Abstract
Due to extensive use in consumer goods, it is important to understand the genotoxicity of silver nanoparticles (AgNPs) and identify susceptible populations. 8-Oxoguanine DNA glycosylase 1 (OGG1) excises 8-oxo-7,8-dihydro-2-deoxyguanine (8-oxoG), a pro-mutagenic lesion induced by oxidative stress. To understand whether defects in OGG1 is a possible genetic factor increasing an individual’s susceptibly to AgNPs, we determined DNA damage, genome rearrangements, and expression of DNA repair genes in Ogg1-deficient and wild type mice exposed orally to 4 mg/kg of citrate-coated AgNPs over a period of 7 d. DNA damage was examined at 3 and 7 d of exposure and 7 and 14 d post-exposure. AgNPs induced 8-oxoG, double strand breaks (DSBs), chromosomal damage, and DNA deletions in both genotypes. However, 8-oxoG was induced earlier in Ogg1-deficient mice and 8-oxoG levels were higher after 7-d treatment and persisted longer after exposure termination. AgNPs downregulated DNA glycosylases Ogg1, Neil1, and Neil2 in wild type mice, but upregulated Myh, Neil1, and Neil2 glycosylases in Ogg1-deficient mice. Neil1 and Neil2 can repair 8-oxoG. Thus, AgNP-mediated downregulation of DNA glycosylases in wild type mice may contribute to genotoxicity, while upregulation thereof in Ogg1-deficient mice could serve as an adaptive response to AgNP-induced DNA damage. However, our data show that Ogg1 is indispensable for the efficient repair of AgNP-induced damage. In summary, citrate-coated AgNPs are genotoxic in both genotypes and Ogg1 deficiency exacerbates the effect. These data suggest that humans with genetic polymorphisms and mutations in OGG1 may have increased susceptibility to AgNP-mediated DNA damage.
Keywords: Silver nanoparticles, oxidative DNA damage, micronucleus, genotoxicity, cancer
Introduction
Silver nanoparticles (AgNPs) are extensively used in consumer and medical products. The main reason for incorporation of AgNPs in household items such as dental care products, food and drink containers, toys and appliances is their unique antibacterial, antifungal, and antimicrobial properties (Ahamed, Alsalhi, and Siddiqui 2010; Tolaymat et al. 2010; Hajipour et al. 2012; Maillard and Hartemann 2013; Duran et al. 2016). For the same reason, AgNPs are used in medical products, including wound dressings, cardiovascular catheters, and bone cement (Ge et al. 2014). In addition, the unique optical properties of AgNPs are also exploited in medical imaging, biosensing, and therapeutic applications (Tolaymat et al. 2010; Pietro et al. 2016). AgNPs are also available as dietary supplements (PEN CPI 2017). As such, AgNP-containing products comprise more than 30% of the consumer goods in the Nanotechnology Consumer Products Inventory (PEN CPI 2017). Oral exposure is a relevant route of exposure to AgNPs as they can be ingested due to their use in dietary supplements, toothpaste and toothbrushes, food and drink containers, possible use in poultry and livestock industry as alternative antibiotics and after clearance of inhaled AgNPs from the respiratory tract by mucociliary escalator (Fondevila et al. 2009; Bergin and Witzmann 2013; Quadros et al. 2013).
Studies in laboratory animals showed that AgNPs are absorbed in the gastrointestinal tract, distribute to different organs and tissues and accumulate in various organs, particularly, in reticuloendothelial organs (liver, spleen, kidney, lung, and other organs) (Loeschner et al. 2011; van der Zande et al. 2012; Bergin et al. 2016; Boudreau et al. 2016; Garcia et al. 2016). AgNP biodistribution analyzes in pregnant mice and developing embryos showed that AgNPs distribute to most maternal organs, extraembryonic tissues, and embryos (Austin et al. 2012). Thus, AgNPs can potentially cause damage to multiple tissues. Most studies failed to observe changes in body and/or organ weights, histology, hematology, and blood chemistry after oral exposure to AgNPs of up to 1000 mg/kg daily doses, although increase in liver enzymes, oxidative stress markers, and inflammatory cytokines were noted in some studies (Bergin et al. 2016; Boudreau et al. 2016; Garcia et al. 2016; Kim et al. 2008; Park et al. 2010; Patlolla, Hackett, and Tchounwou 2015b). Although these data suggest that ingestion of AgNPs results in relatively low general toxicity, they do not specifically address cancer risk. Genotoxicity is a precursor to carcinogenicity and is, therefore, measured as the first step in cancer risk assessment. Studies by both others and us have shown that AgNPs induce genotoxic effects in mice and rats after oral exposure (Kovvuru et al. 2015; Nallanthighal et al. 2017; Patlolla, Hackett, and Tchounwou 2015a), intravenous injection (Dobrzynska et al. 2014; Asare et al. 2016), and intraperitoneal injection (Song et al. 2012). Likewise, AgNPs are genotoxic in cultured cells (AshaRani et al. 2009; Vecchio et al. 2014; Butler et al. 2015; Guo et al. 2016). Micronuclei, indicative of chromosomal damage, and oxidative DNA damage (oxidative base damage and single strand breaks) were commonly observed in vitro in cultured cells and in vivo in rodent animals. However, not all animal studies observed chromosomal damage after AgNP treatment (Kim et al. 2008; Li et al. 2014), and this may be due to differences in study designs and/or the physicochemical properties of AgNPs used. For example, inverse correlations were reported between AgNP size and their cellular uptake, oxidative stress induction, cytotoxicity, and/or genotoxicity (Liu et al. 2010; Prasad et al. 2013; Miethling-Graff et al. 2014; Guo et al. 2016). Likewise, studies demonstrated that surface coatings stabilizing nanoparticles can modulate the genotoxicity of AgNPs (Vecchio et al. 2014; Guo et al. 2016; Nallanthighal et al. 2017). We recently reported that citrate-coated AgNPs induced DNA and chromosomal damage in orally exposed mice, while equivalent doses of polyvinylpyrrolidone (PVP)-coated AgNPs did not (Nallanthighal et al. 2017). Citrate and PVP are the most frequently used coatings of AgNPs (Huynh and Chen 2011; Wang et al. 2014).
Given the abundance of AgNP exposure sources and the ability of AgNPs to enter the tissues and induce genotoxic damage, it is important to identify the susceptible populations. It is possible that inter-individual differences exist in the susceptibility to AgNP-mediated genotoxic damage and thus, the risk of cancer. Genetic polymorphisms and/or mutations in DNA repair genes may be the possible genetic factors underlying such differences.
8-Oxoguanine DNA glycosylase 1 (OGG1) is a base excision repair (BER) enzyme that removes oxidatively damaged guanine from double stranded DNA (Hirano 2008; Sampath 2014). OGG1 is a bifunctional glycosylase that exhibits DNA glycosylase and apurinic/apyrimidinic (AP) lyase activities. DNA glycosylase excises the damaged base to generate an AP site, whereas an AP lyase cleaves the phosphodiester backbone at the AP site to generate a single strand break (SSB). Subsequently, the 5′ sugar fragment is removed, the resulting gap is filled with an undamaged nucleotide and the nick is sealed. Alternatively, during DNA replication, a SSB can result in a double strand break (DSB) when DNA polymerase encounters a SSB on the DNA template and falls off the template. Thus, in proliferating cells, oxidative DNA damage can result in DSBs and genome rearrangements (deletions, translocations or duplications) via intermediate SSBs (Obe et al. 2002). Major substrates of the OGG1 enzyme are 8-oxo-7,8-dihydro-2-deoxyguanine (8-oxoG) and the formamidopyrimidine derivative of guanine (FapyG) (Hazra et al. 2007; Sampath 2014). 8-oxoG and FapyG form from a common intermediate during oxidative modification of guanine and both are pro-mutagenic lesions that can result in point mutations (Greenberg 2012). For example, 8-oxoG often mispairs with adenine (A) leading to C:G to A:T transversion mutations and genomic instability (Hirano 2008).
OGG1 is a highly polymorphic gene and the chromosomal region harboring OGG1 gene (3p26.2) is susceptible to deletions that are frequently found in cancer cells (Weiss et al. 2005). A number of sequence variants, referred to as polymorphisms or mutations in the scientific literature, have been reported for OGG1 (reviewed in Goode, Ulrich, and Potter 2002; Weiss et al. 2005; Ali et al. 2015; Boiteux, Coste, and Castaing 2017). A single nucleotide polymorphism at codon 326 that results in a serine to cysteine amino acid substitution (Ser326Cys) is common in human populations with allelic frequencies of 0.22–0.45 (Goode, Ulrich, and Potter 2002). Epidemiological studies investigated an association between the Ser326Cys polymorphism and the risk of various types of cancer. An increased risk of cancer was found in some studies, whereas other studies failed to find this association (reviewed in Ali et al. 2015; Boiteux, Coste, and Castaing 2017; Goode, Ulrich, and Potter 2002; Hirano 2008; Weiss et al. 2005). An increased risk for lung, digestive system, and head and neck cancers was most consistently observed. Experimental studies showed the Ser326Cys polymorphic variant has reduced 8-oxoG repair capacity and a decreased ability to prevent mutagenesis, particularly, under oxidizing conditions (Yamane et al. 2004; Lee, Hodges, and Chipman 2005; Smart, Chipman, and Hodges 2006; Bravard et al. 2009; Kershaw and Hodges 2012).
Given that AgNPs are known to induce oxidative DNA damage, defects in OGG1 may exacerbate genotoxic effects of AgNPs. To elucidate this possible gene–environmental interaction, we performed a battery of genotoxicity assays and examined the expression of DNA repair and antioxidant genes in two mouse models, Ogg1-deficient and wild type mice. Mice were exposed to AgNPs by oral route, mimicking human exposure to AgNPs. We hypothesized that AgNPs induce more 8-oxoG, but fewer DNA strand breaks and genome rearrangements in Ogg1-deficient mice compared to wild type. The latter hypothesis is based on observations that Ogg1-deficient cells treated with pro-oxidants displayed markedly increased oxidative base damage but reduced frequency of strand breaks, compared to wild type cells (Smart, Chipman, and Hodges 2006). The strand breaks were not a result of direct damage of pro-oxidants, but rather a reflection of the BER process during which they are generated as repair intermediates. These data suggested that Ogg1 deficiency enhances susceptibility to oxidative base damage, while reduce susceptibility to DNA strand breaks and genome rearrangements which arise from BER process.
In the current study, we exposed mice orally to 20-nm citrate-coated AgNPs at a daily dose of 4 mg/kg for 7 consecutive days. This is a significantly lower dose compared to other oral exposure studies performed (Kim et al. 2008; Kovvuru et al. 2015; Patlolla, Hackett, and Tchounwou 2015a). To relate this dose to human exposure levels, we used the data of estimated AgNP intake from AgNP-containing beverage containers and dietary supplements. According to these data, daily oral exposure of AgNPs when drinking milk formula from a sippy cup containing silver nanomaterial is 1.5 µg/kg of silver (Ag) (Tulve et al. 2015) and is 1.4–7 µg/kg when taking AgNP dietary supplements (Munger et al. 2014). Allometric dose conversion of 4 mg/kg in a mouse yields 0.3 mg/kg in an average human (Reagan-Shaw, Nihal, and Ahmad 2008). This dose would correspond to the total Ag intake after use of dietary supplements for 6 weeks. Thus, this is considered a high but possible dose in humans after acute exposure.
Materials and methods
AgNPs
Citrate-coated 20 -nm AgNPs (lot/batch number MGM 1659) were manufactured by nanoComposix (San Diego, CA). Nanoparticles were supplied by the National Institute of Environmental Health Sciences Centers for Nanotechnology Health Implications Research (NCNHIR) consortium and their physiochemical characteristics were described in other studies (Bergin et al. 2016; Munusamy et al. 2015; Wang et al. 2014, 2016). Briefly, ~20 nm diameter particles had been grown on ~7–8 nm gold (Au) seed particles and had a thin layer (~1 nm) of sodium citrate (Munusamy et al. 2015; Wang et al. 2016). AgNPs were supplied as particles dispersed in 2 mM sodium citrate at Ag concentration of 1 mg/ml (Biopure™).
Characterization of AgNPs
Physicochemical characterization of AgNPs was performed by the manufacturer (nanoComposix), the Nanotechnology Characterization Laboratory (NCL) at the National Cancer Institute, the NCNHIR consortium (Bergin et al. 2016; Munusamy et al. 2015; Wang et al. 2014, 2016), and in-house prior to use. AgNP colloidal stability was monitored by UV-visible (UV-vis) spectroscopy with a NanoDrop1000 Spectrophotometer v3.8 (Thermo Fisher Scientific, Franklin, MA). The particle morphology, size, and size distribution were analyzed by scanning transmission electron microscopy (STEM). Samples were prepared by drop casting a drop of undiluted nanoparticle dispersion on a 400 mesh formvar/carbon copper transmission electron microscope (TEM) grid and air dried. STEM images were acquired on a FEI Titan 80–300 microscope operated at an accelerating voltage of 200 keV using the high angular annular dark field (HAADF) detector and/or bright field (BF) detector. STEM images were collected with a convergence angle of 15 mrad and a working distance of 230 mm for both HAADF and BF detectors. Images were obtained using a beam current of approximately 0.5 nA. For particle size distribution analyzes, Fiji image processing program (an open-source ImageJ software focused on biological-image analysis) (Schindelin et al. 2012) was utilized to analyze the size of at least 1000 particles from HAADF images. Au core/Ag shell nanoparticle structure was verified by energy dispersive X-ray spectroscopy (EDS) mapping on a FEI Titan 80–300 microscope. EDS HyperMaps (Spectral Cubes) were obtained with a 0.5 nA beam current with Bruker SuperX EDS system with Esprit software. HyperMaps were collected for 240 s with approximately 9000 counts per second. To form the maps for Au and Ag, the 9.7 and 2.9 keV photons, respectively, were used. Ag and Au data were extracted from the HyperMap and plotted as a single map indicating the Ag and Au location in the particles. Hydrodynamic particle size was measured by dynamic light scattering (DLS) and surface charge was measured by zeta potential measurements with a Zetasizer Nanoseries (Malvern Instruments, Westborough, MA). In addition, particle size and size distribution were determined by single particle inductively coupled mass spectrometry (SP-ICP-MS) using a NexION 350X ICP-MS Spectrometer (PerkinElmer, Waltham, MA), a Meinhard nebulizer, and operated at a radio frequency power of 1600 W. Ag was measured at m/z (molecule mass/number of elementary charges) values of 107 and 109. Data acquisition was performed using PerkinElmer Syngistix Nano Application Module software in the time-resolved analysis mode with a dwell time of 100 µs and an acquisition time of 100 s. Prior to analysis, particle solutions were diluted in deionized water to an estimated concentration of 105 particles per ml. The particle sizes were calculated from the measured Ag masses by assumption of a spherical shape of the particles. Particle size calibration was performed using AgNP standards from nanoComposix.
Mice and AgNP Treatments
C57BL/6 J pun/pun background mice (Jackson Laboratory, Bar Harbor, ME) were used. C57BL/6 J pun/pun mice (congenic to C57BL/6 J strain) contain the recessive 70 kb internal duplication in the pink-eyed dilution (p) gene, termed pink-eyed unstable (pun) allele, resulting in a light gray coat color and pink eyes in homozygous pun/pun mice. Ogg1-deficient (−/−) mice in C57BL/6 J pun/pun background were a gift from Dr. Robert H. Schiestl, University of California Los Angeles, CA. Ogg1−/− mice were originally generated by Klungland et al. (1999). Mouse genotyping was carried out by polymerase chain reaction (PCR) using tail snip DNA (Yamamoto, Chapman, and Schiestl 2013).
Mice were housed in the University at Albany’s Cancer Research Center virus-free vivarium under standard conditions. All procedures were approved by the institutional animal use and care committee. Pregnancy was timed by checking for vaginal plugs, with noon of the day of discovery counted as 0.5 days post coitum (dpc). Similarly, the time of birth of a litter was timed with the noon of discovery counted as 0.5 days post partum (dpp).
AgNPs or vehicle (2 mM sodium citrate) was administered by gavage to 3- to 4-month-old Ogg1−/− mice or wild type mice. Ogg1−/− and wild type mice were treated at different times. Mice received a daily dose of 4 mg/kg of AgNPs (equivalent to 4 mg Ag/kg) between noon and 2 PM for a total of 7 d. Peripheral blood was sampled via submandibular bleeding at 3 and 7 d of treatment (24 h after the last dose), as well as 7 and 14 d after termination of the 7-d treatment. Blood was used immediately for the analyzes of 8-oxoG and phosphorylated histone H2AX (γ-H2AX) foci in mononuclear cells and analyzes of micronuclei in erythrocytes. Livers were harvested after 7 d of treatment (24 h after the last dose) for quantitative real-time PCR (qPCR) analyzes. For the pun reversion/DNA deletion assay, 3- to 4-month-old pregnant dams were treated orally with the same dose of AgNPs (4 mg/kg) for the same duration (7 d) from 9.5 dpc to 15.5 dpc and the resulting offspring were euthanized at 20 dpp for determination of the pun reversions/DNA deletions. The harvested cells/tissues represent possible target organs of AgNPs. For example, blood mononuclear cells can uptake AgNPs following their absorption into blood. Bone marrow, the organ in which micronucleus formation occurs, accumulates high levels of AgNPs that is consistent with the preferential accumulation of AgNPs in reticuloendothelial tissues (Li et al. 2014). Likewise, high levels of AgNPs accumulate in the liver (Bergin et al. 2016; Boudreau et al. 2016; Garcia et al. 2016; Loeschner et al. 2011; van der Zande et al. 2012). Also, AgNPs were detected in developing embryos after exposure of pregnant dams and thus can potentially induce damage to embryos (Austin et al. 2012).
Immunofluorescence of 8-oxoG
8-oxoG, indicative of oxidative DNA damage, was examined in peripheral blood mononuclear cells by immunofluorescence as in our previous studies (Kovvuru et al. 2015; Nallanthighal et al. 2017). Immunofluorescent 8-oxoG detection method allows evaluation of 8-oxoG in the nuclear and mitochondrial DNA in fixed cells and tissues in situ (Beckman and Ames 1997; Soultanakis et al. 2000). It requires a low cell count that can be obtained by nonterminal bleeding and, thus, allows for follow-up observation in the same animals. Unlike chromatography-based 8-oxoG detection methods, this method eliminates the need for DNA isolation that is known to induce artificial DNA oxidation (Beckman and Ames 1997; Collins et al. 2004). However, due to the immunofluorescent 8-oxoG detection method being a semi-quantitative technique, it cannot yield the number of 8-oxoG residues per dG like the chromatography-based method. Instead, it produces a strong fluorescent signal in the nuclei (and/or mitochondria) of the damaged cells, whereas intact cells emit very low, if any, fluorescence. Therefore, data are frequently reported as the percentage of cells/nuclei emitting bright fluorescence, often termed 8-oxoG positive cells.
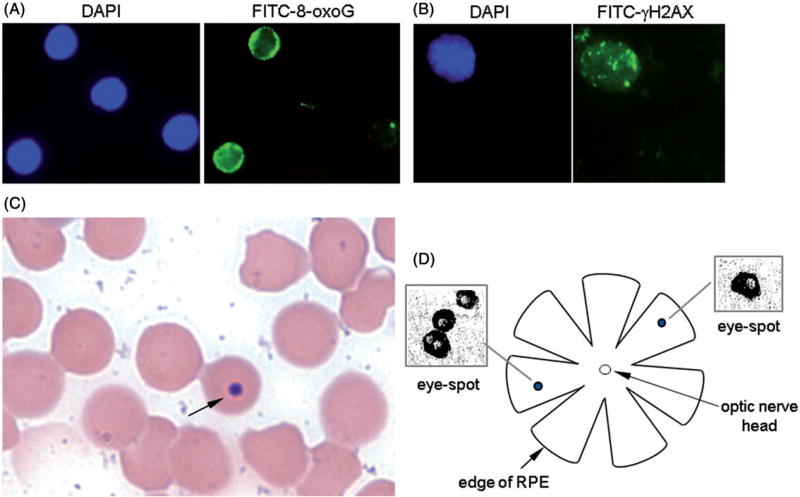
The assay was performed as follows. First, we incubated 50 µl of peripheral blood with 10× (v/v) erythrocyte lysis buffer (8.5 g/l ammonium chloride in 0.01 M Tris-HCl buffer; pH 7.5) to eliminate erythrocytes. The remaining cells were suspended in 50 µl of PBS and pipetted onto poly-d-lysine-coated cover slips to allow attachment to the cover slips. Cells were then fixed with 4% of paraformaldehyde, permeabilized with 0.5% of Triton-X 100 and washed with PBS. After blocking, cells were incubated with mouse anti-8-oxoG antibody clone 438.15 (Millipore # MAB3560) at a dilution of 1:500 followed by incubation with FITC-conjugated anti-mouse IgG secondary antibody (Jackson Immunochemicals, West Grove, PA, Catalog #115-095-008) at a 1:200 dilution. Coverslips were mounted on microscopic slides with 7 µl of DAPI/Vectashield solution (Vector Laboratories, Burlingame, CA) to counterstain nuclei. Slides were visualized under a 100× objective on a Nikon Eclipse TS100 microscope and a minimum of 100 cells were analyzed. Cells exhibiting green fluorescence in the nucleus were considered positive for 8-oxoG. Figure 1(A) shows that out of four nuclei (DAPI staining), two emit green fluorescence, i.e., are positive for 8-oxoG (FITC staining). Data were analyzed blinded to treatment.
Figure 1.
Genotoxicity assays used. (A) Immunofluorescence of 8-oxoG. Cellular nuclei are shown by DAPI staining and 8-oxoG is visualized by FITC staining. (B) Immunofluorescence of γ-H2AX. Cellular nucleus is visualized by DAPI staining and γ-H2AX foci are visualized by FITC staining. (C) Micronucleus assay. A Giemsa-stained peripheral blood smear with a micronucleated erythrocyte highlighted by an arrow is shown. (D) DNA deletion assay. Schematic of RPE and close-up of eye-spots are shown.
Immunofluorescence of γ-H2AX
γ-H2AX foci, indicative of DSBs (Rogakou et al. 1999), were determined in peripheral blood mononuclear cells by immunofluorescent assay as in our previous studies (Kovvuru et al. 2015; Nallanthighal et al. 2017). Briefly, 50 µl of peripheral blood was incubated with 10× (v/v) erythrocyte lysis buffer (8.5 g/l ammonium chloride in 0.01 M Tris-HCl buffer; pH 7.5) to lyse erythrocytes. The remaining cells were suspended in 50 µl of PBS, pipetted onto poly-d-lysine-coated cover slips, fixed with 4% of paraformaldehyde, permeabilized with 0.5% of Triton-X 100 and washed with PBS. After blocking, cells were incubated with mouse anti-phospho-Histone2AX (Ser139) antibody clone JBW301 (EMD Millipore, Billerica, MA, Catalog #05-636) at a 1:400 dilution followed by incubation with FITC-conjugated anti-mouse IgG secondary antibody (Jackson Immunochemicals, West Grove, PA, Catalog #115-095-008) at a 1:200 dilution. Coverslips were mounted on microscopic slides with 7 µl of DAPI/Vectashield solution (Vector Laboratories, Burlingame, CA) to counterstain nuclei. Slides were visualized under a 100× objective on a Nikon Eclipse TS100 microscope. A minimum of 100 cells were analyzed and cells with more than four distinct foci in the nucleus were considered positive for γ-H2AX foci (Figure 1(B)). Data were analyzed blinded to treatment.
In Vivo Micronucleus Assay
Micronuclei, indicative of chromosome fragmentation and/or loss occurring in dividing erythroblasts in bone marrow, were scored in peripheral blood erythrocytes. In mice, micronuclei can be scored in peripheral blood since damaged erythroblasts complete cell division and maturation in the bone marrow and subsequently enter the systemic circulation (Hayashi et al. 2000). Three microliter of peripheral blood was smeared on microscopic glass and stained with modified Giemsa dye (Sigma-Aldrich, St. Louis, MO). Micronuclei were examined under a 100× objective on a Nikon Eclipse TS100 microscope (Figure 1(C)). At least 2000 erythrocytes were scored and the frequency of micronuclei was calculated as the number of micronucleated erythrocytes per 2000 erythrocytes. Data were analyzed blinded to treatment.
Pun Reversion/DNA Deletion Assay
The 70 kb DNA deletions were determined as a measure of large-scale genome rearrangements. Such deletions occur from homologous recombination between two 70 kb tandem repeats in the pink-eyed unstable (pun) locus spanning exons 6–18 of the pink-eye dilution (p) gene, resulting in the deletion of one of the repeats (Reliene et al. 2004a). The pun allele is reverted to the functional p gene by such a deletion event, allowing black pigment accumulation in the cells of the otherwise transparent retinal pigment epithelium (RPE) of the pun/pun mice. The RPE cells proliferate from 9.5 to 15.5 dpc. Thus, the pun reversions/DNA deletions can only be induced during embryonic development, when the RPE cells are proliferating. Deletions are visualized microscopically after birth, as described below.
Dissection of the RPE
Offspring were sacrificed at 20 d of age and their eyes were dissected and processed to expose the RPE layer as previously described (Reliene et al. 2004a). Briefly, the eye was removed from its orbit and immersed in fixative (4% of paraformaldehyde in 0.1 M phosphate buffer; pH 7.4) for 1 h and then in PBS until dissection. An incision was made at the upper corneo-scleral border to allow removal of the cornea and lens. To flatten the eye-cup, 6–8 incisions were made from the corneo-scleral margin toward the centrally positioned optic nerve, and the dissected eyecup was placed on a glass slide with the retina facing up (Figure 1(D)). The retina was then gently removed and the residual specimen consisting of sclera, choroid and RPE, with the RPE facing up, was mounted in 90% of glycerol.
Scoring of DNA Deletions Visualized as Eye-Spots
A pigmented cell or a group of adjacent pigmented cells separated from each other by no more than five unpigmented cells was considered as one eye-spot that resulted from one pun reversion/DNA deletion event (Figure 1(D)) (Reliene et al. 2004a, 2004b). The number of eye-spots in each RPE was counted under a microscope using a 10× objective.
Quantitative Real Time PCR
Total RNA from livers was extracted with TRIzol® reagent (Thermo Fisher Scientific, Franklin, MA). Reverse transcription PCR reactions were performed with 1.5 µg total RNA using MultiScribe™ Reverse Transcriptase (Thermo Fisher Scientific, Franklin, MA, Catalog #4311235) to synthesize cDNA. Quantitative PCR was performed using iTaq™ Universal SYBR® Green Supermix (Bio-Rad Laboratories, Hercules, CA, Catalog #172–5850) on the ABI7900 HT Fast Real-Time PCR system (Applied Biosystems, Carlsbad, CA). Relative expression levels of each gene were analyzed using the 2−ΔΔCT method and presented relative to the expression of the housekeeping gene GAPDH. Each sample was replicated twice from independent cDNA preparations. Primer information is provided below:
Gapdh (5′ TCGTCCCGTAGACAAAATGG, 3′ TTGAG GTCAATGAAGGGGTC)
DNA-PKcs (5′ AGACTGTTGGAGAGCAGGA, 3′ TCCT GCTCTCCAACAGTCT)
Ogg1 (5′ CTCTATTGCACTGTGTACCG, 3′ CACCTT GGAATTTCTGGG)
Neil1 (5′ GAGAACGTACTTCGGAACCT, 3′ GGCCTC TAGAACTGTACGA)
Neil2 (5′ TTTAGTGGTGGTGGCTTC, 3′ GGTCCAA GAGTGTGTAGCA)
Myh (5′ CCAAGCATCTGTGAGGAGT, 3′ TCACGCTT CTCTTGGTCA)
Xrcc4 (5′ CCTAAAATGGCTCCACAGGA 3′ GCTGCT GTTTCTCAGGGTTT)
Gpx1 (5′ AAGGCTCACCCGCTCTTTA, 3′ CCCACCAG GAACTTCTCAAAG)
Cat (5′ CTTTGACAGAGAGCGGATTCC, 3′ CTTTGAC AGAGAGCGGATTCC)
Sod1 (5′TGTTGGAGACCTGGGCAAT, 3′ ACTGCGC AATCCCAATCAC)
Sod2 (5′TCAAGCGTGACTTTGGGTCT, 3′ TGTTCCTT GCAATGGGTCCT)
Hmox-1 (5′ AACCCAGTCTATGCCCCA, 3′ CTCGTGG AGACGCTTTACA)
Statistics
Data were analyzed by two-way ANOVA followed by the post hoc Tukey’s Multiple Comparison test or Student’s t-test, as appropriate, using Statistical Analysis Systems (SAS) studio software and presented as means±SEM. Differences between groups were considered statistically significant if p value was less than 0.05.
Results
Characterization of AgNPs
We characterized nanoparticles using a variety of analytical techniques including STEM, EDS, UV-vis spectroscopy, DLS, zeta potential measurements and SP-ICP-MS. STEM and EDS analyzes showed that particles were nearly spherical in shape with the Au core in the center or off-center of the particle (Figure 2(A,B)). The particle size and size distribution analyzes by STEM indicated that particles exhibited a monomodal size distribution and the mean particle diameter was 19.9 nm (Figure 2(C)). UV-vis spectroscopy measurements showed that AgNP dispersions in deionized water exhibited a Surface Plasmon Resonance (SPR) peak at 400 nm, which is characteristic of AgNPs that are about 20 nm in size (NanoComposix 2017) (Figure 2(D)). DLS characterization revealed that particles had a monomodal size distribution with a mean hydrodynamic diameter of 25.1 nm and PDI of 0.061 (Figure 2(E)). Nanoparticles had a negative surface charge (zeta potential value −43 mV), consistent with the citrate coating. The mean particle diameter determined by SP-ICP-MS was 19.8 nm, which is in accordance with STEM and DLS measurements (Figure 2(F)).
Figure 2.
Characterization of AgNPs. (A) BF STEM image of nanoparticles. Au cores are highlighted by arrows. (B) EDS map indicating Ag and Au location in the particle. Red color indicates Ag and blue color indicates Au. (C) Size distribution analysis by STEM. (D) UV-vis absorbance spectra. (E) Size distribution analysis by DLS. (F) Size distribution analysis by SP-ICP-MS.
Effect of AgNPs on Oxidative DNA Damage
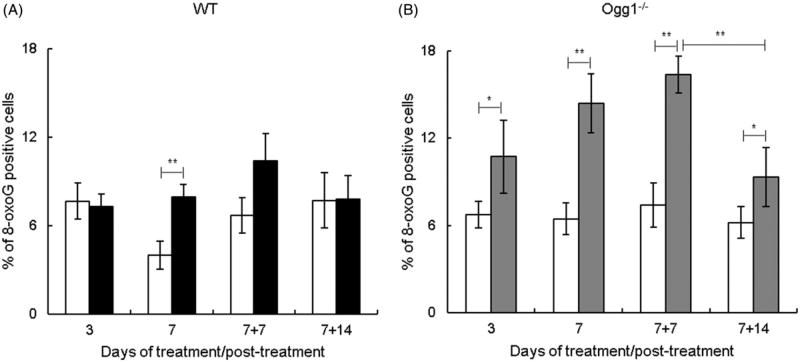
The effect of AgNPs on oxidative DNA damage was assessed by measuring 8-oxoG. In wild type mice, the levels of 8-oxoG were not altered after AgNP treatment for 3 d, but increased significantly after exposure for 7 d, and this increasing trend was observed at 7 d post-exposure (7 + 7 d) with the levels returning to basal at 14 d post-exposure (7 +14 d) (Figure 3(A)). In contrast, in Ogg1−/− mice, 8-oxoG levels increased at 3 d, increased further at 7 d, and remained significantly elevated at 7 + 7 d and 7 + 14 d (Figure 3(B)). Thus, in Ogg1−/− mice, 8-oxoG was induced after shorter exposure duration (3 vs. 7 d) and remained elevated during a 2-week follow-up period. Comparison of data in Ogg1−/− and wild type mice revealed that spontaneous 8-oxoG levels were similar in both genotypes. In comparison, AgNP-induced 8-oxoG levels were significantly higher in Ogg1−/− mice than wild type mice at 3 of the 4 observed time points (3, 7, and 7 + 7 d). The levels of 8-oxoG were higher in Ogg1−/− mice by 1.5-, 1.8-, and 1.6-fold at 3, 7, and 7 + 7 d, respectively. These data indicate that Ogg1−/− mice exhibit elevated susceptibility to AgNP-induced oxidative DNA damage.
Figure 3.
Effect of AgNPs on oxidative DNA damage. (A) Wild type mice. (B) Ogg1-deficient mice. Filled bars show AgNP-treated mice and open bars show controls. 8-oxoG was determined at 3 and 7 d of treatment and at 7 (7 + 7) and 14 (7 + 14) d post-treatment. Mean ± SEM is shown, n = 8–10 mice/group, *p ≤ 0.05, **p ≤ 0.01.
Effect of AgNPs on DSBs
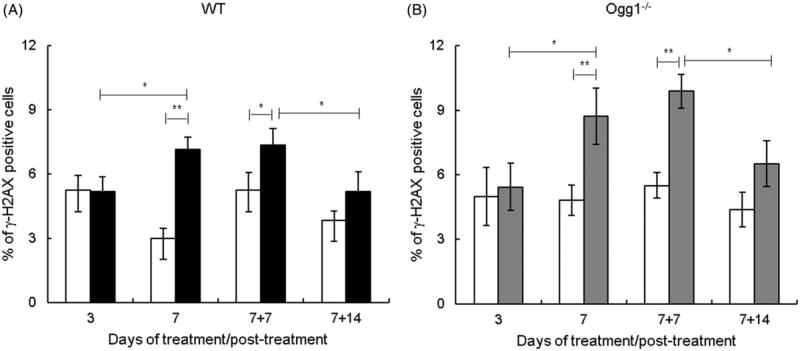
We determined the effect of AgNPs on DSBs by measuring formation of γ-H2AX foci. γ-H2AX foci increased significantly at 7 d, remained elevated at 7 + 7 d, and returned to basal levels at 7 +14 d in both Ogg1−/− and wild type mice (Figure 4(A, B)). Thus, DSBs were induced after AgNP ingestion for 7 d and were elevated up to 1 week post-exposure. Comparison of γ-H2AX foci in Ogg1−/− and wild type mice showed that the levels of spontaneous and AgNP-induced γ-H2AX foci were not significantly different between genotypes.
Figure 4.
Effect of AgNPs on DSBs. (A) Wild type mice. (B) Ogg1-deficient mice. Filled bars show AgNP-treated mice and open bars show controls. γ-H2AX foci were determined at 3 and 7 d of treatment and at 7 (7 + 7) and 14 (7 + 14) d post-treatment. Mean ± SEM is shown, n = 8–10 mice/group, *p ≤ 0.05, **p ≤ 0.01.
Effect of AgNPs on micronucleus formation
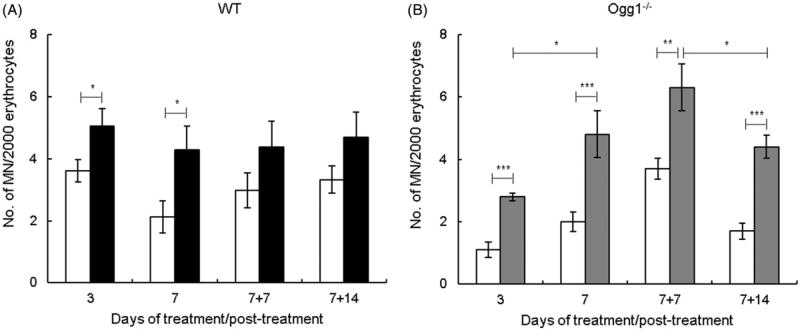
We determined the effect of AgNPs on chromosomal damage by scoring micronuclei. The number of micronucleated cells was increased significantly at 3 and 7 d in both genotypes (Figure 5(A,B)). At 7 + 7 d and 7 +14 d, AgNP-induced micronuclei showed an increasing trend in wild type mice and were significantly increased in Ogg1−/− mice and wild type (Figure 5(A,B)). Comparison of data in Ogg1−/− and wild type mice showed that micronucleus frequency was not significantly different between mutant and wild type mice at any time points with the exception of one. At 3 d, the frequencies of spontaneous and AgNP-induced micronuclei were significantly lower in Ogg1−/− than wild type mice. This is likely due to interexperimental variability, given that this difference was observed at one time point only and that variability of this magnitude is frequently observed with the in vivo micronucleus assay.
Figure 5.
Effect of AgNPs on chromosomal damage. (A) Wild type mice. (B) Ogg1-deficient mice. Filled bars show AgNP-treated mice and open bars show controls. Micronuclei were determined at 3 and 7 d of treatment and at 7 (7 + 7) and 14 (7 + 14) d post-treatment. Mean ± SEM is shown, n = 8–10 mice/group, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001.
Effect of AgNPs on induction of DNA deletions
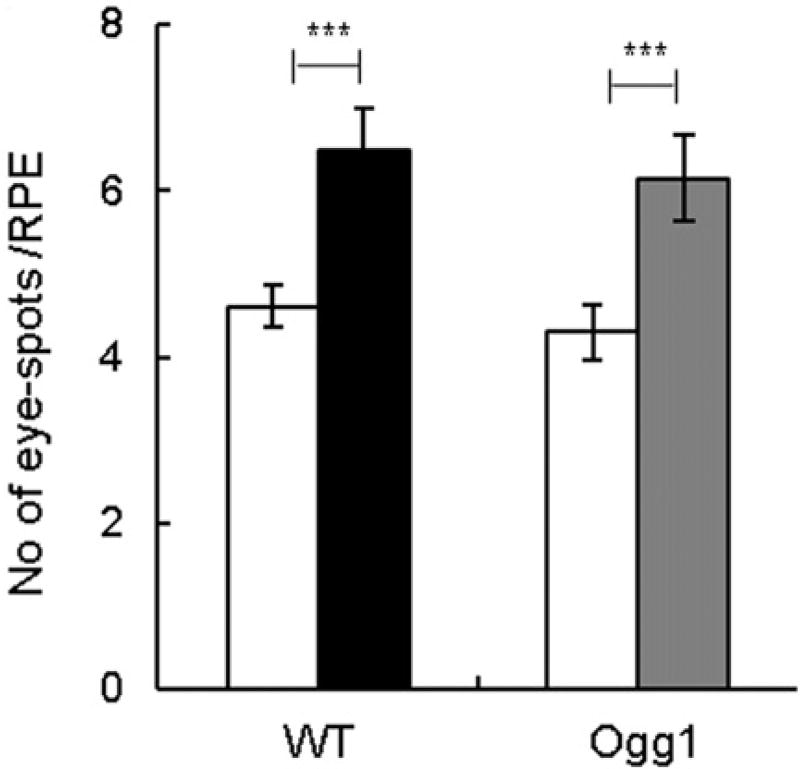
We determined the effect of AgNPs on genome rearrangements by analyzing 70 kb DNA deletions that were scored as pigmented eye-spots. AgNPs induced eye-spots in both Ogg1−/− and wild type mice (Figure 6). Comparison of data between the two genotypes showed that the number of eye-spots was not significantly different between mutant and wild type mice. Thus, AgNPs induced DNA deletions in mice, but a deficiency in Ogg1 did not modulate the effect.
Figure 6.
Effect of AgNPs on DNA deletions. Frequency of eye-spots after AgNP treatment for 7 d. Filled bars show AgNP-treated mice and open bars show controls. Mean ± SEM is shown, n = 20–22 eyes/group, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001.
Effect of AgNPs on expression of DNA repair genes
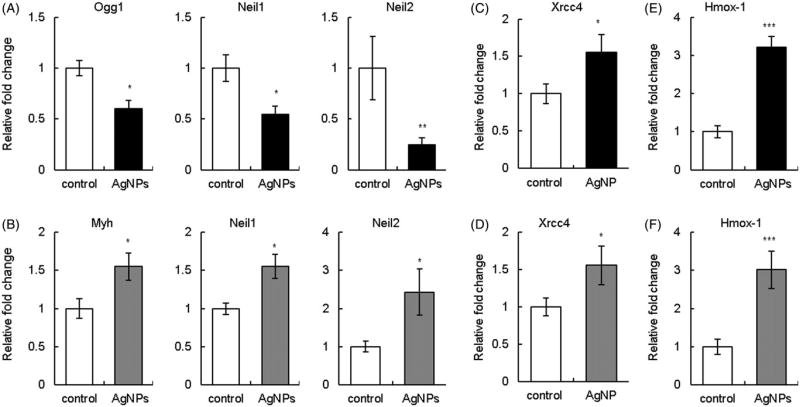
Next, we examined whether AgNPs upregulated DNA repair and antioxidant genes that can be viewed as an adaptive response to AgNP-induced DNA damage. We determined expression levels of relevant DNA glycosylases involved in BER (Ogg1, Neil1, Neil2, and Myh), DSB repair genes (DNA-PKcs and Xrcc4) and antioxidant genes (Cat, Gpx1, Hmox-1, Sod1, and Sod2) in the liver, a major internal organ of AgNP accumulation. AgNP treatment was associated with downregulation of Ogg1, Neil1, Neil2 genes in wild type mice (Figure 7(A)) and Myh levels were unaltered (data not shown). The same genes with the exception of Ogg1 were analyzed in Ogg1−/− mice. In contrast to wild type mice, AgNP exposure resulted in upregulation of Neil1, Neil2, and Myh in Ogg1−/− mice (Figure 7(B)). In comparison, AgNPs modulated DSB repair and antioxidant genes similarly in Ogg1−/− and wild type mice. For example, expression of DNA-PKcs was unaltered (data not shown) and Xrcc4 was upregulated (Figure 7(C,D)). Likewise, an antioxidant response gene Hmox-1 was significantly upregulated in both genotypes (Figure 7(E,F)). Expression levels of Cat, Sod1, Sod2, and Gpx1 were unaltered (data not shown). These data indicate that expression of DNA glycosylases are modulated by AgNPs in a genotype-specific manner.
Figure 7.
Effect of AgNPs on gene expression. (A) BER genes in wild type mice. (B) BER genes in Ogg1-deficient mice. (C) DSB repair gene Xrcc4 in wild type mice. (D) DSB repair gene Xrcc4 in Ogg1-deficient mice. (E) Antioxidant response gene Hmox-1 in wild type mice. (F) Antioxidant response gene Hmox-1 in Ogg1-deficient mice. Filled bars show AgNP-treated mice and open bars show controls. Gene expression levels were determined by qPCR at 7 d of treatment. Data were normalized to GAPDH. Mean ± SEM is shown, n = 5–6 mice/group, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001.
Discussion
We report that AgNP exposure by oral route induced oxidative base damage (8-oxoG), DSBs, chromosomal damage, and DNA deletions in both Ogg1-deficient and wild type mice. The observed genome damage can play a role in carcinogenesis. 8-oxoG is a precursor of C:G to A:T transversion mutations which are the most frequently found mutations in human cancers (Greenman et al. 2007). DSBs can result in chromosomal fragmentation, translocations, and deletions (Kanaar, Hoeijmakers, and van Gent 1998). Deletions are critical in cancer development, given that tumor suppressors, cell cycle regulators and DNA repair genes can be lost through this process. This is further supported by observations that deletions occur in cancer-prone mice spontaneously and in wild type mice after exposure to carcinogens (Reliene et al. 2004a, Reliene, Fischer, and Schiestl 2004c; Reliene et al. 2010).
The levels of 8-oxoG were higher in AgNP-treated Ogg1-deficient than wild type mice, whereas the magnitude of other DNA lesions was similar in both genotypes. Higher 8-oxoG levels in Ogg1-deficient mice can be explained by the lack of functional Ogg1, but similar frequencies of other DNA lesions in both genotypes is an unexpected finding. A study reported that embryonic fibroblasts derived from Ogg1-deficient mice exposed to pro-oxidants (sodium dichromate, potassium bromate, and Ro19-8022), displayed higher levels of oxidative base damage, but exhibited fewer strand breaks than wild type counterparts (Smart, Chipman, and Hodges 2006). This observation was explained by the lack of DNA glycosylase and AP lyase activities in Ogg1-deficient cells that generate AP sites and SSBs, respectively, as repair intermediates. SSBs can result in DSBs, chromosomal damage and genome rearrangements in mitotically active cells (Obe et al. 2002). Thus, our original hypothesis was that AgNP-treated Ogg1-deficient mice will display higher levels of 8-oxoG, but lower levels of strand breaks and genome rearrangements than wild type mice. In support to our hypothesis, we observed that Ogg1-deficient mice exhibited higher 8-oxoG levels during 7-d exposure and even after exposure termination. This implies that a deficiency in Ogg1 increases susceptibility to AgNP-induced pro-mutagenic lesions. However, the magnitude of DSBs, micronuclei, and DNA deletions was similar in both Ogg1-deficient and wild type mice. Thus, the lack of Ogg1 function does not protect against large-scale genomic instability induced by AgNPs. A possible explanation of this is that, unlike pro-oxidant chemicals, AgNPs can employ several mechanisms. For example, AgNPs can induce oxidative base damage via generation of reactive oxygen species (ROS) and directly induce strand breaks in DNA (AshaRani et al. 2009; Piao et al. 2011a).
AgNP treatment was associated with reduced expression levels of DNA glycosylases in wild type mice. We postulate that increased levels of 8-oxoG in AgNP-treated wild type mice is in part due to downregulation of DNA glycosylases that are involved in the repair of 8-oxoG. Ogg1, Neil1, and Neil2 are bifunctional DNA glycosylases with DNA glycosylase and AP lyase activities and have overlapping physiological substrates (Hazra et al. 2007; Jacobs and Schar 2012; Sampath 2014). The common substrates recognized by Ogg1, Neil1, and Neil2 are 8-oxoG and FapyG. No other mammalian DNA glycosylases are known to repair 8-oxoG and FapyG. Thus, AgNP-mediated downregulation of Ogg1 and Neil genes can impair recognition and removal of pro-mutagenic base lesions. In agreement with our findings, studies reported a decrease in OGG1 expression in human Chang liver and Jurkat T cell lines exposed to AgNPs in vitro (Chatterjee, Eom, and Choi 2014; Piao et al. 2011b). Downregulation of OGG1 was associated with reduced expression, nuclear translocation and transcriptional activity of the transcription factor nuclear factor erythroid 2-related factor 2 (NRF2), a regulator of inducible expression of OGG1 (Chatterjee, Eom, and Choi 2014; Piao et al. 2011b). Downregulation of OGG1 is a common mechanism of relevant environmental carcinogens. For example, arsenic, cadmium, and chromium compounds were shown to reduce OGG1 levels and/or enzymatic activity (Hodges and Chipman 2002; Potts, Watkin, and Hart 2003; Ebert et al. 2011). This shared molecular mechanism between well-known human carcinogens and AgNPs raises an additional concern about potential carcinogenicity of AgNPs.
In contrast to wild type mice, AgNP treatment resulted in upregulation of DNA glycosylases in Ogg1-deficient mice. This finding suggests an induction of an adaptive mechanism in Ogg1 mutants. Neil1 and Neil2 are capable of removing 8-oxoG and Myh repairs A:8-oxoG mispairs, which collectively ensures that genomic integrity is maintained (Hazra et al. 2007; Hirano 2008). Although Ogg1, Neil1, and Neil2 act on the same base modifications, they seem to have preferred DNA templates. OGG1 is active on duplex DNA, whereas NEIL glycosylases prefer bubble DNA that occurs during replication and/or transcription (Hazra et al. 2007). On the other hand, and in contrast to Harza et al.’s other studies reported that NEIL1 efficiently removes 8-oxoG only from double stranded DNA (Morland et al. 2002; Vik et al. 2012) or only has minor activity on 8-oxoG (Bandaru et al. 2002). Overall, data reported in the literature suggests Neil enzymes could have repaired some of the AgNP-induced 8-oxoG in Ogg1-deficient mice. However, given that 8-oxoG levels were higher in Ogg1-deficient than wild type mice, Ogg1 is indispensable for the efficient repair of AgNP-induced damage.
Conclusions
Oral exposure to citrate-coated 20 nm AgNPs caused a wide spectrum of genotoxic effects ranging from oxidative base damage to large DNA deletions. These effects were induced in both Ogg1-deficient and wild type mice. However, a defect in Ogg1 exacerbated the effect of AgNP-induced oxidative base damage. In addition, AgNPs reduced expression of DNA glycosylases in wild type mice and, in contrast, increased expression in Ogg1-deficient mice. This may have reduced the difference in oxidative base damage observed between the genotypes, but ultimately, did not fully compensate for the loss of Ogg1. These data suggest that humans with polymorphisms and/or mutations in OGG1 may have elevated susceptibility to AgNP-mediated damage, which, in turn, may increase the risk of cancer.
Acknowledgments
Funding
This study was supported by the National Institute of Environmental Health Sciences (R56ESO24123) and the Dominic Ferraioli Foundation. AgNPs used in this study were procured, characterized and provided by the NCNHIR consortium. The authors would like to thank Dr. Robert Schiestl, University of California Los Angeles, for providing Ogg1-deficient mice and Dr. Dhruba Bharali, Pharmaceutical Research Institute, Albany College of Pharmacy and Health Sciences, for DLS analysis.
Footnotes
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Ahamed M, Alsalhi MS, Siddiqui MK. Silver Nanoparticle Applications and Human Health. Clinica Chimica Acta; International Journal of Clinical Chemistry. 2010;411:1841–1848. doi: 10.1016/j.cca.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Ali K, Mahjabeen I, Sabir M, Mehmood H, Kayani MA. OGG1 Mutations and Risk of Female Breast Cancer: Meta-Analysis and Experimental Data. Disease Markers. 2015;2015:1–16. doi: 10.1155/2015/690878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asare N, Duale N, Slagsvold HH, Lindeman B, Olsen AK, Gromadzka-Ostrowska J, Meczynska-Wielgosz S, et al. Genotoxicity and Gene Expression Modulation of Silver and Titanium Dioxide Nanoparticles in Mice. Nanotoxicology. 2016;10:312–321. doi: 10.3109/17435390.2015.1071443. [DOI] [PubMed] [Google Scholar]
- AshaRani PV, Low Kah Mun G, Hande MP, Valiyaveettil S. Cytotoxicity and Genotoxicity of Silver Nanoparticles in Human Cells. ACS Nano. 2009;3:279–290. doi: 10.1021/nn800596w. [DOI] [PubMed] [Google Scholar]
- Austin CA, Umbreit TH, Brown KM, Barber DS, Dair BJ, Francke-Carroll S, Feswick A, et al. Distribution of Silver Nanoparticles in Pregnant Mice and Developing Embryos. Nanotoxicology. 2012;6:912–922. doi: 10.3109/17435390.2011.626539. [DOI] [PubMed] [Google Scholar]
- Bandaru V, Sunkara S, Wallace SS, Bond JP. A Novel Human DNA Glycosylase that Removes Oxidative DNA Damage and Is Homologous to Escherichia coli Endonuclease VIII. DNA Repair (Amst) 2002;1:517–529. doi: 10.1016/s1568-7864(02)00036-8. [DOI] [PubMed] [Google Scholar]
- Beckman KB, Ames BN. Oxidative Decay of DNA. The Journal of Biological Chemistry. 1997;272:19633–19636. doi: 10.1074/jbc.272.32.19633. [DOI] [PubMed] [Google Scholar]
- Bergin IL, Wilding LA, Morishita M, Walacavage K, Ault AP, Axson JL, Stark DI, et al. Effects of Particle Size and Coating on Toxicologic Parameters, Fecal Elimination Kinetics and Tissue Distribution of Acutely Ingested Silver Nanoparticles in a Mouse Model. Nanotoxicology. 2016;10:352–360. doi: 10.3109/17435390.2015.1072588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergin IL, Witzmann FA. Nanoparticle Toxicity by the Gastrointestinal Route: Evidence and Knowledge Gaps. International Journal of Biomedical Nanoscience and Nanotechnology. 2013;3:1–44. doi: 10.1504/IJBNN.2013.054515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiteux S, Coste F, Castaing B. Repair of 8-Oxo-7,8-Dihydroguanine in Prokaryotic and Eukaryotic Cells: Properties and Biological Roles of the Fpg and OGG1 DNA N-Glycosylases. Free Radical Biology and Medicine. 2017;107:179–201. doi: 10.1016/j.freeradbiomed.2016.11.042. [DOI] [PubMed] [Google Scholar]
- Boudreau MD, Imam MS, Paredes AM, Bryant MS, Cunningham CK, Felton RP, Jones MY, et al. Differential Effects of Silver Nanoparticles and Silver Ions on Tissue Accumulation, Distribution, and Toxicity in the Sprague Dawley Rat Following Daily Oral Gavage Administration for 13 Weeks. Toxicological Sciences: An Official Journal of the Society of Toxicology. 2016;150:131–160. doi: 10.1093/toxsci/kfv318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravard A, Vacher M, Moritz E, Vaslin L, Hall J, Epe B, Radicella JP. Oxidation Status of Human OGG1-S326C Polymorphic Variant Determines Cellular DNA Repair Capacity. Cancer Research. 2009;69:3642–3649. doi: 10.1158/0008-5472.CAN-08-3943. [DOI] [PubMed] [Google Scholar]
- Butler KS, Peeler DJ, Casey BJ, Dair BJ, Elespuru RK. Silver Nanoparticles: Correlating Nanoparticle Size and Cellular Uptake with Genotoxicity. Mutagenesis. 2015;30:577–591. doi: 10.1093/mutage/gev020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee N, Eom HJ, Choi J. Effects of Silver Nanoparticles on Oxidative DNA Damage-Repair as a Function of p38 MAPK Status: A Comparative Approach Using Human Jurkat T Cells and the Nematode Caenorhabditis elegans. Environmental and Molecular Mutagenesis. 2014;55:122–133. doi: 10.1002/em.21844. [DOI] [PubMed] [Google Scholar]
- Collins AR, Cadet J, Moller L, Poulsen HE, Vina J. Are We Sure We Know How to Measure 8-oxo-7,8-Dihydroguanine in DNA from Human Cells? Archives of Biochemistry and Biophysics. 2004;423:57–65. doi: 10.1016/j.abb.2003.12.022. [DOI] [PubMed] [Google Scholar]
- Dobrzynska MM, Gajowik A, Radzikowska J, Lankoff A, Dusinska M, Kruszewski M. Genotoxicity of Silver and Titanium Dioxide Nanoparticles in Bone Marrow Cells of Rats In Vivo. Toxicology. 2014;315:86–91. doi: 10.1016/j.tox.2013.11.012. [DOI] [PubMed] [Google Scholar]
- Duran N, Duran M, de Jesus MB, Seabra AB, Favaro WJ, Nakazato G. Silver Nanoparticles: A New View on Mechanistic Aspects on Antimicrobial Activity. Nanomedicine: Nanotechnology, Biology, and Medicine. 2016;12:789–799. doi: 10.1016/j.nano.2015.11.016. [DOI] [PubMed] [Google Scholar]
- Ebert F, Weiss A, Bultemeyer M, Hamann I, Hartwig A, Schwerdtle T. Arsenicals Affect Base Excision Repair by Several Mechanisms. Mutation Research. 2011;715:32–41. doi: 10.1016/j.mrfmmm.2011.07.004. [DOI] [PubMed] [Google Scholar]
- Fondevila M, Herrer R, Casallas MC, Abecia L, Ducha JJ. Silver Nanoparticles as a Potential Antimicrobial Additive for Weaned Pigs. Animal Feed Science and Technology. 2009;150:259–269. [Google Scholar]
- Garcia T, Lafuente D, Blanco J, Sanchez DJ, Sirvent JJ, Domingo JL, Gómez M. Oral Subchronic Exposure to Silver Nanoparticles in Rats. Food and Chemical Toxicology : An International Journal Published for the British Industrial Biological Research Association. 2016;92:177–187. doi: 10.1016/j.fct.2016.04.010. [DOI] [PubMed] [Google Scholar]
- Ge L, Li Q, Wang M, Ouyang J, Li X, Xing MM. Nanosilver Particles in Medical Applications: Synthesis, Performance, and Toxicity. International Journal of Nanomedicine. 2014;9:2399–2407. doi: 10.2147/IJN.S55015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode EL, Ulrich CM, Potter JD. Polymorphisms in DNA Repair Genes and Associations with Cancer Risk. Cancer Epidemiology, Biomarkers &Amp; Prevention : a Publication of the American Association for Cancer Research, Cosponsored by the American Society of Preventive Oncology. 2002;11:1513–1530. [PubMed] [Google Scholar]
- Greenberg MM. The Formamidopyrimidines: Purine Lesions Formed in Competition with 8-Oxopurines from Oxidative Stress. Accounts of Chemical Research. 2012;45:588–597. doi: 10.1021/ar2002182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, et al. Patterns of Somatic Mutation in Human Cancer Genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Li Y, Yan J, Ingle T, Jones MY, Mei N, Boudreau MD, et al. Size- and Coating-Dependent Cytotoxicity and Genotoxicity of Silver Nanoparticles Evaluated Using In Vitro Standard Assays. Nanotoxicology. 2016;10:1373–1384. doi: 10.1080/17435390.2016.1214764. [DOI] [PubMed] [Google Scholar]
- Hajipour MJ, Fromm KM, Ashkarran AA, Jimenez de Aberasturi D, de Larramendi IR, Rojo T, Serpooshan V, et al. Antibacterial Properties of Nanoparticles. Trends in Biotechnology. 2012;30:499–511. doi: 10.1016/j.tibtech.2012.06.004. [DOI] [PubMed] [Google Scholar]
- Hayashi M, MacGregor JT, Gatehouse DG, Adler ID, Blakey DH, Dertinger SD, Krishna G, et al. In Vivo Rodent Erythrocyte Micronucleus Assay. II. Some Aspects of Protocol Design Including Repeated Treatments, Integration with Toxicity Testing, and Automated Scoring. Environmental and Molecular Mutagenesis. 2000;35:234–252. [PubMed] [Google Scholar]
- Hazra TK, Das A, Das S, Choudhury S, Kow YW, Roy R. Oxidative DNA Damage Repair in Mammalian Cells: A New Perspective. DNA Repair (Amst) 2007;6:470–480. doi: 10.1016/j.dnarep.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. Repair System of 7, 8-Dihydro-8-Oxoguanine as a Defense Line Against Carcinogenesis. Journal of Radiation Research. 2008;49:329–340. doi: 10.1269/jrr.08049. [DOI] [PubMed] [Google Scholar]
- Hodges NJ, Chipman JK. Down-Regulation of the DNA-Repair Endonuclease 8-Oxo-Guanine DNA Glycosylase 1 (hOGG1) by Sodium Dichromate in Cultured Human A549 Lung Carcinoma Cells. Carcinogenesis. 2002;23:55–60. doi: 10.1093/carcin/23.1.55. [DOI] [PubMed] [Google Scholar]
- Huynh KA, Chen KL. Aggregation Kinetics of Citrate and Polyvinylpyrrolidone Coated Silver Nanoparticles in Monovalent and Divalent Electrolyte Solutions. Environmental Science and Technology. 2011;45:5564–5571. doi: 10.1021/es200157h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs AL, Schar P. DNA Glycosylases: In DNA Repair and Beyond. Chromosoma. 2012;121:1–20. doi: 10.1007/s00412-011-0347-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaar R, Hoeijmakers JH, van Gent DC. Molecular Mechanisms of DNA Double Strand Break Repair. Trends in Cell Biology. 1998;8:483–489. doi: 10.1016/s0962-8924(98)01383-x. [DOI] [PubMed] [Google Scholar]
- Kershaw RM, Hodges NJ. Repair of Oxidative DNA Damage Is Delayed in the Ser326Cys Polymorphic Variant of the Base Excision Repair Protein OGG1. Mutagenesis. 2012;27:501–510. doi: 10.1093/mutage/ges012. [DOI] [PubMed] [Google Scholar]
- Kim YS, Kim JS, Cho HS, Rha DS, Kim JM, Park JD, Choi BS, et al. Twenty-Eight-Day Oral Toxicity, Genotoxicity, and Gender-Related Tissue Distribution of Silver Nanoparticles in Sprague-Dawley Rats. Inhalation Toxicology. 2008;20:575–583. doi: 10.1080/08958370701874663. [DOI] [PubMed] [Google Scholar]
- Klungland A, Rosewell I, Hollenbach S, Larsen E, Daly G, Epe B, Seeberg E, et al. Accumulation of Premutagenic DNA Lesions in Mice Defective in Removal of Oxidative Base Damage. Proceedings of the National Academy of Sciences of the United States of America USA96. 1999:13300–13305. doi: 10.1073/pnas.96.23.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovvuru P, Mancilla PE, Shirode AB, Murray TM, Begley TJ, Reliene R. Oral Ingestion of Silver Nanoparticles Induces Genomic Instability and DNA Damage in Multiple Tissues. Nanotoxicology. 2015;9:162–171. doi: 10.3109/17435390.2014.902520. [DOI] [PubMed] [Google Scholar]
- Lee AJ, Hodges NJ, Chipman JK. Interindividual Variability in Response to Sodium Dichromate-Induced Oxidative DNA Damage: Role of the Ser326Cys Polymorphism in the DNA-Repair Protein of 8-oxo-7,8-Dihydro-2′-Deoxyguanosine DNA Glycosylase 1. Cancer Epidemiology, Biomarkers and Prevention : A Publication of the American Association for Cancer Research, Cosponsored by the American Society of Preventive Oncology. 2005;14:497–505. doi: 10.1158/1055-9965.EPI-04-0295. [DOI] [PubMed] [Google Scholar]
- Li Y, Bhalli JA, Ding W, Yan J, Pearce MG, Sadiq R, Cunningham CK, et al. Cytotoxicity and Genotoxicity Assessment of Silver Nanoparticles in Mouse. Nanotoxicology. 2014;8(Suppl 1):36–45. doi: 10.3109/17435390.2013.855827. [DOI] [PubMed] [Google Scholar]
- Liu W, Wu Y, Wang C, Li HC, Wang T, Liao CY, Cui L, et al. Impact of Silver Nanoparticles on Human Cells: Effect of Particle Size. Nanotoxicology. 2010;4:319–330. doi: 10.3109/17435390.2010.483745. [DOI] [PubMed] [Google Scholar]
- Loeschner K, Hadrup N, Qvortrup K, Larsen A, Gao X, Vogel U, Mortensen A. Distribution of Silver in Rats Following 28 Days of Repeated Oral Exposure to Silver Nanoparticles or Silver Acetate. Particle and Fibre Toxicology. 2011;8:1–14. doi: 10.1186/1743-8977-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard JY, Hartemann P. Silver as an Antimicrobial: Facts and Gaps in Knowledge. Critical Reviews in Microbiology. 2013;39:373–383. doi: 10.3109/1040841X.2012.713323. [DOI] [PubMed] [Google Scholar]
- Miethling-Graff R, Rumpker R, Richter M, Verano-Braga T, Kjeldsen F, Brewer J, Hoyland J, et al. Exposure to Silver Nanoparticles Induces Size- and Dose-Dependent Oxidative Stress and Cytotoxicity in Human Colon Carcinoma Cells. Toxicology in Vitro : An International Journal Published in Association with Bibra. 2014;28:1280–1289. doi: 10.1016/j.tiv.2014.06.005. [DOI] [PubMed] [Google Scholar]
- Morland I, Rolseth V, Luna L, Rognes T, Bjoras M, Seeberg E. Human DNA Glycosylases of the Bacterial Fpg/MutM Superfamily: An Alternative Pathway for the Repair of 8-Oxoguanine and Other Oxidation Products in DNA. Nucleic Acids Research. 2002;30:4926–4936. doi: 10.1093/nar/gkf618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger MA, Radwanski P, Hadlock GC, Stoddard G, Shaaban A, Falconer J, Grainger DW, et al. In Vivo Human Time-Exposure Study of Orally Dosed Commercial Silver Nanoparticles. Nanomedicine: Nanotechnology, Biology, and Medicine. 2014;10:1–9. doi: 10.1016/j.nano.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munusamy P, Wang C, Engelhard MH, Baer DR, Smith JN, Liu C, Kodali V, et al. Comparison of 20 nm Silver Nanoparticles Synthesized With and Without a Gold Core: Structure, Dissolution in Cell Culture Media, and Biological Impact on Macrophages. Biointerphases. 2015;10:1–16. doi: 10.1116/1.4926547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallanthighal S, Chan C, Bharali DJ, Mousa SA, Vasquez E, Reliene R. Particle Coatings but not Silver Ions Mediate Genotoxicity of Ingested Silver Nanoparticles in a Mouse Model. NanoImpact. 2017;5:92–100. doi: 10.1016/j.impact.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NanoComposix. 2017 http://nanocomposix.com/collections/silver.
- Obe G, Pfeiffer P, Savage JR, Johannes C, Goedecke W, Jeppesen P, Natarajan AT, et al. Chromosomal Aberrations: Formation, Identification and Distribution. Mutation Research. 2002;504:17–36. doi: 10.1016/s0027-5107(02)00076-3. [DOI] [PubMed] [Google Scholar]
- Park EJ, Bae E, Yi J, Kim Y, Choi K, Lee SH, Park K. Repeated-Dose Toxicity and Inflammatory Responses in Mice by Oral Administration of Silver Nanoparticles. Environmental Toxicology and Pharmacology. 2010;30:162–168. doi: 10.1016/j.etap.2010.05.004. [DOI] [PubMed] [Google Scholar]
- Patlolla AK, Hackett D, Tchounwou PB. Genotoxicity Study of Silver Nanoparticles in Bone Marrow Cells of Sprague-Dawley Rats. Food and Chemical Toxicology : An International Journal Published for the British Industrial Biological Research Association. 2015a;85:52–60. doi: 10.1016/j.fct.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patlolla AK, Hackett D, Tchounwou PB. Silver Nanoparticle-Induced Oxidative Stress-Dependent Toxicity in Sprague-Dawley Rats. Molecular and Cellular Biochemistry. 2015b;399:257–268. doi: 10.1007/s11010-014-2252-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PEN CPI. Consumer Products Inventory. [Accessed 10 August 2017];The Project on Emerging Nanotechnologies (PEN) 2017 http://www.nanotechproject.org/inventories/consumer/
- Piao MJ, Kang KA, Lee IK, Kim HS, Kim S, Choi JY, Choi J, et al. Silver Nanoparticles Induce Oxidative Cell Damage in Human Liver Cells Through Inhibition of Reduced Glutathione and Induction of Mitochondria-Involved Apoptosis. Toxicology Letters. 2011a;201:92–100. doi: 10.1016/j.toxlet.2010.12.010. [DOI] [PubMed] [Google Scholar]
- Piao MJ, Kim KC, Choi JY, Choi J, Hyun JW. Silver Nanoparticles Down-Regulate Nrf2-Mediated 8-Oxoguanine DNA Glycosylase 1 Through Inactivation of Extracellular Regulated Kinase and Protein Kinase B in Human Chang Liver Cells. Toxicology Letters. 2011b;207:143–148. doi: 10.1016/j.toxlet.2011.09.002. [DOI] [PubMed] [Google Scholar]
- Pietro PD, Strano G, Zuccarello L, Satriano C. Gold and Silver Nanoparticles for Applications in Theranostics. Current Topics in Medicinal Chemistry. 2016;16:3069–3102. doi: 10.2174/1568026616666160715163346. [DOI] [PubMed] [Google Scholar]
- Potts RJ, Watkin RD, Hart BA. Cadmium Exposure Down-Regulates 8-Oxoguanine DNA Glycosylase Expression in Rat Lung and Alveolar Epithelial Cells. Toxicology. 2003;184:189–202. doi: 10.1016/s0300-483x(02)00579-6. [DOI] [PubMed] [Google Scholar]
- Prasad RY, McGee JK, Killius MG, Suarez DA, Blackman CF, DeMarini DM, Simmons SO. Investigating Oxidative Stress and Inflammatory Responses Elicited by Silver Nanoparticles Using High-Throughput Reporter Genes in HepG2 Cells: Effect of Size, Surface Coating, and Intracellular Uptake. Toxicology in Vitro: An International Journal Published in Association with Bibra. 2013;27:2013–2021. doi: 10.1016/j.tiv.2013.07.005. [DOI] [PubMed] [Google Scholar]
- Quadros ME, Pierson R, Tulve NS, Willis R, Rogers K, Thomas TA, Marr LC. Release of Silver from Nanotechnology-Based Consumer Products for Children. Environmental Science & Technology. 2013;47:8894–8901. doi: 10.1021/es4015844. [DOI] [PubMed] [Google Scholar]
- Reagan-Shaw S, Nihal M, Ahmad N. Dose Translation from Animal to Human Studies Revisited. FASEB Journal : Official Publication of the Federation of American Societies for Experimental Biology. 2008;22:659–661. doi: 10.1096/fj.07-9574LSF. [DOI] [PubMed] [Google Scholar]
- Reliene R, Bishop AJ, Aubrecht J, Schiestl RH. In Vivo DNA Deletion Assay to Detect Environmental and Genetic Predisposition to Cancer. Methods in Molecular Biology. 2004a;262:125–139. doi: 10.1385/1-59259-761-0:125. [DOI] [PubMed] [Google Scholar]
- Reliene R, Bishop AJ, Li G, Schiestl RH. Ku86 Deficiency Leads to Reduced Intrachromosomal Homologous Recombination In Vivo in Mice. DNA Repair (Amst) 2004b;3:103–111. doi: 10.1016/j.dnarep.2003.09.010. [DOI] [PubMed] [Google Scholar]
- Reliene R, Fischer E, Schiestl RH. Effect of N-Acetyl Cysteine on Oxidative DNA Damage and the Frequency of DNA Deletions in atm-Deficient Mice. Cancer Research. 2004c;64:5148–5153. doi: 10.1158/0008-5472.CAN-04-0442. [DOI] [PubMed] [Google Scholar]
- Reliene R, Yamamoto ML, Rao PN, Schiestl RH. Genomic Instability in Mice is Greater in Fanconi Anemia Caused by Deficiency of Fancd2 than Fancg. Cancer Research. 2010;70:9703–9710. doi: 10.1158/0008-5472.CAN-09-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Boon C, Redon C, Bonner WM. Megabase Chromatin Domains Involved in DNA Double-Strand Breaks In Vivo. The Journal of Cell Biology. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath H. Oxidative DNA Damage in Disease-Insights Gained from Base Excision Repair Glycosylase-Deficient Mouse Models. Environmental and Molecular Mutagenesis. 2014;55:689–703. doi: 10.1002/em.21886. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S. Fiji: An Open-Source Platform for Biological-Image Analysis. Nature Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart DJ, Chipman JK, Hodges NJ. Activity of OGG1 Variants in the Repair of Pro-Oxidant-Induced 8-oxo-2'-Deoxyguanosine. DNA Repair (Amst) 2006;5:1337–1345. doi: 10.1016/j.dnarep.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Song MF, Li YS, Kasai H, Kawai K. Metal Nanoparticle-Induced Micronuclei and Oxidative DNA Damage in Mice. Journal of Clinical Biochemistry and Nutrition. 2012;50:211–216. doi: 10.3164/jcbn.11-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soultanakis RP, Melamede RJ, Bespalov IA, Wallace SS, Beckman KB, Ames BN, Taatjes DJ. Fluorescence Detection of 8-Oxoguanine in Nuclear and Mitochondrial DNA of Cultured Cells Using a Recombinant Fab and Confocal Scanning Laser Microscopy. Free Radical Biology and Medicine. 2000;28:987–998. doi: 10.1016/s0891-5849(00)00185-4. [DOI] [PubMed] [Google Scholar]
- Tolaymat TM, El Badawy AM, Genaidy A, Scheckel KG, Luxton TP, Suidan M. An Evidence-Based Environmental Perspective of Manufactured Silver Nanoparticle in Syntheses and Applications: A Systematic Review and Critical Appraisal of Peer-Reviewed Scientific Papers. The Science of the Total Environment. 2010;408:999–1006. doi: 10.1016/j.scitotenv.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Tulve NS, Stefaniak AB, Vance ME, Rogers K, Mwilu S, LeBouf RF, Schwegler-Berry D, et al. Characterization of Silver Nanoparticles in Selected Consumer Products and Its Relevance for Predicting Children's Potential Exposures. International Journal of Hygiene and Environmental Health. 2015;218:345–357. doi: 10.1016/j.ijheh.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Zande M, Vandebriel RJ, Van Doren E, Kramer E, Herrera Rivera Z, Serrano-Rojero CS, Gremmer ER, et al. Distribution, Elimination, and Toxicity of Silver Nanoparticles and Silver Ions in Rats After 28-Day Oral Exposure. ACS Nano. 2012;6:7427–7442. doi: 10.1021/nn302649p. [DOI] [PubMed] [Google Scholar]
- Vecchio G, Fenech M, Pompa PP, Voelcker NH. Lab-on-a-Chip-Based High-Throughput Screening of the Genotoxicity of Engineered Nanomaterials. Small (Weinheim an Der Bergstrasse, Germany) 2014;10:2721–2734. doi: 10.1002/smll.201303359. [DOI] [PubMed] [Google Scholar]
- Vik ES, Alseth I, Forsbring M, Helle IH, Morland I, Luna L, Bjørås M, et al. Biochemical Mapping of Human NEIL1 DNA Glycosylase and AP Lyase Activities. DNA Repair (Amst) 2012;11:766–773. doi: 10.1016/j.dnarep.2012.07.002. [DOI] [PubMed] [Google Scholar]
- Wang X, Ji Z, Chang CH, Zhang H, Wang M, Liao YP, Lin S, et al. Use of Coated Silver Nanoparticles to Understand the Relationship of Particle Dissolution and Bioavailability to Cell and Lung Toxicological Potential. Small (Weinheim an Der Bergstrasse, Germany) 2014;10:385–398. doi: 10.1002/smll.201301597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YC, Engelhard MH, Baer DR, Castner DG. Quantifying the Impact of Nanoparticle Coatings and Nonuniformities on XPS Analysis: Gold/Silver Core-Shell Nanoparticles. Analytical Chemistry. 2016;88:3917–3925. doi: 10.1021/acs.analchem.6b00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss JM, Goode EL, Ladiges WC, Ulrich CM. Polymorphic Variation in hOGG1 and Risk of Cancer: A Review of the Functional and Epidemiologic Literature. Molecular Carcinogenesis. 2005;42:127–141. doi: 10.1002/mc.20067. [DOI] [PubMed] [Google Scholar]
- Yamamoto ML, Chapman AM, Schiestl RH. Effects of Side-Stream Tobacco Smoke and Smoke Extract on Glutathione- and Oxidative DNA Damage Repair-Deficient Mice and Blood Cells. Mutation Research. 2013;749:58–65. doi: 10.1016/j.mrfmmm.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane A, Kohno T, Ito K, Sunaga N, Aoki K, Yoshimura K, Murakami H, et al. Differential Ability of Polymorphic OGG1 Proteins to Suppress Mutagenesis Induced by 8-Hydroxyguanine in Human Cell In Vivo. Carcinogenesis. 2004;25:1689–1694. doi: 10.1093/carcin/bgh166. [DOI] [PubMed] [Google Scholar]