Abstract
Due to high heterogeneity, molecular characterization of prostate cancer (PCa) based on biopsy sampling is often challenging. Hence, a minimally invasive method to determine the molecular imprints of a patient's tumor for risk stratification would be advantageous. In this study, we employ a novel, digital amplification-free quantification method using the nCounter technology (Nanostring Technologies) to profile exosomal serum miRNAs (ex-miRNA) from aggressive PCa cases, benign prostatic hyperplasia (BPH), and disease-free controls. We identified several dysregulated miRNAs, one of which was the tumor suppressor miR-1246. miR-1246 was downregulated in PCa clinical tissues and cell lines and was selectively released into exosomes. Overexpression of miR-1246 in a PCa cell line significantly inhibited xenograft tumor growth in vivo and increased apoptosis and decreased proliferation, invasiveness, and migration in vitro. miR-1246 inhibited N-cadherin and vimentin activities, thereby inhibiting epithelial-mesenchymal transition. Ex-miR-1246 expression correlated with increasing pathological grade, positive metastasis, and poor prognosis. Our analyses suggest ex-miR-1246 as a promising PCa biomarker with diagnostic potential that can predict disease aggressiveness.
Keywords: microRNA, exosomes, miR-1246, prostate cancer
INTRODUCTION
Prostate cancer (PCa) is a leading cause of male cancer-related mortality in the United States with an estimated 26,730 deaths in 2017 (1). This disease is remarkably heterogeneous (2) with tumors ranging from indolent to very aggressive. Aggressive tumors often metastasize locally or distantly to other organs, causing significant morbidity and mortality (3). A major clinical challenge in PCa clinical management is posed by the inability of current diagnostic tests, such as serum Prostate-specific antigen (PSA) testing, digital rectal examination (DRE) and histopathological grading of tissues, to discern between indolent and aggressive disease (4, 5). Owing to inherent limitations of serum PSA including lack of specificity, PSA screening has led to PCa over diagnosis and overtreatment (4, 6). In view of these limitations, additional risk stratification tools have been developed that incorporates serum PSA levels with currently available clinicopathological parameters such as Gleason score and pathological staging (5). Despite advances, these strategies fail to distinguish between aggressive and indolent tumors and predicting PCa outcome remains a major clinical challenge (7, 8). To address this challenge, molecular biomarkers for improving PCa diagnosis and prognosis are highly sought. Based on molecular characterization of primary and metastatic prostate tumors, several promising alternate tissue-based assays are being developed that show improved sensitivity and specificity over PSA (5). However, these assays are based on biopsy sampling that is an invasive, expensive procedure and does not accurately represent multifocal disease. It is desirable to have an easily accessible, minimally invasive way to accurately determine the molecular imprints of patient’s tumor that can aid in risk stratification.
Exosomes are small extracellular vesicles (30–100nm in size) (9) that are gaining significant interest as alternate disease biomarkers that can be detected non-invasively in biological fluids such as serum, plasma, semen and urine (10) and can be used as a liquid biopsy for PCa (11, 12). Exosomes contain proteins, RNAs (including microRNAs) and lipids and their cargo often varies under various pathological conditions, being reflective of the physiological state of the originating host cell. Hence, exosomes are a promising source of non-invasive biomarkers for early diagnosis and prognosis of various diseases including PCa (13). MicroRNAs (miRNAs), small non-coding RNAs that suppress gene expression post transcriptionally via sequence-specific interactions with the 3’- untranslated regions (UTRs) of cognate mRNA targets (14), are stable biomarkers that are abundantly present in exosomes (15, 16). Exosomes provide an enriched source of miRNAs for biomarker profiling by protecting against RNases as compared to intracellular miRNAs/ miRNAs present in cell-free blood (17).
The primary objective of the current study was to define novel exosomal microRNA (exRNA) biomarkers in aggressive PCa. We sought to identify exRNAs that (i) distinguish between benign disease and aggressive PCa, (ii) differentiate between normal and malignant disease. We employed a novel, digital amplification-free quantification method using the nCounter technology (Nanostring Technologies) (18) to measure the abundance of 800 miRNAs in exosomes from sera of individuals with aggressive PCa and BPH/ disease-free controls and identified several dysregulated miRNAs. We further validated exosomal miR-1246 as a promising PCa biomarker that has diagnostic potential and was associated with disease aggressiveness.
MATERIALS AND METHODS
Cell lines and cell culture
Nonmalignant prostate epithelial cell line RWPE-1 and prostate carcinoma cell lines (LNCaP, Du145, PC3) were obtained from the American Type Culture Collection (ATCC) and cultured under recommended conditions as detailed in supplemental methods. All cell lines were maintained in an incubator with a humidified atmosphere of 95% air and 5% CO2 at 37°C. Prostate cell lines were authenticated by DNA short-tandem repeat analysis. All cell lines were tested and found negative for mycoplasma. The experiments with cell lines were performed within 6 months of their procurement/resuscitation.
Clinical samples
Written informed consent was obtained from all patients and the study was approved by the UCSF Committee on Human Research. Patient studies were conducted in accordance with the ethical guidelines of the Belmont Report. Formalin-fixed, paraffin-embedded (FFPE) PCa samples were obtained from the SFVAMC or Cooperative Human Tissue Network (CHTN). All slides were reviewed by a board-certified pathologist for the identification of PCa foci as well as adjacent normal glandular epithelium. Tissues were micro dissected as described in (19).
Serum samples (0.5ml–1ml) from PCa patients and clinical information were obtained from CHTN/ Prostate Cancer Biorepository Network (PCBN) and stored at −80°C till processed. Controls were selected from age and race-matched normal individuals or men whose prostate glands were free of cancer and underwent transurethral resection of the prostate for benign conditions (e.g., BPH). CHTN samples included cases with no prior radiation therapy or chemotherapy and was divided equally into prognostic risk groups defined by American Joint Committee on Cancer (20) as low, intermediate and high categories.
Isolation of exosomes from serum samples
Serum derived exosomes (EVs) were isolated from 250µL of serum using the Total exosome isolation reagent (Life Technologies, Cat. No. 4478360) as per manufacturer’s instructions. Briefly, serum samples were spun at 2000× g for 30 minutes to remove cells and debris. Next, 0.2 volumes of exosome isolation reagent were added to clarified supernatants and samples were incubated at 2°C to 8°C for 30 min. The precipitated exosomes were recovered by centrifugation at 10,000× g for 10 minutes at room temperature. Exosome pellets were resuspended in PBS.
Exosome quantitation and size determination
To confirm the integrity of exosomal preparations, the purity of exosomes was verified by evaluation of particle size and concentration using Nanoparticle Tracking Analysis (NTA). A NanoSight LM10 instrument (Malvern Instruments) equipped with a 405nm laser-equipped sample chamber was employed as per manufacturer’s instructions.
Exosomal RNA extraction
Exosomal RNA was prepared using a Plasma/Serum Exosome Purification kit (Norgen Biotek, Catalog no. 57400) as per manufacturer’s instructions with minor modifications. To control for variance in starting material and RNA extraction efficiency, after adding lysis buffer, 1000 attomoles each of RNA spike-in controls (cel-miR-248, cel-miR-254, osa-miR-414, osa-miR-442) were added. The extracted RNA was eluted with 20µL of RNase-free water. The quantity and quality of the RNA was determined by a Agilent Bioanalyzer 2100 (Agilent Technologies) with a nano RNA chip as per the manufacturer’s instructions.
Nanostring nCounter analyses
For each sample analyzed, 10µl exosomal RNA was concentrated using a vacuum concentrator to a volume of 3µL that was used for miRNA profiling by a Nanostring nCounter® microRNA platform (version 3) (Nanostring Technologies) as per the manufacturer’s instructions (detailed in supplemental methods).
Quantitative real-time PCR
Mature miRNAs were assayed using the TaqMan MicroRNA Assays (Applied Biosystems) in accordance with the manufacturer's instructions. Taqman assays used were hsa-miR-1246 (assay ID CSFARKI) and RNU6A (assay ID 001973). The comparative Ct method was used to calculate the relative changes in gene expression on the 7500 Fast Real Time PCR System.
Xenograft tumors
Animal studies were approved by IACUC and were performed in accordance with institutional guidelines under an approved protocol. PC3 cells (1.8 × 106) stably overexpressing control miRNA or miR-1246- clone 1 and clone 2- were injected subcutaneously into nude mice (4–5-week-old, Simonsen Laboratories, n=6 for control, n=5/ group for each miR-1246 clone). Cells were injected in the right or left flanks of mice in a volume of 100 µl mixed with 50% matrigel. Once palpable tumors developed, caliper measurements were taken once a week and tumor volumes were calculated on the basis of width (x), length (y), height/depth (z).
Statistics
All quantified data represents an average of triplicate samples or as indicated. Data are represented as mean ± S.E.M or as indicated. Statistical analyses were performed using MedCalc version 10.3.2. Results were considered statistically significant at P ≤ 0.05.
RESULTS
Exosomal miRNAs are dysregulated in aggressive PCa
In a preliminary screening, exosomal miRNAs were extracted from patients with aggressive PCa (n=6), age and race-matched normal cases (n=3) and patients with benign prostate hyperplasia (BPH) (n=3). The integrity of exosomal preparations was confirmed by nanoparticle tracking analysis (NTA) (Fig. S1A–C). NTA analyses showed that the average exosome size (Fig. S1B) and concentration (Fig. S1C) were not significantly different between normal, BPH and aggressive PCa though number of particles in aggressive PCa were ~2-fold higher than from those with benign disease (Fig. S1C). To further validate our exosomal preparations, we performed Western blot analyses for exosomal markers CD63 and TSG101 (Fig. S1D). RNA was extracted from characterized exosomes followed by determination of the quantity and quality of the exosomal RNA by a Agilent Bioanalyzer 2100 (Agilent Technologies). miRNA profiling was performed utilizing nCounter miRNA arrays v3.0a to digitally measure the abundance of 800 exRNAs in individuals with aggressive PCa versus BPH/ disease- free controls (Fig. 1). This preliminary screening identified four miRNAs (miR-1283, miR-1246, miR-26b-5p, miR-302c-3p) that are significantly upregulated in aggressive PCa compared to BPH (Fig. 1A, Tables S1). In addition, 16 miRNAs (miR-766-3p, miR-1304-5p, miR-500a-5p+501-5p, miR-105-5p, miR-1285-3p, miR-217, miR-1269a, miR-215-5p, miR-34a-5p, miR-1268b, miR-802, miR-138-5p, miR-30a-3p, miR-98-3p, miR-1469, and miR-542-5p) were found to be downregulated in serum exosomes derived from these diseased cases as compared to BPH controls (Fig. 1A, Table S1). Comparison of exRNAs between normal and aggressive PCa showed three significantly upregulated miRNAs (miR-1246, miR-302c-3p, miR-509-5p) and 21 downregulated miRNAs (miR-25-3p, miR-7-5p, miR-491-5p, miR-127-5p, miR-2110, miR-663a, miR-3928-3p, miR-450a-2-3p, miR-766-3p, miR-590-5p, miR-519 family, miR-3151-5p, miR-3605-5p, miR-378h, miR-103a-3p, miR-23c, miR-331-3p, miR-485-5p, miR-514a-5p, miR-592, miR-320a) (Fig. 1A, Table S2). Three miRNAs- miR-1246, miR-766-3p and miR-302c-3p were common miRNAs that were dysregulated in aggressive PCa relative to normal and benign disease (Fig. 1A–B). Significantly, ex-miR-1246 was identified as the common upregulated miRNA that could distinguish between normal, benign and aggressive PCa (Fig. 1A and 1C). ex-miR-1246 was upregulated ~31-fold in aggressive PCa as compared to normal (P=0.026) and ~23-fold in aggressive PCa as compared to BPH (P=0.035). Importantly, normal vs BPH showed a non-significant increase of 1.35 (P=0.361) suggesting that this exRNA is not significantly altered in benign conditions. This suggests that ex-miR-1246 is a potential marker to distinguish between aggressive PCa and BPH.
Fig. 1. Dysregulated exosomal microRNAs in aggressive PCa.
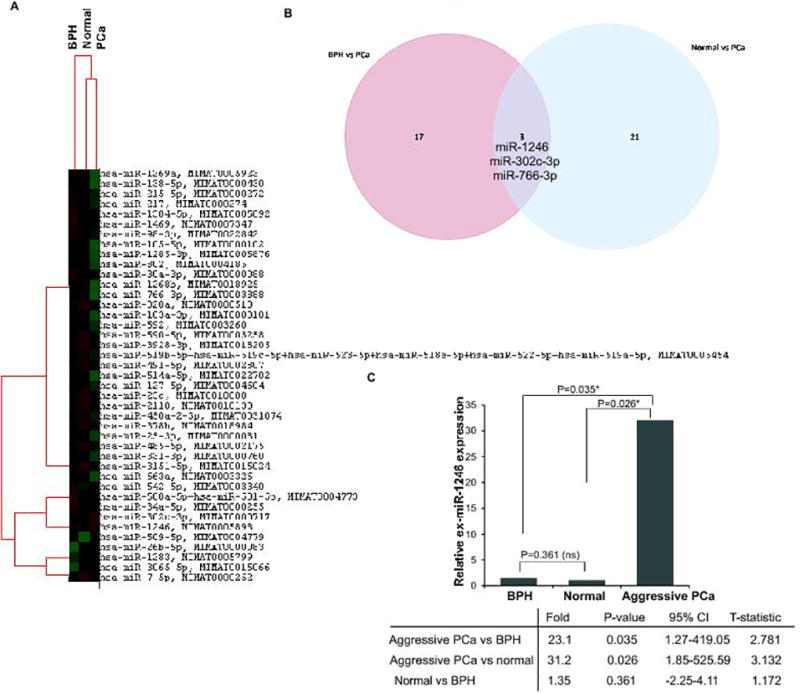
A. MicroRNA profiling was performed using Nanostring miRNA (v3a) platform in aggressive prostate tumors (n=6), normal individuals (n=3) and cases with BPH (n=3). Significantly dysregulated miRNAs are represented in the heatmap. Top 100 genes and four spike in controls were used for normalization.
B. Venn diagram showing dysregulated miRNAs in aggressive PCa as compared to BPH and normal controls.
C. Grouped data analyses for ex-miR-1246 signal intensities using Nanostring miRNA platform.
Biological pathways potentially influenced by dysregulated miRNAs in aggressive PCa
We performed in silico analyses to identify the biological pathways that are potentially influenced by dysregulated exosomal miRNAs in aggressive PCa. In silico pathway analyses of dysregulated exRNAs using FunRich (Functional Enrichment Analyses tool)8 showed dysregulation of key biological signaling and cell adhesion pathways that have been implicated in metastatic PCa such as ErbB receptor signaling1, 7, c-Met/HGF (mesenchymal epithelial transition factor/ hepatocyte growth factor)signaling4, 6, IGF1 signaling and Nectin adhesion pathway10. Additional top significantly dysregulated signaling pathways included plasma membrane estrogen receptor signaling, Insulin-like Growth Factor 1 (IGF1) signaling, LKB1 signaling, TRAIL signaling and IFN-gamma signaling pathway (Fig. S2 and Table S3).
Ex-miR-1246 is specifically upregulated in aggressive PCa
Further, to validate our preliminary ex-miR-1246 data, we performed real-time PCR based expression profiling in a training cohort of PCa patients (Cohort 1, n=44) (Fig. 2A). Clinicopathological characteristics of this cohort are represented in Table S4A. Exosomal miRNA were extracted from sera of patients with PCa (n=44), race-matched normal (n=8) and BPH (n=4) followed by ex-miR-1246 profiling. This cohort also included the samples that were used for miRNA profiling in Fig. 1. As compared to normal controls, ex-miR-1246 levels were significantly upregulated (relative expression >1.25, P= 0.0001) in 41/44 PCa cases (93%) while 3/44 (7%) cases showed no change in expression (Fig. 2A). Importantly, the average expression of ex-miR-1246 in PCa was found to be significantly high as compared to those with BPH (P=0.0041) (Fig. 2B).
Fig. 2. Ex-miR-1246 is specifically upregulated in aggressive PCa and is a potential diagnostic marker.
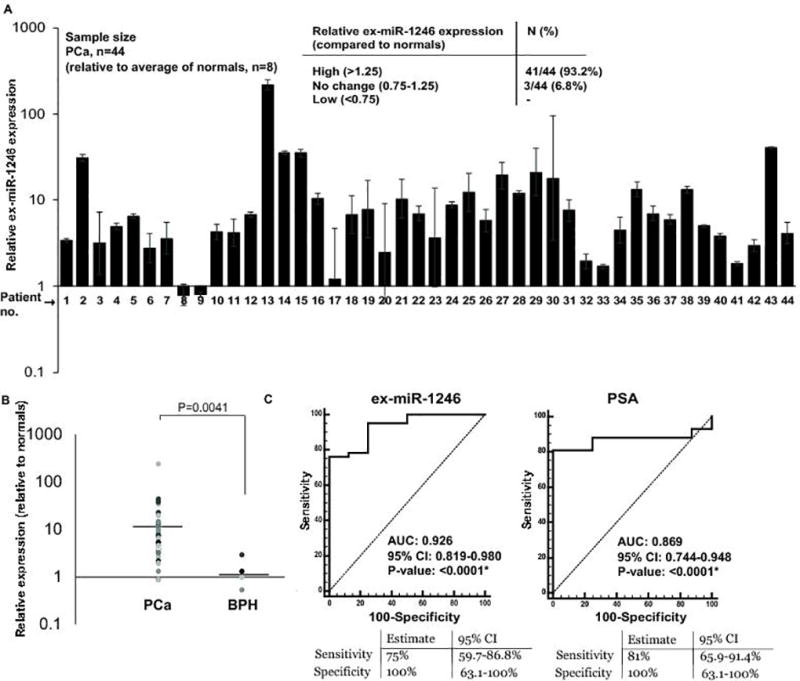
A. Relative miR-1246 expression in exosomes derived from sera of PCa patients (n=44) as compared to normal individuals (n=8) as assessed by real-time PCR. RNU6A was used as a control. Error bars represent SEM.
B. Relative ex-miR-1246 expression in BPH and PCa cases. Horizontal lines represent the average in each group. C. ROC curve analyses for ex-miR-1246 (left panel) and serum PSA (right panel) as parameters to discriminate between tumor and normal samples.
Ex-miR-1246 is a potential diagnostic marker for PCa
Next, we examined the potential diagnostic significance of ex-miR-1246. ROC curve analyses based on dCt values of PCa (n=44) and normal (n=8) showed that ex-miR-1246 expression can be a significant parameter to discriminate between normal and PCa cases with an area under the ROC curve (AUC) of 0.926 (P<0.0001), 100% specificity and 75% sensitivity (Fig. 2C, left panel). Comparison with serum PSA showed that the difference between the diagnostic abilities of miR-1246 vs serum PSA were statistically insignificant (P = 0.3299), with PSA showing an AUC of 0.869 (P<0.0001) (Fig. 2C, right panel).
Ex-miR-1246 is specifically upregulated in aggressive PCa
We further sought to determine if ex-miR-1246 is specifically upregulated in aggressive PCa. Since our training cohort 1 primarily included PCa cases with Stage IV PCa cases, we included PCa cases with disease stages IIA-III as training cohort 2 (n=46; 21 BPH and 25 PCa cases) (Fig. 3A). Clinical characteristics of cohort 2 are summarized in Table S4A. As compared to BPH, PCa cases showed ex-miR-1246 upregulation in 13/25 (52%) cases, downregulation in 9/25 PCa cases (36%) while no significant change was observed in 3/25 (12%) cases. In view of these data, we examined the correlation of ex-miR-1246 with clinicopathological parameters of PCa (cohort 1+2) (Fig. 3B). Interestingly, we observed a significant inverse correlation between high ex-miR-1246 and pathological stage (P=0.0020) suggesting that this exosomal marker increases with advancing PCa stage. Significantly, we observed a correlation between positive lymph node metastasis and high expression of this exosomal marker (P=0.0436) pointing to this miRNA’s potential to predict aggressiveness/ localized metastasis. No significant correlations were observed between ex-miR-1246 and age, Gleason score, race or serum PSA (Fig. 3B). Correlation of ex-miR-1246 expression with prognostic risk groups (IIA-IV) (as defined by American Joint Committee on Cancer (20)) suggests that high ex-miR-1246 expression is significantly correlated with poor prognostic groups (Fig. 3C). Overall, our data suggests that ex-miR-1246 expression possess significant prognostic potential.
Fig. 3. High expression of serum exosomal miR-1246 is associated with advanced pathological stage and localized PCa metastasis.
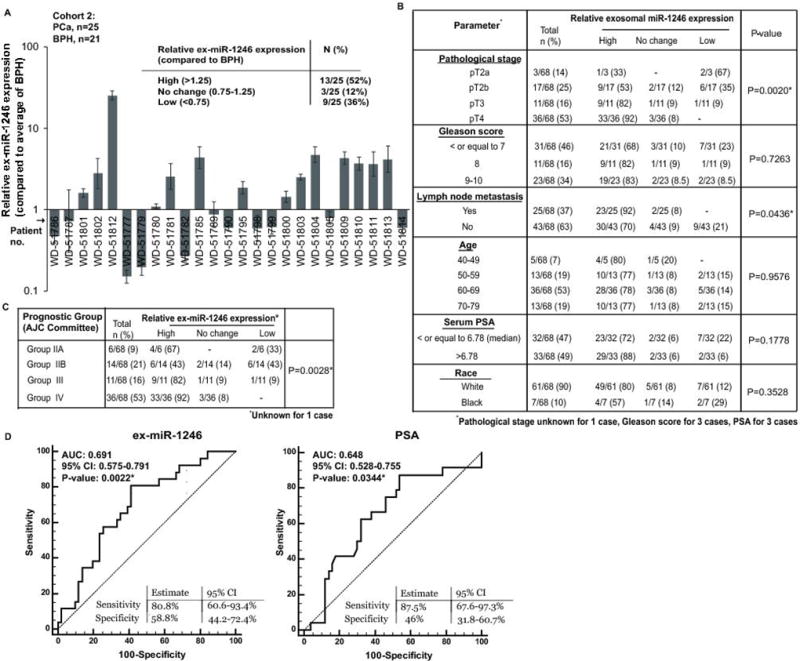
A. Relative ex-miR-1246 expression in exosomes derived from the sera of PCa patient training cohort (cohort 2) as assessed by real-time PCR. RNU6A was used as an endogenous control. Patients with BPH (n=21) were used as calibrators.
B. Correlation of serum exosomal miR-1246 expression with clinicopathological parameters in PCa patients. P-values are based on Chi square test.
C. Correlation of serum exosomal miR-1246 expression with prognostic risk groups IIA-IV as defined by American Joint Committee on Cancer. P-values are based on Chi square test.
D. ROC curve analyses for ex-miR-1246 expression (left panel) and PSA (right panel) as parameters to discriminate between non-metastatic and localized metastatic PCa cases Analyses was based on dCT values of ex-miR-1246 in training cohort (1 and 2, n=77) that included lymph node metastatic PCa (n=26), non-metastatic PCa (n=43) and normal (n=8) individuals.
miR-1246 is a potential PCa exosomal biomarker that can predict localized metastasis
In view of our data suggesting significant correlation of ex-miR-1246 expression with lymph node metastasis, we next asked if ex-miR-1246 can be used to predict localized metastasis in PCa. To address this, we performed ROC curve analyses on our training cohort (1 and 2) (n=77) based on dCt values of ex-miR-1246 (Fig. 3D, left panel). This analyses included lymph node metastatic PCa (n=26), non-metastatic PCa (n=43) and normal (n=8) individuals. Our analyses showed that ex-miR-1246 expression can be a significant parameter to discriminate between non-metastatic and localized metastatic PCa cases with an AUC of 0.691 (P=0.0022). We compared these results with that of PSA (Fig. 3D, right panel). PSA showed an AUC of 0.648 (P=0.0344). While PSA has a 46% specificity, 87.5% sensitivity as a metastasis predictor, ex-miR-1246 exhibits higher specificity (~59%) with ~81% sensitivity in our clinical cohort.
Validation of ex-miR-1246 as an exosomal biomarker for aggressive PCa
In view of our preceding data with the training cohort demonstrating the value of ex-miR-1246 as a biomarker for aggressive PCa, we extended our analyses to an independent validation cohort. This cohort included metastatic castration-resistant PCa cases (n=43, Table S4B). Ex-miR-1246 expression was high in 37/43 (86%) of cases, downregulated in 5/43 PCa cases (12%) while no significant change in ex-miR-1246 was observed in 1/43 (2%) cases (Fig. 4A). ROC curve analyses validated the diagnostic ability of ex-miR-1246 to discriminate between normal and aggressive PCa cases with an AUC of 0.933 (P<0.0001), 100% specificity and 88.37% sensitivity. The difference between diagnostic abilities of ex-miR-1246 vs serum PSA were statistically insignificant (P = 0.5161), with PSA showing an AUC of 0.973 (P<0.0001) (Fig. 2C, right panel). Correlation with clinicopathological parameters showed a significant correlation between metastasis and high expression of ex-miR-1246 (P<0.0001) pointing to this miRNA’s potential to predict aggressiveness/ localized metastasis. No significant correlations were observed between ex-miR-1246 and Gleason score, race or serum PSA though a significant association was seen with age in this cohort. These data validate ex-miR-1246 as a biomarker for aggressive PCa.
Fig. 4. Validation of ex-miR-1246 as a PCa exosomal biomarker for aggressive PCa.
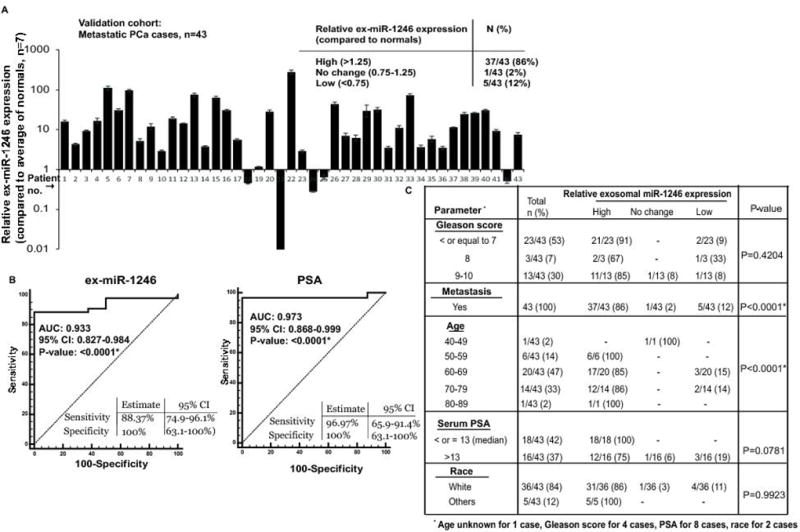
A. Relative ex-miR-1246 expression in exosomes derived from sera of validation cohort of aggressive PCa patients as assessed by real-time PCR. RNU6A was used as an endogenous control. Normals (n=7) were used as calibrators.
B. ROC curve analyses for ex-miR-1246 (left panel) and serum PSA (right panel) as parameters to discriminate between tumor and aggressive, metastatic PCa.
C. Correlation of serum exosomal miR-1246 expression with clinicopathological parameters in validation cohort. P-values are based on Chi square test.
miR-1246 is a tumor suppressor miRNA that is downregulated in PCa clinical tissues and is selectively released in exosomes
We next sought to determine the role of miR-1246 in PCa. We profiled the expression of miR-1246 in microdissected PCa clinical tissues and matched adjacent normal tissues (n=36) by real-time PCR (Fig. 5A). miR-1246 expression was downregulated in 26/36 PCa cases (72%) while 4/36 cases (~11%) showed no change and 6/36 cases (17%) showed higher expression. Clinicopathological characteristics of the patients are summarized in Table S4C. These data suggest that miR-1246 expression is commonly downregulated in PCa (Wilcoxon Signed Rank test, P< 0.001). Further, miR-1246 expression analyses in prostate cell lines showed that its expression is attenuated in PCa cell lines (PC3, DU145, LNCaP) as compared to normal immortalized prostate epithelial cell line RWPE-1 (Fig. 5B). However, low miR-1246 expression was also observed in BPH1 cells. Collectively, these data confirm the downregulated expression of miR-1246 in PCa. In view of this, we hypothesized that this miRNA may be a tumor suppressor in PCa that is released via exosomes from tumor cells, causing its upregulated expression in body fluids. To see if miR-1246 is released in PCa exosomes, we treated PC3 and LNCaP cells with exosome inhibitor GW4869 followed by miR-1246 expression profiling in cellular and exosomal fractions (Fig. 5C). Ex-miR-1246 expression was decreased upon GW4869 treatment of both cell lines suggesting that this miRNA is selectively released in PCa exosomes. To unequivocally test our hypothesis, we also examined the tissue and corresponding serum exosome levels of miR-1246 from the same patients in a small subset of our clinical cohort (Fig. 5D) and observed an inverse correlation. We also examined the levels of circulating miR-1246 in a subset of our PCa clinical cohort that was used for ex-miR-1246 profiling (Fig. S3A). We found no significant correlation between expression of circulating and exosomal miR-1246 levels (Fig. S3B).
Fig. 5. miR-1246 is a tumor suppressor miRNA that is downregulated in PCa clinical tissues and is selectively released in exosomes.
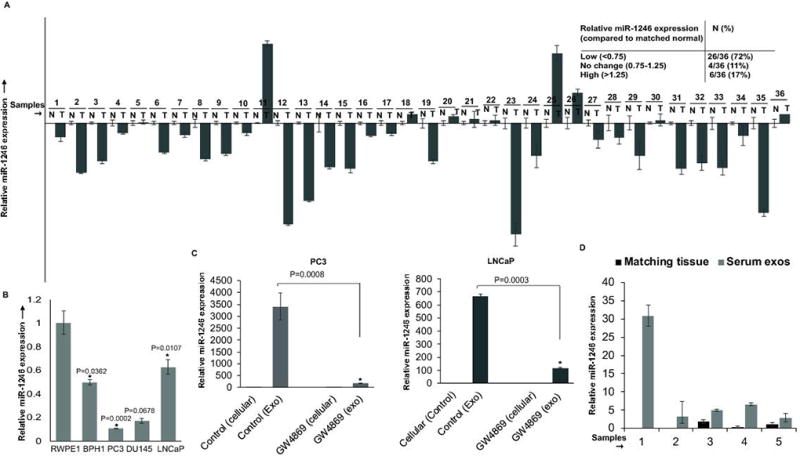
A. Relative miR-1246 expression in microdissected PCa tissues (n=36) and matched adjacent normal regions as assessed by real-time PCR. Data were normalized to RNU48 control and represented as mean ± SEM.
B. Relative miR-1246 expression in prostate cell lines as assessed by real-time PCR. Data were normalized to RNU48 control and represented as mean ± SEM.
C. PCa cell lines PC3 (left panels) and LNCaP (right panels) were treated with GW4869 for 48 hrs followed by real time PCR analyses of miR-1246 expression in cellular and exosomal fractions. Data were normalized to RNU6A control and represented as mean ± SEM.
D. Relative miR-1246 expression in PCa tissues and corresponding serum exosomes from the same patients in a training cohort subset. Data were normalized to RNU6A for exosomes and RNU48 for tissues and represented as mean ± SEM.
miR-1246 is a tumor suppressor miRNA with pleiotropic roles in PCa
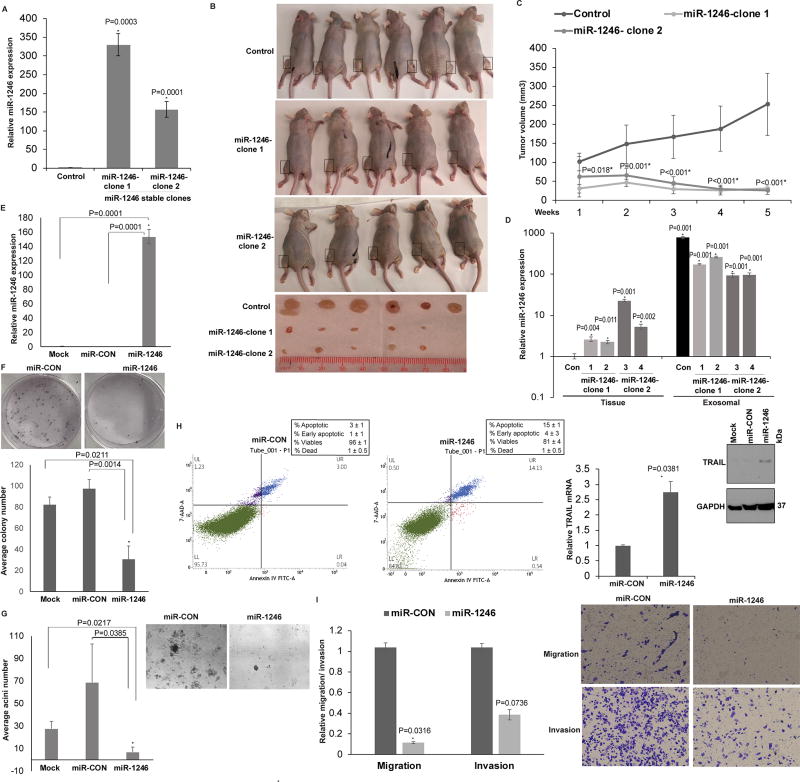
Next, we evaluated the functional significance of miR-1246 in PCa by performing a series of in vivo and in vitro assays (Fig. 6). We tested the functional impact of miR-1246 overexpression in an in vivo PCa xenograft mouse model (Fig. 6A–C). PC3 cells were stably transfected with miR-CON/miR-1246 overexpression construct (Fig. 6A). Two of the miR-1246 overexpression clones- miR-1246-clone 1 and miR-1246-clone 2- were used for further studies. Control miR or miR-1246 expressing cells (clone 1 or 2) were subcutaneously injected into three groups of nude mice (n=6 for control group, n=5 each for two miR-1246 clones) to generate PCa xenograft tumors (Fig. 6B–C). Our results show that miR-1246 overexpression led to significant inhibition of xenograft tumor growth over time, validating its role as a tumor suppressor miRNA. Further, to unequivocally validate our hypothesis that miR-1246 is a PCa tumor suppressor miRNA that is released via exosomes in blood, we investigated miR-1246 expression in xenograft tissues and corresponding exosomes derived from sera of xenograft mouse models (Fig. 6D) and found an inverse correlation. Serum exosomes and tissues were collected from xenograft mouse models (control/ miR-1246 overexpressing, n=4 each) followed by miR-1246 expression profiling. Ex-miR-1246 expression was abundant in serum exosomes in control tumor bearing mice with low tissue miR-1246 levels while in miR-1246 overexpressing xenografts, exosomal release was decreased (Fig. 6D). These data lend credence to our hypothesis that miR-1246 is a tumor suppressor miRNA that is selectively released in PCa exosomes, leading to its high levels in serum and low cellular levels. Further, to gain insights into functional role of miR-1246 in PCa, we performed in vitro functional assays after transient transfection of miR-1246 mimic/control miRNA mimic (miR-CON) in PC3 cells (Fig. 6D–I). miR-1246 overexpression (Fig. 6E) significantly suppressed the proliferation (Fig. 6F) and anchorage-independent growth (Fig. 6G) of PC3 cells as compared to controls. Further, miR-1246 overexpression led to ~5-fold increase in the average apoptotic cells (early apoptotic + apoptotic) as compared to controls (Fig. 6H) accompanied by induction of TNF-related apoptosis-inducing ligand (TRAIL) mRNA and protein. This data suggests that miR-1246 impacts the TRAIL signaling pathway (Fig. 6H, right panels). miR-1246 overexpression led to reduced invasive and migratory abilities of PC3 cells as compared to controls (Fig. 6I). Collectively, these data suggest that miR-1246 overexpression plays a tumor suppressor role in PCa via its pleiotropic effects on cellular proliferation, apoptosis, invasion and migration.
Fig. 6. miR-1246 is a tumor suppressor miRNA with pleiotropic roles in PCa.
A. Relative miR-1246 expression levels in PC3 cells stably transfected with control miR/miR-1246 expression construct (clone-1 and -2) as assessed by RT-PCR. Data were normalized to RNU48 control.
B. Control miR or miR-1246 expressing cells (clone 1 or 2) were subcutaneously injected into the right or left flank of nude mice (n=6 for control group, n=5 each for two miR-1246 clones) to generate PCa xenograft tumors. Images of mice and extracted tumors from miR-CON/ miR-1246 groups at 5 weeks are shown.
C. Tumor volumes of xenograft tumors from miR-CON and miR-1246 groups, at the indicated time points. Data represent the mean of each group ±SD.
D. Real-time PCR analyses of miR-1246 expression in xenograft tumors (left panels) and corresponding serum exosomes (right panels) in control/ miR-1246 PCa xenografts (1–4). Data were normalized to RNU6A control.
E. Relative miR-1246 expression in PC3 cells transiently transfected with either miR-CON/miR-1246 or mock transfected cells as assessed by real time PCR. Data were normalized to RNU48 control and are represented as mean ± SEM.
F. Colony formation assay in mock/miR-CON/miR-1246 transfected PC3 cell line. Representative pictures from miR-CON/miR-1246 transfected cells are shown.
G. Matrigel assay using mock/miR-CON/miR-1246 transfected PC3 cells. Representative pictures from miR-CON/miR-1246 transfected cells are shown above.
H. Apoptosis assay in PC3 cells after miR-CON/ miR-1246 transfections as assessed by ANNEXIN V-FITC/7-AAD staining. Right panels: Real time PCR and immunoblot analyses of TRAIL in transfected PC3 cells. RNU48 and GAPDH were used as respective controls.
I. Transwell migration and invasion assay in PC3 cells transfected with miR-CON/ miR-1246. Representative pictures are shown in left panels (* P< .05).
miR-1246 inhibits EMT by direct repression of mesenchymal genes in PCa
Further, we examined miR-1246 potential target genes by in silico analyses using miRANDA (21) and TargetScan algorithms (22) and identified that miR-1246 potentially targets epithelial to mesenchymal transition (EMT) related genes including CDH2, VIM and ZEB1 (Fig. 7A). The 3’UTR regions of VIM and ZEB1 possess one potential miR-1246 binding site while that of CDH2 has two binding sites. miR-1246 overexpression resulted in a significant downregulation of CDH2, VIM and ZEB1 mRNA suggesting that these EMT genes are under the regulatory control of miR-1246 (Fig. 7B). We performed luciferase reporter assays (Fig. 7C) with control/CDH2-1/ CDH2-2/ VIM/ ZEB1 3’UTR constructs in miR-CON/ miR-1246 overexpressing PC3 cells. A significant repression of luciferase reporter activity was observed upon transfection with CDH2-1 and VIM 3’ UTR constructs suggesting that vimentin and CDH2/ N-cadherin are direct miR-1246 targets and that miR-1246 affects CDH2 primarily via binding to site 1. ZEB1 was not found to be a direct miR-1246 target (Fig. 7C, right panel). To verify that these effects are due to direct miR-1246 interaction with the corresponding binding sites, we mutated the putative binding site/sites in CDH2/VIM 3’ UTRs (represented in Fig. 7A). Mutation of the miR-1246 binding site prevented the repression of luciferase activity observed upon miR-1246 overexpression in PC3 cells (Fig. 7C). Immunoblotting analyses of miR-CON/miR-1246 overexpressing PC3 cells confirmed that miR-1246 expression represses N-cadherin and vimentin protein expression (Fig. 7D). Further, in view of our analyses on the biological pathways that are potentially influenced by dysregulated exosomal miRNAs in aggressive PCa using FunRich (Functional Enrichment Analyses tool) (23) (Fig. S2 and Table S3), we examined the potential regulation of these biological pathways by miR-1246. Immunoblotting analyses of PC3 cells stably overexpressing miR-1246 led to downregulation of Epidermal Growth Factor Receptor (EGFR) and phosphor-Akt levels (Fig. 7E) suggesting that miR-1246 regulates the ErbB receptor signaling pathway.
Fig. 7. miR-1246 inhibits EMT by direct repression of mesenchymal genes in PCa.
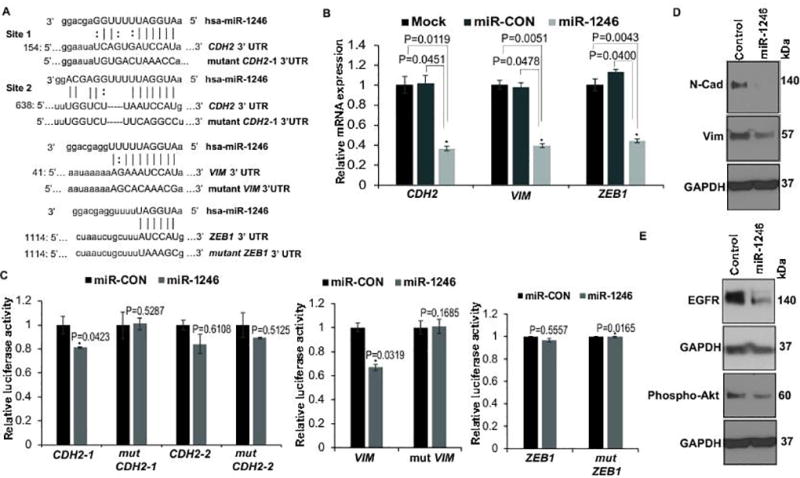
A. Schematic representation of the CDH2, VIM and ZEB1 3’-UTRs showing the putative miR-1246 binding sites. Mutant 3’ UTRs used in luciferase reporter assays are represented below.
B. Real-time PCR analyses of relative transcript levels of CDH2, VIM and ZEB1 in PC3 cells transfected as indicated. Data were normalized to GAPDH.
C. Luciferase reporter assays with the indicated wt and mutated 3’ UTR constructs or control luciferase construct co-transfected with miR-CON/ miR-1246 in PC3 cells. Firefly luciferase values were normalized to Renilla luciferase activity and plotted as relative luciferase activity to control 3’ UTR construct (* P< .05).
D. Immunoblots of endogenous N-cadherin and Vimentin in PC3 cells stably transfected with miR-CON/miR-1246. E. Immunoblot analyses of EGFR and phospho Akt in PC3 cells stably expressing control miRNA/miR-1246. GAPDH was used a loading control for D and E.
DISCUSSION
A significant interest has been generated in exosomal miRNA as alternate PCa biomarkers that can be detected non-invasively in body fluids (10) and can be used as liquid biopsies for early diagnosis and prognosis (13). In this direction, a few studies have examined exosomal miRNA profiles in PCa (11, 29). Huang et al. performed NGS analyses of exosomal miRNA in plasma of CRPC patients employing Illumina HiSeq2000 platform and reported high exosomal miR-1290 and miR-375 as prognostic markers associated with poor overall survival (30). In another study, an analysis of miRNAs in serum-derived exosomes from metastatic PCa showed an association of miR-141 and miR-375 with metastasis (31). In our present study, we employed a digital amplification-free quantification method using nCounter technology to perform exosomal miRNA profiling analyses from sera of aggressive PCa patients and identified novel, dysregulated exosomal miRNAs. It has been shown that exosomal miRNAs are selectively sorted into exosomes via mechanisms that are yet to be fully understood (11). A few miRNAs have been shown to possess short sequence motifs called EXOmotifs that guide and controls their loading into exosomes via recognition by the cellular enzyme heterogeneous nuclear nucleoprotein A2B1 (hnRNPA2B1) (32). We hypothesize that miRNAs enriched in exosomes in aggressive PCa, as exemplified by miR-1246, may be selectively released by tumor cells. Conversely, downregulated miRNAs such as miR-766-3p, miR-34a-5p may be selectively retained by tumor cells leading to their low expression in tumor exosomes. Considering the small sample size employed in the present study, validation of identified exosomal miRNA candidates in a larger cohort are warranted.
Importantly, our study demonstrates that ex-miR-1246 is a potential PCa exosomal biomarker with diagnostic and predictive/prognostic potential. Our analyses of ex-miR-1246 with training and validation cohorts show that this miRNA has significant potential as a marker to predict PCa aggressiveness/ metastasis. High ex-miR-1246 expression was specifically observed in Stage IV metastatic PCa patients as compared to Stage II/III. Our analyses suggest correlation of ex-miR-1246 with increasing pathological grade, positive lymph node metastasis, positive distant metastasis and poor prognostic groups. Importantly, the median expression of ex-miR-1246 was found to increase with metastatic aggressiveness/metastasis (Fig. S4), with the highest expression in distant metastatic group. These data highlight the potential of this exosomal marker to discern aggressive disease from benign/ indolent disease. Huang et al. reported miR-1246 to be upregulated in plasma exosomes from CRPC patients though no significant association was observed with overall survival (30). We could not perform survival analyses in the present study due to lack of follow up data.
While miR-1246 expression was high in exosomes from PCa clinical samples and cell lines, its cellular/tissue expression was lower as compared to corresponding normals. Our in vivo and in vitro data demonstrates that miR-1246 is a potent PCa tumor suppressor. Based on our present results, we hypothesize that miR-1246 is selectively released into PCa exosomes, leading to its high expression in serum samples and contributing to its utility as an exRNA marker. The following lines of data lend credence to our hypothesis:
Ex-miR-1246 expression was decreased upon treatment of PCa cell lines with exosome inhibitor GW4869 suggesting that this miRNA is selectively released in PCa exosomes;
An inverse correlation was observed between serum exosomal and tissue miR-1246 expression in a subset of our clinical cohort;
An inverse correlation was observed in miR-1246 expression in control/miR-1246 overexpressing xenograft mouse models and corresponding exosomes derived from their sera. In line with our study, several recent studies indicate that cells employ exosomes as vehicles to get rid of tumor suppressor miRNAs (33, 34). For example, miR-23b is disposed from bladder cancer cells via exosomes, leading to its low cellular levels that promoted metastasis (34). Similarly, let-7 has been reported to be discarded via exosomes resulting in high let-7 levels in exosomes derived from metastatic gastric cancer cell line compared to the non-metastatic counterpart (33). These findings suggest that exosomal miRNAs and other contents are selectively sorted into exosomes via mechanisms that are yet to be fully understood (11).
Restoration of miR-1246 expression in PC3 cells led to reduced cellular proliferation, anchorage-independent growth, invasiveness and migration pointing to an important pleiotropic role of this miRNA in regulating these attributes of tumorigenicity. Increased apoptosis observed upon miR-1246 overexpression suggests a pro-apoptotic role of this miRNA that may play a mechanistic role in TRAIL signaling pathway. Further, we found that miR-1246 plays an important regulatory role in controlling EGFR and phospho-Akt levels. EGFR, a tyrosine kinase receptor of the ErbB (erythroblastic leukemia viral oncogene homologue) transmembrane growth factor receptor family, is frequently overexpressed in PCa and is an important therapeutic target (35). EGFR signals through Akt and other pathways to regulate cell proliferation, migration, differentiation, apoptosis, and cell motility (35). Akt, an integral component of Phophatidylinositol 3-kinase (PI3K)/AKT signaling pathway, is aberrantly activated in PCa contributing to tumorigenesis (36). Our data suggests that miR-1246-mediated regulation of EGFR and Akt may underlie the observed effects of miR-1246 overexpression on cellular proliferation, apoptosis and invasion. In view of the lack of potential miR-1246 binding sites within the 3’ UTRs of EGFR/ Akt, we hypothesize that miR-1246 impacts these kinases indirectly. While the role of miR-1246 has not been examined in PCa, it has been reported to play a tumor suppressor role in cervical cancer (37) and oncogenic roles in non-small cell lung, pancreatic, colorectal, liver cancers (38–42). miR-1246 along with miR-1290, was reported as a crucial driver of tumor initiation and cancer progression in human non-small cell lung cancer (42).
Significantly, we found that miR-1246 is a crucial regulator of mesenchymal genes including N-cadherin and vimentin in PCa and thereby regulates EMT. EMT is the initiating step in invasion and metastasis that is characterized by decreased expression of epithelial genes (such as E-cadherin) and increased expression of mesenchymal genes (such as vimentin and N-cadherin) (43–45). EMT involves multiple signaling pathways, is coordinated by EMT-transcription factors (TFs) (such as ZEB family) and is regulated by miRNAs (46). Prominent examples are the miR-200 family and miR-205 that regulate EMT through direct targeting of ZEB1, ZEB2 (47, 48) amongst other targets. We showed that miR-203 and miR-3622a regulates PCa EMT and metastasis (49, 50). Our study suggests that miR-1246 is another crucial miRNA regulator of PCa EMT.
In conclusion, our data suggest that miR-1246 is a PCa tumor suppressor miRNA that plays a pleiotropic role by inhibiting EMT, cellular proliferation, survival and promoting apoptosis. In cancer cells, this miRNA is released in exosomes, leading to its high levels in serum and low cellular levels. The latter contributes to tumor progression by promoting EMT. Selective release of this miRNA in PCa exosomes may underlie its diagnostic and predictive potential. Importantly, we validated ex-miR-1246 as an exosomal miRNA marker that can be valuable for predicting/diagnosing aggressive PCa. Future studies with larger cohorts are warranted to validate these findings. Such validation has the potential to yield a simple test that can be combined with other clinical risk calculators to predict PCa aggressiveness non-invasively.
Supplementary Material
SIGNIFICANCE STATEMENT.
Dysregulation of exosomal miRNAs in aggressive prostate cancer leads to alteration of key signaling pathways associated with metastatic prostate cancer.
Acknowledgments
This work was supported by the National Cancer Institute at the National Institutes of Health (Grant Number RO1CA177984, U01CA184966). We thank Dr. Roger Erickson for his support and assistance with preparation of the manuscript.
Footnotes
Conflicts of interest: The authors declare no potential conflicts of interest
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Boyd LK, Mao X, Lu YJ. The complexity of prostate cancer: genomic alterations and heterogeneity. Nat Rev Urol. 2012;9:652–64. doi: 10.1038/nrurol.2012.185. [DOI] [PubMed] [Google Scholar]
- 3.Loberg RD, Logothetis CJ, Keller ET, Pienta KJ. Pathogenesis and treatment of prostate cancer bone metastases: targeting the lethal phenotype. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2005;23:8232–41. doi: 10.1200/JCO.2005.03.0841. [DOI] [PubMed] [Google Scholar]
- 4.Cary KC, Cooperberg MR. Biomarkers in prostate cancer surveillance and screening: past, present, and future. Ther Adv Urol. 2013;5:318–29. doi: 10.1177/1756287213495915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saini S. PSA and beyond: alternative prostate cancer biomarkers. Cell Oncol (Dordr) 2016;39:97–106. doi: 10.1007/s13402-016-0268-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walter LC, Bertenthal D, Lindquist K, Konety BR. PSA screening among elderly men with limited life expectancies. JAMA : the journal of the American Medical Association. 2006;296:2336–42. doi: 10.1001/jama.296.19.2336. [DOI] [PubMed] [Google Scholar]
- 7.Cooperberg MR, Broering JM, Carroll PR. Risk assessment for prostate cancer metastasis and mortality at the time of diagnosis. J Natl Cancer Inst. 2009;101:878–87. doi: 10.1093/jnci/djp122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D'Amico AV, Whittington R, Malkowicz SB, Schultz D, Blank K, Broderick GA, et al. Biochemical outcome after radical prostatectomy, external beam radiation therapy, or interstitial radiation therapy for clinically localized prostate cancer. JAMA : the journal of the American Medical Association. 1998;280:969–74. doi: 10.1001/jama.280.11.969. [DOI] [PubMed] [Google Scholar]
- 9.Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. Journal of proteomics. 2010;73:1907–20. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 10.Giusti I, Dolo V. Extracellular vesicles in prostate cancer: new future clinical strategies? BioMed research international. 2014;2014:561571. doi: 10.1155/2014/561571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hessvik NP, Sandvig K, Llorente A. Exosomal miRNAs as Biomarkers for Prostate Cancer. Front Genet. 2013;4:36. doi: 10.3389/fgene.2013.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valentino A, Reclusa P, Sirera R, Giallombardo M, Camps C, Pauwels P, et al. Exosomal microRNAs in liquid biopsies: future biomarkers for prostate cancer. Clin Transl Oncol. 2017 doi: 10.1007/s12094-016-1599-5. [DOI] [PubMed] [Google Scholar]
- 13.Duijvesz D, Luider T, Bangma CH, Jenster G. Exosomes as biomarker treasure chests for prostate cancer. Eur Urol. 2011;59:823–31. doi: 10.1016/j.eururo.2010.12.031. [DOI] [PubMed] [Google Scholar]
- 14.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cannistraci A, Di Pace AL, De Maria R, Bonci D. MicroRNA as New Tools for Prostate Cancer Risk Assessment and Therapeutic Intervention: Results from Clinical Data Set and Patients' Samples. BioMed research international. 2014;2014:146170. doi: 10.1155/2014/146170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids--the mix of hormones and biomarkers. Nature reviews Clinical oncology. 2011;8:467–77. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng L, Sharples RA, Scicluna BJ, Hill AF. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. Journal of extracellular vesicles. 2014;3 doi: 10.3402/jev.v3.23743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–25. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 19.Bucay N, Shahryari V, Majid S, Yamamura S, Mitsui Y, Tabatabai ZL, et al. miRNA Expression Analyses in Prostate Cancer Clinical Tissues. J Vis Exp. 2015 doi: 10.3791/53123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–4. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 21.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–53. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pathan M, Keerthikumar S, Ang CS, Gangoda L, Quek CY, Williamson NA, et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15:2597–601. doi: 10.1002/pmic.201400515. [DOI] [PubMed] [Google Scholar]
- 24.Angelucci A, Gravina GL, Rucci N, Millimaggi D, Festuccia C, Muzi P, et al. Suppression of EGF-R signaling reduces the incidence of prostate cancer metastasis in nude mice. Endocr Relat Cancer. 2006;13:197–210. doi: 10.1677/erc.1.01100. [DOI] [PubMed] [Google Scholar]
- 25.Lu X, Kang Y. Epidermal growth factor signalling and bone metastasis. Br J Cancer. 2010;102:457–61. doi: 10.1038/sj.bjc.6605490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hass R, Jennek S, Yang Y, Friedrich K. c-Met expression and activity in urogenital cancers - novel aspects of signal transduction and medical implications. Cell Commun Signal. 2017;15:10. doi: 10.1186/s12964-017-0165-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Humphrey PA, Zhu X, Zarnegar R, Swanson PE, Ratliff TL, Vollmer RT, et al. Hepatocyte growth factor and its receptor (c-MET) in prostatic carcinoma. Am J Pathol. 1995;147:386–96. [PMC free article] [PubMed] [Google Scholar]
- 28.Takai Y, Miyoshi J, Ikeda W, Ogita H. Nectins and nectin-like molecules: roles in contact inhibition of cell movement and proliferation. Nat Rev Mol Cell Biol. 2008;9:603–15. doi: 10.1038/nrm2457. [DOI] [PubMed] [Google Scholar]
- 29.Junker K, Heinzelmann J, Beckham C, Ochiya T, Jenster G. Extracellular Vesicles and Their Role in Urologic Malignancies. Eur Urol. 2016;70:323–31. doi: 10.1016/j.eururo.2016.02.046. [DOI] [PubMed] [Google Scholar]
- 30.Huang X, Yuan T, Liang M, Du M, Xia S, Dittmar R, et al. Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. Eur Urol. 2015;67:33–41. doi: 10.1016/j.eururo.2014.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bryant RJ, Pawlowski T, Catto JW, Marsden G, Vessella RL, Rhees B, et al. Changes in circulating microRNA levels associated with prostate cancer. Br J Cancer. 2012;106:768–74. doi: 10.1038/bjc.2011.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Villarroya-Beltri C, Gutierrez-Vazquez C, Sanchez-Cabo F, Perez-Hernandez D, Vazquez J, Martin-Cofreces N, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohshima K, Inoue K, Fujiwara A, Hatakeyama K, Kanto K, Watanabe Y, et al. Let-7 microRNA family is selectively secreted into the extracellular environment via exosomes in a metastatic gastric cancer cell line. PLoS One. 2010;5:e13247. doi: 10.1371/journal.pone.0013247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ostenfeld MS, Jeppesen DK, Laurberg JR, Boysen AT, Bramsen JB, Primdal-Bengtson B, et al. Cellular disposal of miR23b by RAB27-dependent exosome release is linked to acquisition of metastatic properties. Cancer Res. 2014;74:5758–71. doi: 10.1158/0008-5472.CAN-13-3512. [DOI] [PubMed] [Google Scholar]
- 35.Zhang H, Berezov A, Wang Q, Zhang G, Drebin J, Murali R, et al. ErbB receptors: from oncogenes to targeted cancer therapies. The Journal of clinical investigation. 2007;117:2051–8. doi: 10.1172/JCI32278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Y, Xie YJ, Xu Q, Chen JX, Shan NC, Zhang Y. Down-regulation of miR-1246 in cervical cancer tissues and its clinical significance. Gynecol Oncol. 2015;138:683–8. doi: 10.1016/j.ygyno.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 38.Hasegawa S, Eguchi H, Nagano H, Konno M, Tomimaru Y, Wada H, et al. MicroRNA-1246 expression associated with CCNG2-mediated chemoresistance and stemness in pancreatic cancer. Br J Cancer. 2014;111:1572–80. doi: 10.1038/bjc.2014.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim G, An HJ, Lee MJ, Song JY, Jeong JY, Lee JH, et al. Hsa-miR-1246 and hsa-miR-1290 are associated with stemness and invasiveness of non-small cell lung cancer. Lung Cancer. 2016;91:15–22. doi: 10.1016/j.lungcan.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 40.Sun Z, Meng C, Wang S, Zhou N, Guan M, Bai C, et al. MicroRNA-1246 enhances migration and invasion through CADM1 in hepatocellular carcinoma. BMC Cancer. 2014;14:616. doi: 10.1186/1471-2407-14-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang S, Zeng Y, Zhou JM, Nie SL, Peng Q, Gong J, et al. MicroRNA-1246 promotes growth and metastasis of colorectal cancer cells involving CCNG2 reduction. Mol Med Rep. 2016;13:273–80. doi: 10.3892/mmr.2015.4557. [DOI] [PubMed] [Google Scholar]
- 42.Zhang WC, Chin TM, Yang H, Nga ME, Lunny DP, Lim EK, et al. Tumour-initiating cell-specific miR-1246 and miR-1290 expression converge to promote non-small cell lung cancer progression. Nat Commun. 2016;7:11702. doi: 10.1038/ncomms11702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. The Journal of clinical investigation. 2009;119:1420–8. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nature reviews Cancer. 2002;2:442–54. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 45.Zhang J, Ma L. MicroRNA control of epithelial-mesenchymal transition and metastasis. Cancer Metastasis Rev. 2012;31:653–62. doi: 10.1007/s10555-012-9368-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sekhon K, Bucay N, Majid S, Dahiya R, Saini S. MicroRNAs and epithelial-mesenchymal transition in prostate cancer. Oncotarget. 2016 doi: 10.18632/oncotarget.11708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 48.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bucay N, Bhagirath D, Sekhon K, Yang T, Fukuhara S, Majid S, et al. A novel microRNA regulator of prostate cancer epithelial-mesenchymal transition. Cell Death Differ. 2017;24:1263–74. doi: 10.1038/cdd.2017.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saini S, Majid S, Yamamura S, Tabatabai L, Suh SO, Shahryari V, et al. Regulatory Role of mir-203 in Prostate Cancer Progression and Metastasis. Clin Cancer Res. 2011;17:5287–98. doi: 10.1158/1078-0432.CCR-10-2619. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.