 led to a staining that remained restricted to the lysosomal compartment, whereas it was diffuse in the cytosol of untreated and AM9-treated HMLER CD24low cells (Supplementary Fig. 12). Additionally, the accumulation of soluble iron in PDX 1 treated with Sal or AM5 reflected a cellular response to iron homeostasis targeting in vivo in a Doc-resistant PDX (Fig. 2f). Next, we investigated whether Sal and AM5 can directly interact with iron(II). Nuclear magnetic resonance (NMR) revealed that addition of 0.5 mol equiv. of FeCl2 to a methanolic solution of AM5 induced broadening and flattening of specific proton signals (Fig. 2g), including that of the C18–C19 vinylic protons at 6.6 and 6.3 ppm. These data, characteristic of the effect of a paramagnetic metal on the relaxation of protons in close proximity indicated interactions between AM5 and iron(II) in solution. Interestingly, previous X-ray crystallography studies had revealed that the vinylic protons are topologically close to the sodium ion inside the cavity formed by Sal around the metal in a co-crystal10. Thus, the pronounced effect of iron(II) onto the signals of these two protons suggested that iron(II) may occupy a similar position inside the folded molecule. The sub-stoichiometric amount of FeCl2 required to promote this effect was consistent with a 2:1 AM5:iron(II) stoichiometry. However, this data could also reflect a time-averaged set of signals between bound and free AM5, indicating a fast exchange that cannot be resolved within the timescale of the NMR experiment. In comparison, the proton signals of naphthalene (Napht), an organic small molecule devoid of heteroatoms and therefore unable to chelate iron, remained unaffected, and thus could be used as internal standard (for example, unaltered signal at 7.8 ppm) to compare the intensity of signals of AM5 between samples. Strikingly, addition of a slight excess of the iron(II) chelator 2,2'-bipyridine (Bipy) to a mixture of AM5, Napht and FeCl2, led to the occurrence of new proton signals of free and bound Bipy along with the concomitant recovery of the signals previously observed for the unbound AM5. These data indicated that under these conditions, Bipy displaces the metal from AM5, in line with the fact that AM5 is a looser iron interacting partner than Bipy. It is noteworthy that a similar trend was observed for Sal (Supplementary Fig. 13), although the shielding effect of iron(II) was more pronounced on the proton signals of AM5. Moreover, while iron(II) promoted the formation of byproducts of Sal over time, AM5 was found to be stable under these conditions. These properties of AM5 provide a rationale for the higher potency of this synthetic derivative compared to Sal. Overall, these data support a model whereby the lipophilic natural product accumulates in the lysosomal compartment and interacts with iron(II), thereby competing with the effective translocation of the metal into the cytosol, which in turn initiates the appropriate response to replenish the available pool of cellular iron. Lysosomal iron accumulation likely perturbs the proteolytic activity of cysteine and carboxyl proteases (for example, cathepsins), which comes in strong agreement with the reported effect of Sal on autophagic flux and the partial inhibition of cathepsins after 24 h treatment12. The depletion of iron likely contributes to the induction of endoplasmic reticulum stress previously reported for Sal (ref. 30).
Figure 2. Sal and AM5 sequester iron in lysosomes and trigger ferritin degradation in response to iron depletion.
a, Chemical strategy to label small molecules in cells. b, Fluorescence microscopy images showing the subcellular localization of labelled Sal derivatives in cells treated as indicated (10 μM, 6 h). LysoTracker Deep Red stains the lysosomes and 4′,6-diamidino-2-phenylindole (DAPI) stains nuclear DNA. Scale bar, 10 μm. Sal derivatives were labelled by means of click chemistry as described in Methods. c, Immunoblotting showing levels of iron homeostasis regulatory proteins in cells treated as indicated. d,e, Immunoblotting showing levels of ferritin in cells treated as indicated. Data quantified against loading controls. f, Detection of soluble iron using Perls’ Prussian Blue in tumours treated as in Fig. 1e. Data representative of PDX 1. Scale bar, 100 μm. g, 1H-NMR spectra of (I) AM5 (2 mM) and Napht (1.0 mol equiv.); (II) AM5 and Napht in the presence of FeCl2 (0.5 mol equiv.); (III) AM5, Napht and Bipy (1.6 mol equiv.) in the presence of FeCl2 (Bipy added after FeCl2); (IV) AM5 and Bipy. Samples prepared in CD3OD, spectra recorded at 298 K, 5 min following sample preparation (600 MHz). Blue stars indicate proton signals shielded by iron(II), green and red boxes highlight signals of free Napht and free/bound Bipy, respectively.
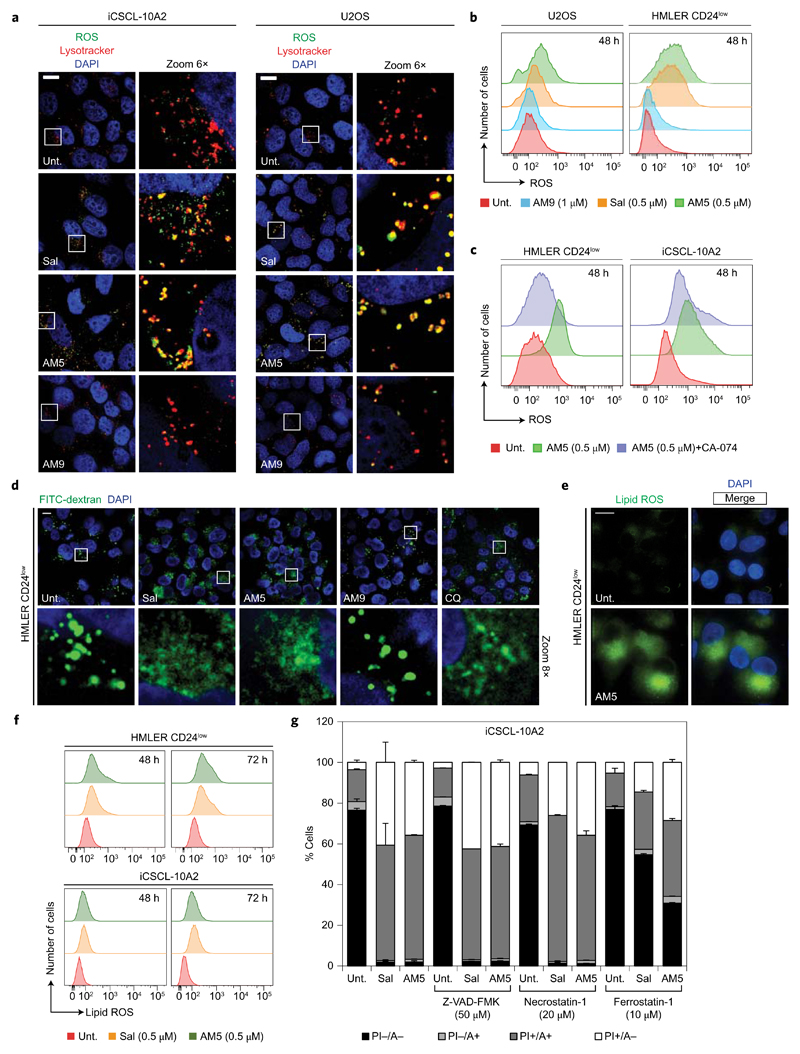
Iron can catalyse the production of reactive oxygen species (ROS) via Fenton chemistry31. Treatment of HMLER CD24low, iCSCL-10A2 and U2OS cells with Sal or AM5 led to the production of lysosomal ROS after 48 h treatment, which mirrored the accumulation of iron in this organelle (Fig. 3a,b and Supplementary Fig. 14). The production of ROS induced by Sal or AM5 was partially reduced upon inhibition of cathepsin B in HMLER CD24low and iCSCL-10A2 cells (Fig. 3c), linking the lysosomal degradation of ferritin to the production of ROS in this organelle through the release of additional soluble redox-active iron. Translocation of bulky lysosomal dextran into the cytosol of HMLER CD24low cells treated with Sal or AM5 was characteristic of lysosomal membrane permeabilization (LMP)32, presumably occurring as a result of ROS production and peroxidation of the lipid components of the lysosomal membrane (Fig. 3d). LMP has been shown to activate distinct cell death pathways, including apoptosis and necrosis, which in this context are commonly referred to as lysosomal cell death33,34. The implication of iron and ROS in the phenotype induced by Sal and AM5 hinted towards the activation of a regulated form of necrotic cell death, termed ferroptosis35–39, detected in HMLER CD24low and iCSCL-10A2 cells after 72 h treatment (Supplementary Fig. 15). In support to this, Bodipy-C11 staining indicated the presence of lipid peroxidation in HMLER CD24low and iCSCL-10A2 cells treated with Sal or AM5 (Fig. 3e,f). In addition, cell death induced by Sal or AM5 could be partially prevented by the ferroptosis inhibitor ferrostatin-1, whereas the apoptosis and necrosis inhibitors Z-VAD-FMK and necrostatin-1, respectively, had no effect on the cell death profiles (Fig. 3g and Supplementary Fig. 15). Furthermore, diminution of endogenous levels of the ROS scavenger glutathione (GSH), a hallmark of ferroptosis, could be detected in HMLER CD24low and iCSCL-10A2 cells treated with Sal or AM5 (Supplementary Fig. 15). Further investigation revealed that inhibition of cathepsin B also partially protected HMLER CD24low cells from Sal and AM5-induced death (Supplementary Fig. 16). Scavenging ROS with N-acetyl-l-cysteine (NAC) or ascorbate (Asc) partly prevented Sal and AM5 from killing these cells (Supplementary Fig. 16), and DFO reduced ROS levels in treated cells, exhibiting a protective effect against Sal and AM5 (Supplementary Fig. 16). Importantly, DFO favourably competed with Sal binding to iron in solution to form a redox-inactive iron complex, whereas Sal had no effect on the catalytic turnover of the metal under the same reaction conditions (Supplementary Fig. 16). This emphasizes distinct binding properties of Sal and DFO and rationalizes how DFO can antagonize the effect of Sal and AM5. While tight iron-chelating ligands can have the ability to alter the catalytic activity of metals in some cases, looser binders forming less-stable metal complexes may neither exacerbate nor inhibit the redox activity of iron. Collectively, these data indicated that the accumulation of iron and ROS in lysosomes of treated cells triggers lysosomal membrane dysfunction and cell death (Supplementary Fig. 16).
Figure 3. Accumulation of iron in lysosomes promotes ROS production and lysosomal dysfunction.
a, Fluorescence microscopy images showing the subcellular localization of ROS (green) by means of fluorogenic reaction with CM-H2DCFDA in cells treated with Sal (0.5 μM), AM5 (0.5 μM) or AM9 (1 μM) for 48 h. Scale bar, 10 μm. b,c, Flow cytometry analysis of ROS in cells treated as indicated. d, Fluorescence microscopy images showing the subcellular localization of FITC-dextran (green) in HMLER CD24low cells treated as in a or with chloroquine (CQ) control (100 μM, 3 h). Scale bar, 10 μm. e, Fluorescence microscopy images showing lipid ROS (green) by means of fluorogenic reaction with BODIPY 581/591 C11 in cells treated with AM5 (0.5 μM) at 48 h. Scale bar, 10 μm. f, Flow cytometry analysis of lipid ROS in cells treated as indicated. g, Flow cytometry analysis of Annexin V-FITC (A) and propidium iodide (PI) fluorescence in iCSCL-10A2 cells treated with Sal (0.5 μM) or AM5 (0.5 μM) for 72 h, in the presence or absence of the indicated inhibitors. Living cells are A–/PI– and ferroptotic cells (regulated necrosis) exhibit a positive PI+ staining. Bars and error bars, mean values and s.d. of two biological replicates. For flow cytometry profiles, see Supplementary Fig. 15.
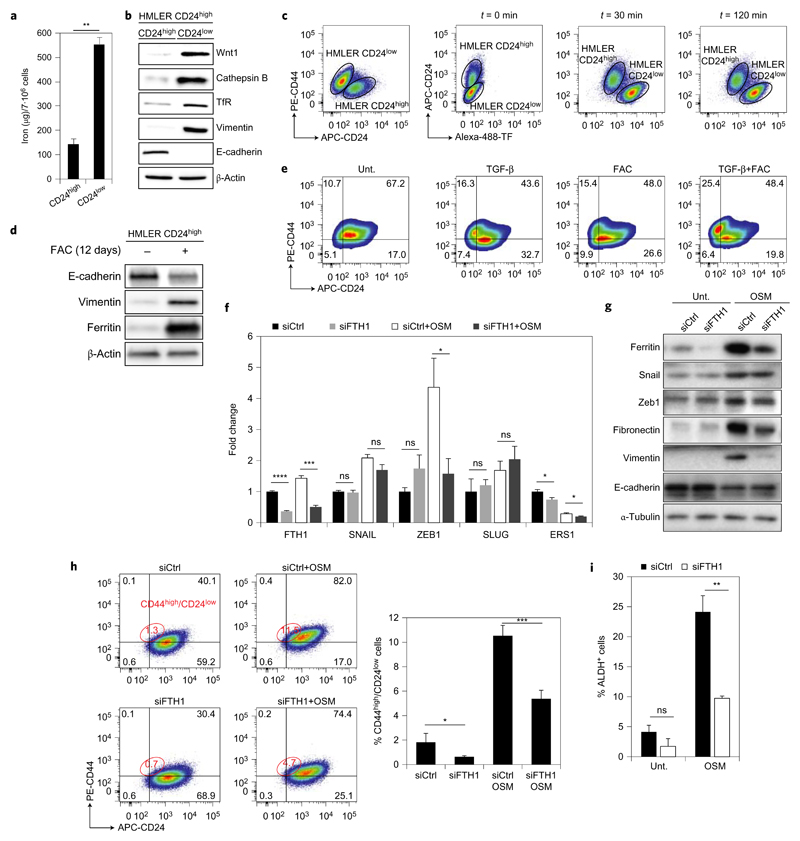
Next, we sought to shed light on the selective effect of Sal and AM5 against CSCs and to identify a role of iron metabolism in these cells. Strikingly, HMLER CD24low cells contained significantly higher levels of iron (Fig. 4a), TfR involved in iron-uptake and active cathepsin B compared with control cells (Fig. 4b), underlying the selective effect of Sal on HMLER CD24low cells and raising a putative function of iron in the maintenance of CSCs. Along these lines, Wnt signalling has previously been implicated in carcinogenesis, requiring iron to repress E-cadherin expression40,41, and the Wnt1 protein level was higher in HMLER CD24low compared to control cells (Fig. 4b). Consistent with a higher level of TfR and intracellular iron, endocytosis of fluorescently labelled-transferrin (TF) was more pronounced in breast CSCs (Fig. 4c and Supplementary Fig. 17). Supplementing HMLER CD24high cells with ferric ammonium citrate (FAC) promoted a phenotype characteristic of mesenchymal cells (Fig. 4d), and FAC and transforming growth factor (TGF)-β synergized to increase the population of HMLER CD24low cells (Fig. 4e). These results suggested that the presence of iron either directly altered cell differentiation or selected for proliferation in favour of a subpopulation exhibiting a pronounced CSC phenotype. Additionally, the cytokine oncostatin M (OSM)42 increased the level of ferritin in MCF-7 cells (Fig. 4f,g), and knocking down ferritin impaired the ability of OSM to induce a stem-like phenotype as defined by levels of zeb1, fibronectin and vimentin as well as ratios of CD44high/CD24low and ALDH+ cells (Fig. 4f–i). These data were consistent with, and extend the scope of, recent findings showing that iron trafficking characterizes glioblastoma stem-like cells43.
Figure 4. Iron is involved in the maintenance of CSCs.
a, Quantification of cellular iron by electrothermal atomic absorption spectrometry in HMLER cells. Bars and error bars, mean values and s.d. of three biological replicates. b, Comparative immunoblotting analysis of endogenous levels of EMT markers, TfR and cathepsin B in HMLER cells. c, Flow cytometry analysis of Alexa-488-TF uptake in mixed HMLER CD24high/low cells. d, Immunoblotting showing levels of EMT markers in HMLER cells supplemented with FAC for 12 days. e, Flow cytometry analysis of subpopulations of HMLER cells treated as indicated for 12 days. f, qPCR quantification of levels of mRNA transcripts of the indicated genes in control and ferritin knocked down conditions in MCF-7 cells supplemented with OSM for 48 h. Bars and error bars, mean values and s.d. of two biological replicates. g, Comparative immunoblotting analysis of ferritin and EMT markers in MCF-7 cells treated as in f. h, Flow cytometry analysis of subpopulations of MCF-7 cells treated as indicated for 48 h and corresponding quantification. i, Quantification of the ALDH+ population in MCF-7 cells treated as indicated measured by flow cytometry. Bars and error bars, mean values and s.d. of two biological replicates. In a,f,i *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, Student’s t-test. ns, not significant.
Ferroportin, a protein that mediates the export of intracellular iron, has been shown to be upregulated at the transcriptional level in breast cancer tissues of patients with good prognosis44, which supports the contention that iron is required for phenotypic transitions occurring during metastasis. This study raises the question of the functional role(s) of iron in metastatic cancers. Dynamic processes of iron-dependent oxidative demethylation mediated by Jumonji (JmjC) family of enzymes have been linked to the epigenetic regulation of cancer4. In particular, the JmjC-domain-containing protein KDM5B, which selectively demethylates H3K4me3, is upregulated in breast cancers45,46. It is noteworthy that H3K4 methylation status is a key component of epithelial–mesenchymal plasticity and loss of H3K4me3 correlates with poor survival in breast cancer patients47. Moreover, ten–eleven translocation (Tet) enzymes have been shown to promote iron-dependent oxidative demethylation of 5-methylcytosine (5mC) and to regulate the EMT and its reverse counterpart48–50. It is conceivable that iron is directly involved in the nuclear compartment to promote selective oxidative demethylation of key DNA or histone residues throughout chromatin to control the epithelial–mesenchymal status in a dynamic manner. This hypothesis may explain the changes in CSC-gene expression previously reported for Sal (ref. 4), which could take place as a direct consequence of lysosomal iron targeting we have observed. Hijacking iron with small molecules such as ironomycin (AM5) may thus provide the means to alter the epigenetic landscape for therapeutic benefits. Targeting metabolic pathways in CSCs, such as those reliant on iron homeostasis, represents a valuable strategy where conventional treatments designed to target highly proliferating cells, but not quiescent CSCs, are notoriously ineffective.
Methods
Reagents
Acridine Orange (AO, A6014, Sigma Aldrich, 2 μg ml–1 soln. for 20 min), ALDH (ALDEFLUOR, 01700, Stemcell Technologies), Alexa-488-TF (Molecular Probes, 50 μg ml−1), Alexa-647-TF (Molecular Probes, 50 μg ml−1), artesumycin (in-house, 10 μM for 30 min), bafilomycin A1 (Baf, B1793, Sigma Aldrich, 100 nM for 2 h), 2,2'-bipyridine (Bipy, D216305-10G, Sigma Aldrich), CA-074 Me (CA-074, BML-PI126-0001, Enzo Life Science, 10 μM for 30 min pre-treatment), chloroquine diphosphate salt (CQ, C6628, Sigma Aldrich, 100 μM for 3 h), N,N-diethylaminobenzaldehyde (DEAB, 01700, Stemcell Technologies), deferoxamine mesylate salt (DFO, D9533, Sigma Aldrich, 10–100 μM for 30 min pre-treatment), docetaxel (Doc, 11885, Sigma Aldrich, 10 mg kg−1 week−1), ferric ammonium citrate (FAC, F5879, Sigma Aldrich, 500 μg ml–1, 12 days), Ferrostatin-1 (SML0583, Sigma Aldrich, 10 μM for 72 h), FITC-dextran (FD10S, 10 kDa, Sigma Aldrich, 1 mg ml–1 for 2 h), Iron(II) chloride anhydrous (FeCl2, 429368-1G, Sigma Aldrich), N-acetyl-l-cysteine (NAC, A9165, Sigma Aldrich, 5 mM, 2 h pre-treatment), Necrostatin-1 (N9037, Sigma Aldrich, 20 μM for 72 h), Oncostatin M (OSM, 295-OM-010, R&D, 100 ng ml–1, 48 h), salinomycin (Sal, S4526, Sigma Aldrich), sodium l-ascorbate (Asc, A7631, Sigma Aldrich, 5 μM, 2 h pre-treatment), TGF-β (TGF-β, D1183, Selleckchem, 15 ng ml–1, 12 days), TR-dextran (D1863, 10 kDa, Thermo Fisher Scientific, 1 mg ml–1, 30 min), Z-VAD-FMK (550377, BD biosciences, 50 μM for 72 h).
Antibodies
α-Tubulin (T5168, Sigma Aldrich, WB 1:1000), β-actin (ab8226, Abcam, WB 1:20000), cathepsin B (sc-6490-R, Santa Cruz Biotechnology, WB 1:1000), CD24-APC (2155590, Sony Biotechnology, FC 1:100), CD44-PE (FAB4948P, R&D Systems, FC 1:100), cytochrome c (12963, Cell Signaling Technology, IF 1:200), E-cadherin (20023195, Cell Signaling Technology, WB 1:1000), EEA1 (ALX-210-239, Enzo Life Science, IF 1:200), ferritin (200-401-090-0100, Rockland, IF 1:200), ferritin (75973, Abcam, WB 1:1000), fibronectin (F0791, Sigma Aldrich, WB 1:1000), IRP2 (sc-33682, Santa Cruz Biotechnology, WB 1:1000), lysosomal-associated membrane protein 1 (Lamp1, 555798, BD Biosciences, IF 1:200), protein disulfide-isomerase A3 (PDIA3, AMAB90988, Sigma, IF 1:200), Ras-related protein Rab-7a (RAB7, 9367, Cell Signaling Technology, IF 1:200), receptor-binding cancer antigen expressed on SiSo cells (Rcas1, 12290, Cell Signaling Technology, IF 1:200), Snail (3895, Cell Signaling Technology, WB 1:1000), transferrin receptor (TfR, 13-6800, Life Technologies, WB 1:1000), vimentin (3932, Cell Signaling Technology, WB 1:1000), Wnt1 (ab15251, Abcam, WB 1:1000), Zeb1 (sc-81428, Santa Cruz Biotechnology, WB 1:1000).
Secondary antibodies for WB
HRP anti-Mouse (A90-116P, Bethyl Laboratories, WB 1:30000) and HRP anti-Rabbit (A120-108P, Bethyl Laboratories, WB 1:30000).
Secondary antibodies for IF
Alexa Fluor 488 conjugate (A-11017 Mouse, A-11008 Rabbit, Life Technologies, IF 1:500), Alexa Fluor 594 conjugate (A-11032 Mouse, A-11072 Rabbit, Life Technologies, IF 1:500), Alexa Fluor 647 conjugate (A-20991 Rabbit, A-20990 Mouse, Life Technologies, IF 1:500). All antibodies were diluted in blocking solution (2 or 5% BSA, 0.1% Tween-20/TBS).
Chemical labelling of Sal derivatives and fluorescence microscopy
Cells were cultured at ~80% confluence and were treated for 6 h with 10 μM compounds unless stated otherwise. LysoTracker Deep Red (L12492, Molecular Probes, 100 nM) was added 1 h prior to cell fixation. Cells were fixed with absolute methanol (at −20 °C for 15 min) or formaldehyde (2% in PBS, 12 min) depending on specific antibodies, prior to permeabilization (Triton X-100, 0.1% in PBS, 5–10 min) and washed three times with 1% BSA/PBS. The click reaction cocktail was prepared from Click-iT EdU Imaging kits (C10337, Life Technologies) according to the manufacturer’s protocol. Briefly, mixing 430 μl of 1 × Click-iT reaction buffer with 20 μl of CuSO4 solution, 1.2 μl Alexa Fluor azide, 50 μl reaction buffer additive (sodium ascorbate) to reach a final volume of ~500 μl. Coverslips were incubated with the click reaction cocktail in the dark at room temperature for 30 min then washed three times with PBS. For immunofluorescence, cells were blocked with 5% BSA, 0.2% Tween-20/PBS (blocking buffer) for 10 min at room temperature. Coverslips were incubated with 50–100 μl of diluted primary antibodies in blocking buffer (for example, cytochrome c, Rcas1, PDIA3, RAB7, Lamp1) 1 h at room temperature. Coverslips were then washed three times with blocking buffer and incubated as described above with the appropriate secondary antibodies for 30 min to 1 h. Coverslips were washed three times with PBS and mounted using Vectashield Mounting Medium with DAPI (H-1200, VECTOR Laboratories). High-resolution fluorescence images were acquired using a Deltavision real-time microscope (Applied Precision). 60×/1.4NA and 100×/1.4NA objectives were used for 2D and 3D acquisitions that were deconvoluted with SoftWorx (Ratio conservative: 15 iterations, Applied Precision) and processed with ImageJ.
Measurement of ROS production
Reactive oxygen species (ROS) levels were measured by flow cytometry or by confocal scanning immunofluorescence microscopy using CM-H2DCF-DA (C6827, Invitrogen). Briefly, cells were treated as indicated in the main figure. Then, cells were trypsinized and incubated with 5 μM CM-H2DCF-DA at 37 °C for 40 min, washed once with PBS and were counterstained with DAPI (0.5 μg ml–1) to exclude non-viable cells. The mean fluorescence intensity was determined as ROS production by flow cytometry with a LSRFortessa cytometer (BD Biosciences, San Jose, California). For immunofluorescence microscopy analysis, cells were seeded on coverslips and were treated as indicated in the main text. LysoTracker Red DND-99 (L-7528, Life Technologies, 1 μM) was used to visualize lysosomes. Then, cells were fixed with 4% PFA/PBS. DAPI was used to visualize nuclear DNA. Cell images were obtained using a Deltavision real-time microscope (Applied Precision) or an ApoTome.2 microscope (Zeiss). ImageJ was used for further image processing.
Supplementary Material
Acknowledgements
We thank the CNRS, INSERM and SATT IDF Innov for generous funding. Research in the R.R. laboratory is supported by the European Research Council (grant number 647973), Fondation pour la Recherche Médicale (grant reference AJE20141031486), Emergence Ville de Paris and Ligue Contre le Cancer. A.Ha. is funded by the Fondation de France. We acknowledge the PICT-IBiSA@Pasteur Imaging Facility of Institut Curie, member of the France-BioImaging national research infrastructure. We thank P. Le Bacon for assistance with high-resolution microscopy, J.-F. Gallard, N. Birlirakis and Christine Gaillet for assistance with NMR spectroscopy and J. Poupon for electrothermal atomic absorption spectrometry experiments. We thank A. Puisieux for providing us with HMLER cells and V. Mitz for mammary tissues obtained from reduction mammoplasty. A patent application (number WO 2016 038223) on the small molecules derived from Sal for the treatment of cancers has been filed.
Footnotes
Data availability. All relevant data are available from the authors, and/or are included with the manuscript.
Author contributions
R.R. conceptualized the study. R.R., T.T.M., M.M., A.Ha. and P.C. designed the experiments and analysed the data. T.T.M., A.Hi., A.Ha. and M.M. performed the experiments unless stated otherwise. A.Hi. synthesized Sal derivatives. T.C. performed NMR experiments. J.C., O.C., C.G., D.B. and E.C.-J. provided PDX data. A.Ha., C.L. and A.D. provided MCF-7 tumour data. S.M. and V.A. provided assistance with cell imaging. A.R. provided iCSCL-10A2 cells. R.R. wrote the manuscript. R.R., M.M., A.Ha. and S.M. prepared the figures. All the authors commented on the manuscript.
Competing financial interests
The authors declare no competing financial interests.
Additional information
Reprints and permissions information is available online at www.nature.com/reprints. Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Nieto MN, Huang RY-J, Jackson RA, Thiery JP. EMT: 2016. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 2.Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med. 2013;19:1438–1449. doi: 10.1038/nm.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pattabiraman DR, Weinberg RA. Tackling the cancer stem cells–what challenges do they pose? Nature Rev Drug Discov. 2014;13:497–512. doi: 10.1038/nrd4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelly PN, Dakic A, Adams JM, Nutt SL, Strasser A. Tumor growth need not be driven by rare cancer stem cells. Science. 2007;317:337. doi: 10.1126/science.1142596. [DOI] [PubMed] [Google Scholar]
- 5.Quintana E, et al. Efficient tumour formation by single melanoma cells. Nature. 2008;456:593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta PB, et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morel A-P, et al. Generation of breast cancer stem cells through epithelial–mesenchymal transition. PLoS ONE. 2008;3:e2888. doi: 10.1371/journal.pone.0002888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Germain AR, et al. Identification of a selective small molecule inhibitor of breast cancer stem cells. Bioorg Med Chem Lett. 2012;22:3571–3574. doi: 10.1016/j.bmcl.2012.01.035. [DOI] [PubMed] [Google Scholar]
- 9.Hartwell KA, et al. Niche-based screening identifies small-molecule inhibitors of leukemia stem cells. Nat Chem Biol. 2013;9:840–848. doi: 10.1038/nchembio.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paulus EF, Kurz M, Matter H, Vértesy L. Solid-state and solution structure of the salinomycin-sodium complex: stabilization of different conformers for an ionophore in different environments. J Am Chem Soc. 1998;120:8209–8221. [Google Scholar]
- 11.Lu D, et al. Salinomycin inhibits Wnt signaling and selectively induces apoptosis in chronic lymphocytic leukemia cells. Proc Natl Acad Sci USA. 2011;108:13253–13257. doi: 10.1073/pnas.1110431108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yue W, et al. Inhibition of the autophagic flux by salinomycin in breast cancer stem-like/progenitor cells interferes with their maintenance. Autophagy. 2013;9:714–729. doi: 10.4161/auto.23997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naujokat C, Steinhart R. Salinomycin as a drug for targeting human cancer stem cells. J Biomed Biotechnol. 2012;2012:950658. doi: 10.1155/2012/950658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huczyński A, et al. Antiproliferative activity of salinomycin and its derivatives. Bioorg Med Chem Lett. 2012;22:7146–7150. doi: 10.1016/j.bmcl.2012.09.068. [DOI] [PubMed] [Google Scholar]
- 15.Borgström B, et al. Synthetic modification of salinomycin: selective O-acylation and biological evaluation. Chem Commun. 2013;49:9944–9946. doi: 10.1039/c3cc45983g. [DOI] [PubMed] [Google Scholar]
- 16.Huang X, et al. Semisynthesis of SY-1 for investigation of breast cancer stem cell selectivity of C-ring-modified salinomycin analogues. ACS Chem Biol. 2014;9:1587–1594. doi: 10.1021/cb5002153. [DOI] [PubMed] [Google Scholar]
- 17.Borgström B, Huang X, Chygorin E, Oredsson S, Strand D. Salinomycin hydroxamic acids: synthesis, structure, and biological activity of polyether ionophore hybrids. ACS Chem Med Lett. 2016;7:635–640. doi: 10.1021/acsmedchemlett.6b00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi Q, et al. Discovery of a 19F MRI sensitive salinomycin derivative with high cytotoxicity towards cancer cells. Chem Commun. 2016;52:5136–5139. doi: 10.1039/c6cc01508e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borgström B, Huang X, Hegardt C, Oredsson S, Strand D. Structure-activity relationships in salinomycin: cytotoxicity and phenotype selectivity of semi-synthetic derivatives. Chem Eur J. 2017;23:2077–2083. doi: 10.1002/chem.201603621. [DOI] [PubMed] [Google Scholar]
- 20.Nishi M, et al. Induction of cells with cancer stem cell properties from nontumorigenic human mammary epithelial cells by defined reprogramming factors. Oncogene. 2014;33:643–652. doi: 10.1038/onc.2012.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minta A, Tsien RY. Fluorescent indicators for cytosolic sodium. J Biol Chem. 1989;264:19449–19457. [PubMed] [Google Scholar]
- 22.Charafe-Jauffret E, et al. ALDH1-positive cancer stem cells predict engraftment of primary breast tumors and are governed by a common stem cell program. Cancer Res. 2013;73:7290–7300. doi: 10.1158/0008-5472.CAN-12-4704. [DOI] [PubMed] [Google Scholar]
- 23.Rodriguez R, et al. Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat Chem Biol. 2012;8:301–310. doi: 10.1038/nchembio.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larrieu D, Britton S, Demir M, Rodriguez R, Jackson SP. Chemical inhibition of NAT10 corrects defects of laminopathic cells. Science. 2014;344:527–532. doi: 10.1126/science.1252651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abell NS, Mercado M, Cañeque T, Rodriguez R, Xhemalce B. Click quantitative mass spectrometry identifies PIWIL3 as a mechanistic target of RNA interference activator enoxacin in cancer cells. J Am Chem Soc. 2017;139:1400–1403. doi: 10.1021/jacs.6b11751. [DOI] [PubMed] [Google Scholar]
- 26.Cañeque T, et al. Synthesis of marmycin A and investigation into its cellular activity. Nat Chem. 2015;7:744–751. doi: 10.1038/nchem.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pantopoulos K, Porwal SK, Tartakoff A, Devireddy L. Mechanisms of mammalian iron homeostasis. Biochemistry. 2012;51:5705–5724. doi: 10.1021/bi300752r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Asano T, et al. Distinct mechanisms of ferritin delivery to lysosomes in iron-depleted and iron-replete cells. Mol Cell Biol. 2011;10:2040–2052. doi: 10.1128/MCB.01437-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirayama T, Okuda K, Nagasawa H. A highly selective turn-on fluorescent probe for iron(II) to visualize labile iron in living cells. Chem Sci. 2013;4:1250–1256. [Google Scholar]
- 30.Li T, et al. Salinomycin induces cell death with autophagy through activation of endoplasmic reticulum stress in human cancer cells. Autophagy. 2013;9:1057–1068. doi: 10.4161/auto.24632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dixon SJ, Stockwell BR. The role of iron and reactive oxygen species in cell death. Nat Chem Biol. 2014;10:9–17. doi: 10.1038/nchembio.1416. [DOI] [PubMed] [Google Scholar]
- 32.Boya P, Kroemer G. Lysosomal membrane permeabilization in cell death. Oncogene. 2008;27:6434–6451. doi: 10.1038/onc.2008.310. [DOI] [PubMed] [Google Scholar]
- 33.Aits S, Jäättelä M. Lysosomal cell death at a glance. J Cell Sci. 2013;126:1905–1912. doi: 10.1242/jcs.091181. [DOI] [PubMed] [Google Scholar]
- 34.Galluzzi L, Bravo-San Pedro JM, Kroemer G. Organelle-specific initiation of cell death. Nat Cell Biol. 2014;16:728–736. doi: 10.1038/ncb3005. [DOI] [PubMed] [Google Scholar]
- 35.Dixon SJ, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang WS, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Conrad M, Angeli JPF, Vandenabeele P, Stockwell BR. Regulated necrosis: disease relevance and therapeutic opportunities. Nat Rev Drug Discov. 2016;15:348–366. doi: 10.1038/nrd.2015.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang WS, Stockwell BR. Ferroptosis: death by lipid peroxidation. Trends Cell Biol. 2016;26:165–176. doi: 10.1016/j.tcb.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cao JY, Dixon SJ. Mechanisms of ferroptosis. Cell Mol Life Sci. 2016;73:2195–2209. doi: 10.1007/s00018-016-2194-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Torti SV, Torti FM. Iron and cancer: more ore to be mined. Nat Rev Cancer. 2013;13:342–355. doi: 10.1038/nrc3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takebe N, et al. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: clinical update. Nature Rev Clin Oncol. 2015;12:445–464. doi: 10.1038/nrclinonc.2015.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.West NR, Murray JI, Watson PH. Oncostatin-M promotes phenotypic changes associated with mesenchymal and stem cell-like differentiation in breast cancer. Oncogene. 2014;33:1485–1494. doi: 10.1038/onc.2013.105. [DOI] [PubMed] [Google Scholar]
- 43.Schonberg DL, et al. Preferential iron trafficking characterizes glioblastoma stem-like cells. Cancer Cell. 2015;28:441–455. doi: 10.1016/j.ccell.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pinnix ZK, et al. Ferroportin and iron regulation in breast cancer progression and prognosis. Sci Transl Med. 2010;2:43ra56. doi: 10.1126/scisignal.3001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamane K, et al. PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol Cell. 2007;25:801–812. doi: 10.1016/j.molcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 46.Yamamoto S, et al. JARID1B is a luminal lineage-driving oncogene in breast cancer. Cancer Cell. 2014;25:762–777. doi: 10.1016/j.ccr.2014.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen L, et al. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell. 2013;153:692–706. doi: 10.1016/j.cell.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsai Y-P, et al. TET1 regulates hypoxia-induced epithelial-mesenchymal transition by acting as a co-activator. Genome Biol. 2014;15:513. doi: 10.1186/s13059-014-0513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu X, et al. Tet and TDG mediate DNA demethylation essential for mesenchymal-to-epithelial transition in somatic cell reprogramming. Cell Stem Cell. 2014;14:512–522. doi: 10.1016/j.stem.2014.01.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.