Abstract
The proteoglycan versican (VCAN) promotes tumor progression and enhances metastasis in several cancers; however, its role in clear cell renal cell carcinoma (ccRCC) remains unknown. Recent evidence suggests that VCAN is an important target of chromosomal 5q gain, one of the most prevalent genetic abnormalities in ccRCC. Thus, we investigated whether VCAN expression is associated with the pathogenesis of ccRCC. VCAN expression was analyzed using three RCC and normal kidney cell lines as well as a clinical cohort of 84 matched ccRCC and normal renal tissues. Functional analyses on growth and progression properties were performed using VCAN depleted ccRCC cells. Microarray expression profiling was employed to investigate the target genes and biological pathways involved in VCAN-mediated ccRCC carcinogenesis. ccRCC had elevated VCAN expression in comparison with normal kidney in both cell lines and clinical specimens. The elevated expression of VCAN was significantly correlated with metastasis (P<0.001) and worse 5-year overall survival after radical nephrectomy (P=0.014). In vitro, VCAN knockdown significantly decreased cell proliferation and increased apoptosis in Caki-2 and 786-O cells, and this was associated with alteration of several TNF signaling-related genes such as TNF-α, BID, and BAK. Furthermore, VCAN depletion markedly decreased cell migration and invasion which correlated with reduction of MMP7 and CXCR4. These results demonstrate that VCAN promotes ccRCC tumorigenesis and metastasis and thus, is an attractive target for novel diagnostic, prognostic and therapeutic strategies.
Keywords: versican, renal cell carcinoma, tumorigenesis, metastasis
Introduction
Renal cell carcinoma (RCC) is the ninth most common cancer in the world of which about 70% is clear cell RCC (ccRCC) with a high rate of invasiveness and metastasis (1). Although recent advances in imaging technology and increased use of screening modalities may contribute to the detection of ccRCC at an early stage, 30% of patients with this disease present with metastasis at the time of diagnosis (2). Metastasis is the major cause of cancer death and metastatic ccRCC patients have a 5-year survival rate of less than 10% (3, 4). The discovery that somatic mutations of tumor suppressor von-Hippel Lindau (VHL) gene, located at 3p25.3, occur in about 90% of sporadic ccRCCs (5, 6) led to the development of new therapies targeting the vascular endothelial growth factor (VEGF) and mammalian target of rapamycin (mTOR) genes against advanced ccRCC. These novel drugs targeting VEGF and mTOR have improved ccRCC patient survival to some extent, however all tumors ultimately develop resistance to these therapies and progress (7, 8). Therefore new molecular targets are needed that regulate invasion and/or metastasis and act as reliable prognostic biomarkers to develop better therapeutic and diagnostic interventions.
Cytogenetic studies have revealed that sporadic ccRCC has characteristic chromosomal losses and gains such as loss of 3p (60%) and gain of 5q (33%) that are the most prevalent genetic abnormalities for this cancer (9, 10). The VHL gene which inhibits the hypoxia-inducible factors HIF1α and HIF2α, is well known to be a major target of 3p loss (10). Although specific targets on 5q have not been well elucidated, a recent study has shown that the versican (VCAN) gene, located at 5q14.3, may be an important target of 5q gain in sporadic ccRCC (11).
The stroma around a solid tumor consists of specific extracellular matrix (ECM) components and plays important roles for activating the microenvironment of primary and secondary tumor sites (12). VCAN, a large aggregating chondroitin sulfate proteoglycan, is an important ECM component and associated with tumorigenesis (13). In vitro and in vivo studies have shown that VCAN enhances cancer cell survival, growth, migration, invasion, angiogenesis, drug resistance, and metastasis (14–17). Indeed increased expression of VCAN has been reported in many types of malignancies, i.e. brain tumors, leukemia, breast, prostate, colon, lung, and ovarian cancers, and is associated with poor prognosis (16–21). Recently we also identified that VCAN expression is implicated in the development of docetaxel-resistance in prostate cancer (22). However, the clinical significance and the role of VCAN in ccRCC remain largely unknown.
In this study we hypothesized that overexpression of VCAN is associated with the pathogenesis of ccRCC. We assessed whether VCAN was upregulated in specimens of ccRCC and whether its expression was associated with clinical parameters or prognosis in cases following radical nephrectomy. Also, the biological role of VCAN in tumorigenesis was determined using ccRCC cell lines. Further, microarray analyses were performed to identify key genes involved in the transformation and tumorigenesis of ccRCC due to VCAN.
Materials and Methods
Clinical ccRCC samples
Eighty-four matched sporadic ccRCC and normal renal tissues were obtained from a urology tissue bank at the Shimane University Hospital (Izumo, Japan). Median (range) patient age at surgery was 63 (16–87) years. Of the 84 patients, 28 (33.3%) were women and 35 (41.7%) of the lesions were located on the right side. Twenty-seven (32.1%) patients had tumor grade 1, 48 (57.1%) were grade 2 and 9 (10.7%) were grade 3. Fifty-eight (69.0%) patients were stage I or II, and 26 (31.0%) were stage III or IV. Six (7.1%) patients had lymph node metastasis and 10 (11.9%) had systematic metastasis. Half of each normal renal and ccRCC tissue specimens were fixed in 10% buffered formalin (pH 7.0) and embedded in paraffin wax. Five µm thick sections were subjected to hematoxylin and eosin staining for histologic evaluation. The remaining half of each normal and ccRCC sample was immediately frozen and stored at −80°C for RNA extraction. Written informed consent was obtained from each patient for molecular analysis of the resected specimen and the study (#20160229-2) was approved by the local ethical committee of Shimane University Faculty of Medicine.
Cell lines and reagents
The human renal proximal tubule epithelial cell line HK-2, and renal cancer lines Caki-2, ACHN and 786-O were obtained from the American Type Culture Collection (ATCC, Manassas, VA). Integrity of the cell lines was confirmed by ATCC using DNA profiling. Keratinocyte serum-free medium (K-SFM), bovine pituitary extract (BPE) and recombinant epidermal growth factor (rEGF) were purchased from Invitrogen (Carlsbad, CA). McCoy’s 5A, Eagle’s MEM (EMEM), RPMI 1640, Opti-MEM and penicillin/streptomycin mixture were obtained from the UCSF Cell Culture Facility (San Francisco, CA). Fetal bovine serum (FBS) was a product of Atlanta Biologicals (Lawrenceville, GA). HK-2 cells were cultured in K-SFM with BPE and rEGF, Caki-2 in McCoy’s 5A with 10% FBS, ACHN in EMEM with 10% FBS, and 786-O in RPMI 1640 with 10% FBS. All cultures contained 100 µg/ml of penicillin/streptomycin and were maintained at 37°C in a humidified atmosphere composed of 5% CO2 in air.
cDNA preparation and gene quantification
Total RNA from clinical samples and cell lines were extracted using TRI reagent (Molecular Research Center, Cincinnati, OH) and RNeasy Mini kit (Qiagen, Valencia, CA), respectively. Extracted RNA was reverse-transcribed into cDNA using iScript cDNA Synthesis kit (Bio-Rad). Quantitative real-time PCR analysis was performed with an Applied Biosystems (Foster City, CA) Prism 7500 Fast Sequence Detection System using TaqMan Universal PCR master mix, probes and primers for target genes. The mRNA transcript levels of target genes were determined using the 7500 Fast System SDS software version 1.3.1 (Applied Biosystems). For clinical samples, a standard curve was generated using a serial dilution of external standards. The level of expression was calculated as the ratio of the target gene to that of the reference glyceraldehyde-3-phosphate dehydrogenase (GAPDH). For cell lines, the data were analyzed by the delta-delta Ct method to calculate fold-change.
Western blot analysis
Whole cell extracts were prepared using radioimmunoprecipitation assay buffer (RIPA; Thermo Scientific, Rockford, IL) containing protease inhibitor cocktail (Roche Diagnostics, Basel, Switzerland). Protein quantification was done using a BCA protein assay kit (Pierce). Total cell protein (20 µg) was loaded onto 4–12% bis–tris gels with 3-(N-morpholino) propanesulfonic acid buffer and separated by a NuPAGE electrophoresis system (Invitrogen). Protein was transferred to Invitrogen™ polyvinylidene difluoride membrane and immunoblotting was carried out according to standard protocols. Antibody against VCAN (monoclonal #ab177480, Abcam), TNF-α (tumor necrosis factor) (polyclonal #3707, Cell Signaling Technology), Caspase-8 (monoclonal #9746, Cell Signaling Technology), BID (BH3 interacting-domain death agonist) (polyclonal #2002, Cell Signaling Technology), BCL-2 (B-cell lymphoma-2) (polyclonal #sc-492, Santa Cruz Biotechnology), BAK (BCL2-antagonist killer) (polyclonal #3814, Cell Signaling Technology), Caspase-9 (polyclonal #9502, Cell Signaling Technology), Cleaved Caspase-9 (polyclonal #9501, Cell Signaling Technology), Caspase-3 (polyclonal #9662, Cell Signaling Technology), Cleaved Caspase-3 (monoclonal #9664, Cell Signaling Technology), MMP7 (matrix metalloproteinase 7) (monoclonal #ab5706, Abcam), and CXCR4 (C-X-C chemokine receptor type 4) (monoclonal #60042-1-lg, Proteintech) were utilized to detect gene expression and antibody against GAPDH (monoclonal #sc-32233, Santa Cruz Biotechnology) was used to confirm equal loading. Blots were washed with TBS containing 0.1% Tween20 and labeled with horseradish peroxidase-conjugated, secondary anti-rabbit or -mouse antibody (Cell Signaling Technology). Specific protein complexes were visualized with an enhanced chemiluminescence detection system (GE Healthcare, Little Chalfont, UK) using the Chemidoc imaging system (Bio Rad, CA).
Knockdown of VCAN
For inhibition of VCAN, two siRNA oligonucleotides (siRNA-VCAN SASI_Hs02_00328457 (designated #1) and SASI_Hs02_00328458 (designated #2)) and mismatch siRNA control oligonucleotides were utilized (Sigma-Aldrich). Five µl of either siRNA or control oligonucleotides and 5 µl of lipofectamine RNAiMAX reagent (Invitrogen, Carlsbad, CA) were diluted with 250 µl of Opti-MEM. Cells were then transfected with either siRNA-VCAN#1, #2, or siRNA control treatments.
MTS assay
Cells were plated in triplicate in 96-well microplates at a density of 3×103 cells per well. The number of viable cells was determined by adding CellTiter 96 Aqueous One Solution reagent (Promega, Madison, WI) to each well and measuring the absorbance at 490 nm on SPECTRA MAX 190 plate reader (Molecular Devices, Sunnyvale, CA).
Migration and Invasion assays
Cell migration was evaluated by a wound-healing assay. Cells were plated in six-well dishes and after VCAN siRNA treatment for 48 hours, monolayers were scraped using a P-20 micropipette tip. The width of the initial gap (0 hour) and the residual gap 16 or 24 hours after wounding were calculated using a Nikon Eclipse TS100 microscope (Technical Instruments, Burlingame CA). Cell invasion assay was carried out using modified Boyden chambers consisting of transwell-precoated Matrigel membrane filter inserts with 8 µm pores in 24-well tissue culture plates (BD Biosciences, Bedford, MA). Transfected cells were re-suspended in culture medium without FBS and placed in the upper chamber. After 24 hours incubation at 37°C, cells migrating through the membrane were stained. The results were expressed as invaded cells quantified at OD 560 nm using a SPECTRA MAX 190 plate reader.
Apoptosis assay
Fluorescence-activated cell-sorting (FACS) analysis for apoptosis was done 48 and 72 hours post-transfection using an annexin V-fluorescein isothiocyanate (FITC)/7-amino-actinomycin D (7-AAD) staining system and a BD FACSVerse™ flow cytometer (BD Biosciences, San Jose, CA). Briefly, RCC cells were harvested and resuspended in binding buffer at a concentration of 1 × 106 cells/ml. For each assay, 1 × 105 cells were incubated with 5 µl of annexin V-FITC and 5 µl of 7-AAD in the dark for 15 min at room temperature. After adding 400 µl of binding buffer, samples were analyzed within an hour. Cells were stained with annexin V-FITC only (early apoptotic) or both annexin V-FITC and 7-AAD (apoptotic) which was considered to be the total apoptotic cell fraction.
Array analyses of apoptosis and metastasis-related genes
cDNAs from control and VCAN siRNA#2 treated 786-O cells were evaluated for gene expression using the RT2 Profiler™ PCR Array PAHS-012ZC (Human Apoptosis) and PAHS-028ZC (Human Tumor Metastasis) on the ABI Fast 7500 real-time PCR system with RT2 Real-time SYBR Green PCR master mix. Expression of genes in wells was determined using SDS software as mentioned above.
Immunohistochemical analyses
Of the 84 patients with ccRCC who underwent radical nephrectomy, 56 were available for the evaluation of VCAN by immunostaining. Among normal kidney control tissues, only 44 of 84 were available. Immunostaining of VCAN was performed using the UltraVision Detection System (Thermo Scientific). After 12 hours incubation with rabbit monoclonal antibody for VCAN (#ab177480, Abcam), diaminobenzidine substrate was added as chromogen followed by counterstaining with hematoxylin. For VCAN levels, cytoplasmic expression was analyzed by the intensity of positive cells using Image J software (http://rsb.info.nih.gov/ij) and was ranked on an overall scale from 0 to 3; with 0 indicating the absence of staining; 1, weak staining; 2, moderate staining; and 3, strong staining according to our previous study (23).
Statistical analyses
Values are presented as the mean ± standard error based on results obtained from at least three independent experiments. All data were analyzed using StatView 5 statistical software (SAS Institute, Inc., Cary, NC). The relationship between two variables and the numerical values were analyzed using the nonparametric Mann-Whitney U test or two-tailed unpaired Student’s t-test. Chi-square test was used for analyzing the correlation between clinicopathologic parameters and VCAN expression. Survival curves were conducted using the Kaplan-Meier method and the differences between 2 curves were analyzed by a log-rank test. Univariate and multivariate analyses for 5-year overall survival (OS) were performed using a Cox proportional hazards regression model. A P-value of less than 0.05 was considered to be statistically significant.
Results
VCAN is upregulated in ccRCC tissues and cell lines
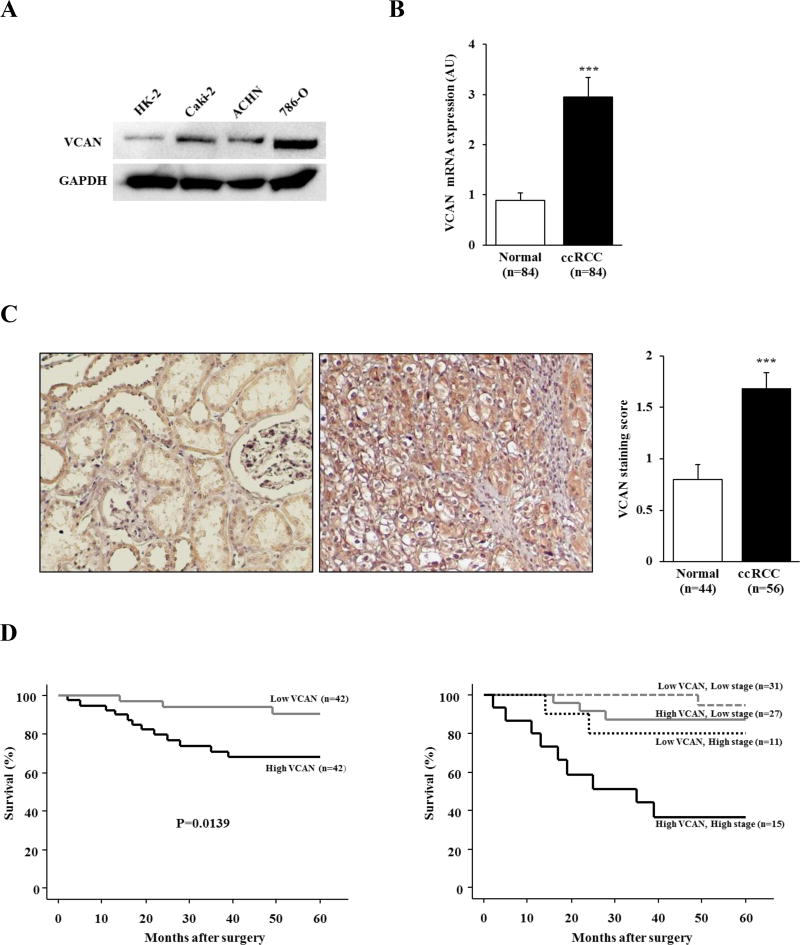
Initially we evaluated the expression level of VCAN using RCC cell lines (Caki-2, ACHN and 786-O) and HK-2 cells by Western blot analysis. As shown in Figure 1A, VCAN protein expression was upregulated in all 3 RCC lines compared with nonmalignant HK-2 cells. Next we investigated the expression of VCAN in 84 ccRCC and matched normal kidney tissues. The level of VCAN mRNA expression was significantly higher in ccRCC (2.94±0.40) than in normal kidney (0.88±0.16) (Figure 1B, P<0.001). In addition, IHC revealed stronger cytoplasmic and/or membrane staining of VCAN that was more common in 56 available ccRCC (1.68±0.16) than in 44 available normal tissues (0.80±0.14) (Figure 1C, P<0.001). There was a significant positive correlation between mRNA transcription and protein level of VCAN in the 56 ccRCC tissues (data not shown). Thus, VCAN is upregulated in both ccRCC tissues and cell lines.
Figure 1. VCAN expression and it’s correlation with prognosis in ccRCC.
(A) Representative immunoblot displaying VCAN expression in HK-2, Caki-2, ACHN and 786-O cells. VCAN protein was upregulated in RCC cell lines in comparison with that of HK-2. GAPDH used as loading control. (B) mRNA expression of VCAN in 84 matched clinical samples. ccRCC samples showed a significantly higher level of VCAN expression in comparison with normal renal tissues. ***, P<0.001. (C) Representative VCAN staining (left) and scoring (right) of available clinical samples. VCAN protein expression in ccRCC samples was significantly higher than that of normal kidney tissues. ***, P<0.001. (D) Kaplan-Meier curves for 5-year overall survival (OS) after radical nephrectomy. Left: Patients with high VCAN expression were significantly associated with lower 5-year OS (P=0.0139). Right: Stratification of VCAN expression with tumor stage identified a stepwise reduction of 5-year OS rate.
VCAN is correlated with poor prognosis in ccRCC patients
To evaluate the association between VCAN expression and clinicopathological findings, we divided the 84 ccRCC patients into 2 groups based on VCAN mRNA levels; namely high (above median level) and low (below median level) VCAN groups that consisted of 42 patients each. As shown in Table 1, higher expression of VCAN mRNA was significantly correlated with systematic metastasis (P=0.007). Also, more male patients had higher expression of VCAN than female patients (P=0.0206). However, no significant association of VCAN expression was found with tumor grade, infiltrative growth pattern, tumor stage, and lymph node involvement.
Table. 1.
Association between VCAN expression and clinicopathological features of patients
| Variables | VCAN expression | P-value | |
|---|---|---|---|
|
|
|||
| Low (n=42) | High (n=42) | ||
| Median age (yrs) | 61.5 | 65.5 | 0.7422 |
| Gender | 0.0206 | ||
| female | 19 (45.2%) | 9 (21.4%) | |
| male | 23 (54.8%) | 33 (78.6%) | |
| Tumor grade | 0.1240 | ||
| less than 2 | 4 (9.6%) | 5 (11.9%) | |
| more than 3 | 38 (90.4%) | 37 (88.1%) | |
| Infiltrative growth pattern (inf) | 0.6563 | ||
| alpha | 22 (52.4%) | 24 (57.1%) | |
| beta or gamma | 19 (45.2%) | 17 (40.5%) | |
| unknown | 1 (2.4%) | 1 (2.4%) | |
| Stage (Robson's classification) | 0.3451 | ||
| I–II | 31 (73.8%) | 27 (64.3%) | |
| III–IV | 11 (26.2%) | 15 (35.7%) | |
| Lymph node involvment | 0.3968 | ||
| present | 2 (4.8%) | 4 (9.6%) | |
| absent | 40 (95.2%) | 38 (90.4%) | |
| Systematic metastasis | 0.0070 | ||
| present | 1 (2.4%) | 9 (22.4%) | |
| absent | 41 (97.6%) | 33 (78.6%) | |
Kaplan-Meier curves for 5-year OS after radical nephrectomy are shown in Figure 1D. Forty-two patients with high VCAN expression among the 84 patients with ccRCC had a significantly worse 5-year OS probability than those with low expression (P=0.0139). The results of univariate and multivariate analysis for 5-year OS are shown in Table 2. Univariate analysis indicated that VCAN expression (P=0.024), tumor stage (P=0.0008), and tumor grade (P=0.0002) were significantly associated with survival. When applied to a multivariate model consisting of these variables, VCAN expression and tumor stage were identified to be significant predictors of 5-year OS in the 84 ccRCC patients (P=0.042 and P=0.0124, respectively). Interestingly we found that VCAN expression in combination with tumor stage could predict patient prognosis more accurately than tumor stage alone, wherein the highest 5-year OS probability was found in ccRCC cases with both low VCAN and low stage, whereas the lowest was in cases having both high VCAN and high stage (Figure 1D). Thus, the combination of tumor stage and VCAN expression level may be an attractive stratification for risk of 5-year OS in ccRCC cases.
Table. 2.
Uni- and multivariate analyses for predicting 5 year overall survival rate of patients
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
|
| ||||
| Variables | Hazard ratio (95% confience interval) |
P-value | Hazard ratio (95% confience interval) |
P-value |
| Age (years) | 1.022 (0.978–1.067) | 0.3391 | — | — |
| Gender (female vs. male) | 0.473 (0.133–1.677) | 0.2463 | — | — |
| VCAN expression (higher vs. lower) | 4.304 (1.211–15.290) | 0.0240 | 3.777 (1.049–13.599) | 0.0420 |
| Tumor stage (I or II vs. III or greater) | 0.141 (0.045–0.444) | 0.0008 | 0.199 (0.056–0.705) | 0.0124 |
| Tumor grade (1 or 2 vs. 3) | 0.123 (0.041–0.372) | 0.0002 | 0.380 (0.110–1.311) | 0.1256 |
VCAN knockdown inhibits cell viability and induces apoptosis in ccRCC cells
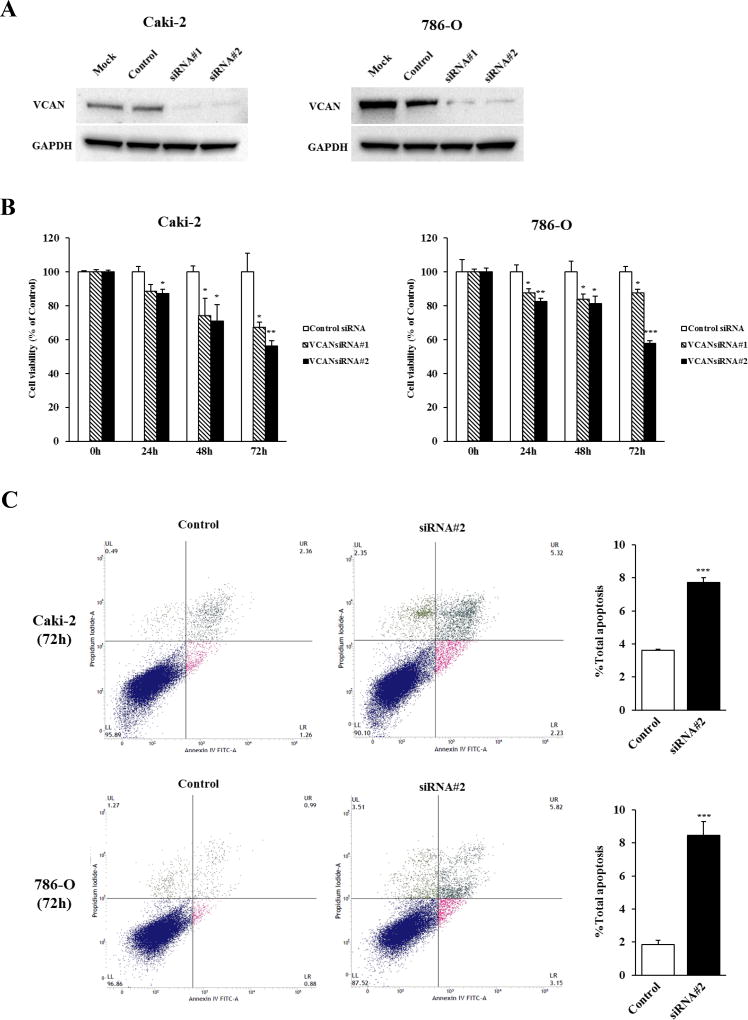
The significant correlation between higher expression levels of VCAN and poor prognosis in clinical samples led us to determine the functional significance of VCAN in ccRCC. We thus examined whether VCAN knockdown affects the growth of ccRCC cells using two different VCAN siRNAs. Caki-2 and 786-O that are both ccRCC cells and that show the highest levels of VCAN (Figure 1A) were selected. As shown in Figure 2A, significant reduction of VCAN protein was observed in both cell lines after transfection with VCAN siRNAs. MTS assay showed that knockdown of VCAN caused significant inhibition of cell proliferation in both cell lines in a time-dependent manner as compared with controls (Figure 2B).
Figure 2. Effect of VCAN knockdown on cell proliferation and apoptosis in ccRCC cells.
(A) Knockdown of VCAN in Caki-2 and 786-O cells was determined by immunoblot analysis at 72 hours after transfection with two different VCAN siRNAs (#1 and #2). GAPDH used as loading control. (B) Cell viability was analyzed by the MTS cell proliferation assay at 0, 24, 48 and 72 hours after treatment with siRNAs. The attenuation of VCAN significantly inhibited cell viability in a time-dependent manner in both cell lines. *, P<0.05. **, P<0.01. ***, P<0.001. (C) Apoptosis assays with Caki-2 and 786-O cells were done 72 hours post siRNA#2 transfection by flow cytometric analyses. Left: Representative biparametric histograms showing cell population in early apoptotic (bottom right quadrant), apoptotic (top right quadrant), and viable (bottom left quadrant) states for each treatment. Right: Bar chart indicates the total apoptotic cell fraction percent (early plus apoptotic) in VCAN siRNA#2 transfectants compared with controls. ***, P<0.001.
Apoptosis was then measured using VCAN siRNA#2-treated cells. As shown in Figure 2C, the early apoptotic and apoptotic fractions (lower right and upper right quadrants of biparametric histograms, respectively) were significantly greater in VCAN-depleted Caki-2 (2.35% + 5.36%) and 786-O (3.26% + 5.54%) cells compared to control Caki-2 (1.27% + 2.36%) and 786-O (0.94% + 0.90%) cells after 72 hours of treatment (total apoptosis, P<0.001 for both cell lines). These results show an oncogenic effect for VCAN in ccRCC cells.
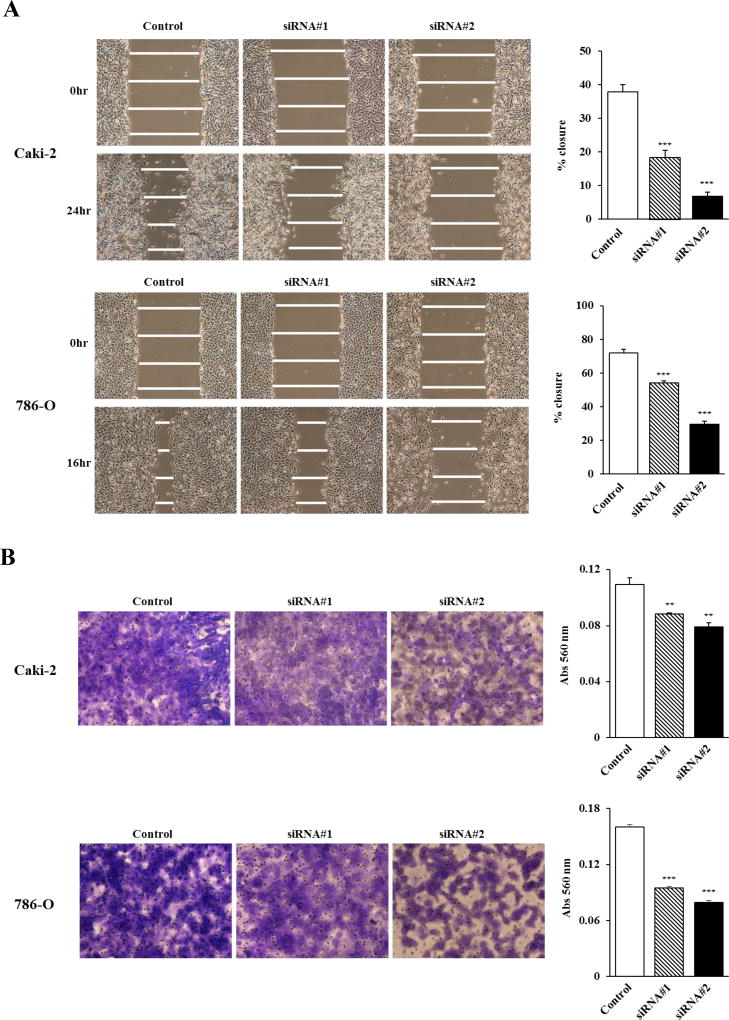
VCAN knockdown inhibits ccRCC cell migration and invasion
Higher expression of VCAN was significantly associated with metastasis in clinical samples as demonstrated in Table 1. We thus examined whether reducing VCAN expression can affect cell migration and invasion properties of ccRCC cell lines. Wound healing was significantly inhibited by both VCAN siRNA transfectants in both Caki-2 and 786-O cells compared with control siRNA (Figure 3A). In addition, matrigel invasion assay demonstrated that the number of invading cells was significantly decreased in VCAN siRNA transfectants compared with their control counterparts (Figure 3B). Thus VCAN plays an important role in ccRCC cell migration and invasion.
Figure 3. Effect of VCAN knockdown on cell migration and invasion in ccRCC cells.
(A) After transfection of cells with siRNA’s for 48 hours, a wound was formed by scraping with a micropipette tip and closure measured after 16 (786-O) or 24 (Caki-2) hours by microscopy. Left: Representative images of wound healing assay. Right: Attenuation of VCAN significantly inhibited cell migration of both ccRCC cell lines. ***, P<0.001. (B) Invasive ability of cells was measured using Matrigel membrane. VCAN knockdown significantly decreased invasiveness of both ccRCC cell lines after 24 hours. Left: Representative images of invasion assay. Right: Invading cells were measured by absorbance at 560 nm. **, P<0.01. ***, P<0.001.
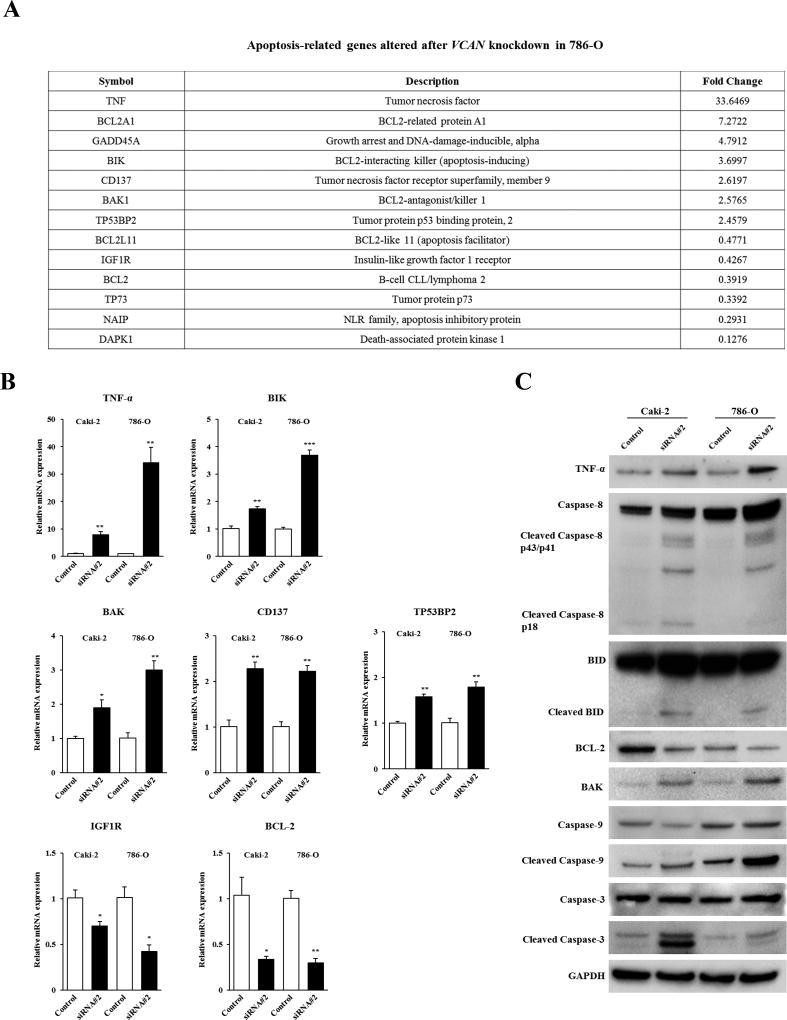
VCAN knockdown induces TNF signaling-mediated apoptosis in ccRCC cells
To further understand the precise mechanism of the pro-apoptotic effect on ccRCC cells induced by VCAN knockdown, we examined gene targets in VCAN siRNA#2-treated 786-O cells using an apoptosis-related gene array. As shown in Figure 4A, 7 genes were upregulated and 6 genes downregulated two-fold or greater after VCAN depletion. Among these altered genes, TNF-α, BIK, BAK, CD137, and TP53BP2 were confirmed to be significantly increased whereas IGF1R and BCL-2 were decreased due to siRNA#2 in both cell lines by real-time PCR (Figure 4B). We also examined protein expression of these genes to validate the association with their mRNA expression levels by Western blot analysis. A consistent, robust increase was observed for TNF-α and BAK and a decrease of BCL-2 protein in both VCAN siRNA#2-treated cell lines (Figure 4C). These results led us to explore other apoptotic genes involved in the TNF signaling pathway. As shown in Figure 4C, VCAN knockdown also caused an increase in cleaved Caspase-8, 9, 3, and cleaved BID in both Caki-2 and 786-O cells. Our data thus indicates that attenuation of VCAN expression inhibits cell viability via induction of TNF signaling-mediated apoptosis in ccRCC.
Figure 4. Apoptosis-related genes altered by VCAN knockdown in ccRCC cells.
(A) Thirteen candidate apoptosis-related genes that were increased or decreased two-fold or greater after VCAN siRNA#2 treatment for 72 hours were identified by gene array analysis in 786-O cells. (B) Verification of gene array data by real-time PCR in Caki-2 and 786-O cells. Five upregulated genes (TNF-α, BIK, BAK, CD137, and TP53BP2) and 2 downregulated genes (IGF1R and BCL-2) were confirmed by real-time PCR using Taqman probes. *, P<0.05. **, P<0.01. ***, P<0.001. (C) Immunoblot analysis for apoptotic markers described in (B) and other TNF signaling pathway-related caspase genes in control and VCAN siRNA#2-transfected Caki-2 and 786-O cells. Shown are immunoblots with marked increase of decrease due to treatment. GAPDH used as a loading control.
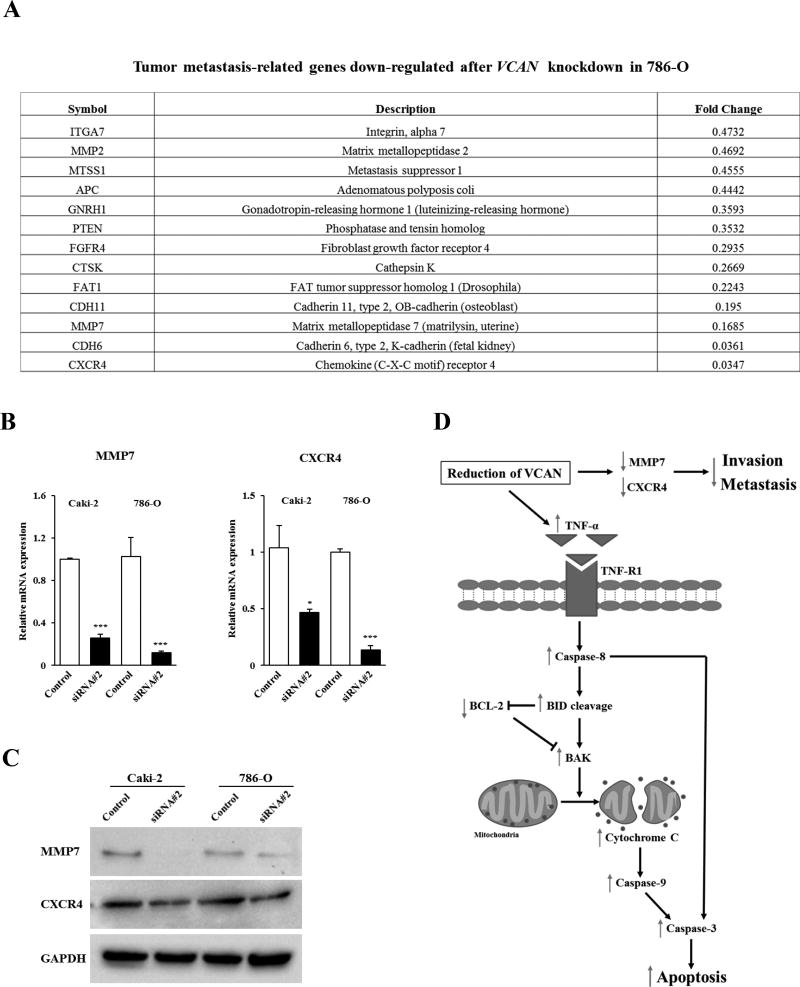
Metastatic Potential of VCAN in ccRCC is associated with MMP7 and CXCR4
We also determined which genes related to metastasis were affected by VCAN in ccRCC using 786-O cells. Figure 5A shows genes that were diminished two-fold or greater after VCAN knockdown using a tumor metastasis-related gene array. Among 13 downregulated genes, the reduction of MMP7 and CXCR4 were confirmed in both Caki-1 and 786-O cells by real-time PCR for RNA (Figure 5B) and Western analyses for protein (Figure 5C) after siRNA#2 treatment. Thus these data indicate that attenuation of VCAN inhibits ccRCC cell migration and invasion via downregulation of MMP7 and CXCR4. A simplified schematic representation of the effect of VCAN silencing on cell apoptosis, invasion and metastasis in ccRCC is shown in Figure 5D.
Figure 5. Metastasis-related genes altered by VCAN knockdown in ccRCC cells.
(A) Thirteen candidate metastasis-related genes that were decreased two-fold or greater after VCAN siRNA#2 treatment for 72 hours were identified by gene array analysis in 786-O cells. (B) Verification of gene array data by real-time PCR in Caki-2 and 786-O cells. Among genes, MMP7 and CXCR4 were confirmed to be downregulated in both cells. *, P<0.05. ***, P<0.001. (C) Immunoblot analysis of MMP7 and CXCR4 in control and VCAN siRNA#2-transfected Caki-2 and 786-O cells. Protein levels were reduced due to treatment. GAPDH used as a loading control. (D) Simplified schematic representation of the proposed effect of VCAN downregulation in ccRCC. Reduction of VCAN results in the induction of TNF-α which can activate initiator Caspase-8, leading to cleavage of BID. Cleaved BID can activate BAK directly and inhibit BCL-2, thereby allowing the release of cytochrome c from mitochondria which in turn activates Caspase-9, Caspase-3, and subsequently promoting programed cell death. In addition, VCAN knockdown results in the inactivation of two metastasis-related genes, MMP7 and CXCR4. In this manner, reduction of VCAN thus results in increased apoptosis and the inhibition of cancer invasion and metastasis.
Discussion
VCAN is a large ECM proteoglycan with an apparent mass of more than 1000 kDa. Several isoforms of VCAN have been identified and each contains the N-terminal (G1) and C-terminal (G3) globular domains, which may play a vital role in tumorigenesis (13, 14, 24). The antibody that we utilized in our study detects all four isoforms as a whole. Prior studies have shown that raised levels of VCAN are strongly associated with poor prognosis of patients in a wide range of malignant tumors (16–21). Therefore we sought to identify the role of VCAN in ccRCC tumorigenesis that has never been studied. We report for the first time that VCAN expression is significantly higher in ccRCC patients with worse 5-year OS probability after radical nephrectomy. In addition, multivariate analysis clearly demonstrated the ability of VCAN levels and tumor stage status to predict 5-year OS probability after surgery and interestingly, interaction between these parameters identified a stepwise reduction of 5-year OS probability. Thus VCAN expression in combination with tumor stage may allow for accurate prediction of cancer progression in ccRCC and may contribute to the development of enhanced diagnostic and prognostic strategies following radical nephrectomy.
Dysregulation of the balance between cellular proliferation and apoptosis is a major factor that leads to tumorigenesis. VCAN has been shown to promote tumor progression and malignant transformation by stimulating cell proliferation and preventing cells from apoptosis in several types of malignancies (25–27). We show here that knocking VCAN expression down strongly inhibited cell proliferation via induction of apoptosis in Caki-2 and 786-O cells. Also, we found that VCAN attenuation affected the expression of several apoptosis-related genes and among them, TNF-α was confirmed to be significantly affected. As a pleiotropic cytokine produced by activated macrophages, natural killer cells, T-lymphocytes, and several types of cancer cells, TNF-α is known to have both antitumor activity and tumor promoting effects via two distinct receptor (TNFR) pathways (28). The actual role of TNF-α in cancer including ccRCC remains controversial (29); however, it is apparent that TNF-α has cytotoxic effects on ccRCC cells (30, 31). The antitumor activity of TNF-α is associated with death receptor-mediated programmed cell death, which is initiated by the binding of TNF-α to TNFR1 (32). As shown in our proposed pathway in Figure 5D, pro-apoptotic BID is activated by initiator Caspase-8 cleavage following the activation of the TNFR1. Cleaved BID has the capacity to either block BCL-2 or activate BAK/BAX that induces the release of cytochrome C from mitochondria (33) which ultimately leads to cell death via downstream caspases such as Caspase-9 and -3. In this study, we clearly demonstrate that protein expression of TNF signaling-associated apoptotic genes were significantly altered after VCAN depletion by Western blot analyses. Our results thus suggest that VCAN is critical for RCC tumorigenesis and by silencing this gene, growth of cancer cells may be abrogated by activation of the TNF signaling pathway.
Generating a microenvironment conducive for cancer cell migration, invasion, and metastasis is one of the critical effects of VCAN aberration (13, 14). Results of our functional studies in cell lines and gene expression levels in clinical tissues show that VCAN can facilitate these activities in ccRCC as well. The mechanism by which VCAN can promote ccRCC progression is not known. Previous studies have shown that together with hyaluronan and CD44, VCAN can form a macromolecular complex in the ECM which is thought to lead to invasion and metastasis through promoting cancer cell motility (34, 35). In our study, we demonstrate that MMP7 which is identified as a target gene of VCAN, can generate VCAN proteolytic fragments (36) and interestingly, these fragments containing the G1 or G3 domains generated by proteases were found to play an important role in cancer progression (14). Although MMPs are predominantly made by stromal cells, MMP7 is synthesized by cancer cells (37) and its overexpression is associated with cancer progression by increasing cancer cell invasiveness and metastatic ability in many types of cancers including ccRCC (38). VCAN can thus promote ccRCC development and metastasis by induction of MMP7 expression, which in turn leads to catabolism of VCAN into protein fragments. In addition to MMP7, chemokines and their respective receptors have recently been found to regulate cancer progression and metastasis. We demonstrate that VCAN can upregulate the α-chemokine receptor, CXCR4. Studies have shown that this receptor is overexpressed in ccRCC and contributes to carcinogenesis and metastasis (39, 40). A meta-analysis review of CXCR4 expression in RCC show high levels predicted poor OS and progression-free survival (41). Other studies demonstrate CXCR4 to be present only in a subpopulation of RCC cells that were more tumorigenic (42) and to enhance cell migration and wound healing of renal cancer cells (43). In vivo, RCC derived spheres developed into tumors and interestingly, expressed high levels of CXCR4 (44). Taken together, we speculate that VCAN can thus promote ccRCC progression and metastases by supporting a suitable ECM environment where MMP7 is activated leading to proteolytic VCAN fragments and CXCR4 induction.
In summary, VCAN expression is upregulated in ccRCC patients and associated with poor prognosis. VCAN induces tumorigenesis in ccRCC cells by reducing TNF signaling-mediated apoptosis. In addition, VCAN promotes ccRCC development and metastasis by inducing MMP7 and CXCR4 expression. Thus, VCAN may be an attractive target for novel diagnostic, prognostic and therapeutic strategies for ccRCC.
Implications.
This study highlights the oncogenic role of VCAN in renal cell carcinogenesis and suggests that this gene has therapeutic and/or biomarker potential for renal cell cancer.
Acknowledgments
This study was supported by the Department of Veterans Affairs (Merit Review) and National Cancer Institute (#R01CA199694) grants. We thank Dr. Roger Erickson for his support and assistance with the preparation of this manuscript.
Footnotes
Conflict of interest: All authors declare no conflict of interest
References
- 1.Jonasch E, Gao J, Rathmell WK. Renal cell carcinoma. BMJ. 2014;349:g4797. doi: 10.1136/bmj.g4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Janzen NK, Kim HL, Figlin RA, Belldegrun AS. Surveillance after radical or partial nephrectomy for localized renal cell carcinoma and management of recurrent disease. Urol Clin North Am. 2003;30:843–52. doi: 10.1016/s0094-0143(03)00056-9. [DOI] [PubMed] [Google Scholar]
- 3.Cohen HT, McGovern FJ. Renal-cell carcinoma. N Engl J Med. 2005;353:2477–90. doi: 10.1056/NEJMra043172. [DOI] [PubMed] [Google Scholar]
- 4.Motzer RJ, Hutson TE, Tomczak P, Michaelson MD, Bukowski RM, Rixe O, et al. Sunitinib versus interferon alfa in metastatic renal-cell carcinoma. N Engl J Med. 2007;356:115–24. doi: 10.1056/NEJMoa065044. [DOI] [PubMed] [Google Scholar]
- 5.Young AC, Craven RA, Cohen D, Taylor C, Booth C, Harnden P, et al. Analysis of VHL Gene Alterations and their Relationship to Clinical Parameters in Sporadic Conventional Renal Cell Carcinoma. Clin Cancer Res. 2009;15:7582–92. doi: 10.1158/1078-0432.CCR-09-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nickerson ML, Jaeger E, Shi Y, Durocher JA, Mahurkar S, Zaridze D, et al. Improved identification of von Hippel-Lindau gene alterations in clear cell renal tumors. Clin Cancer Res. 2008;14:4726–34. doi: 10.1158/1078-0432.CCR-07-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kroeger N, Choueiri TK, Lee JL, Bjarnason GA, Knox JJ, MacKenzie MJ, et al. Survival outcome and treatment response of patients with late relapse from renal cell carcinoma in the era of targeted therapy. Eur Urol. 2014;65:1086–92. doi: 10.1016/j.eururo.2013.07.031. [DOI] [PubMed] [Google Scholar]
- 8.Rini BI, Atkins MB. Resistance to targeted therapy in renal-cell carcinoma. Lancet Oncol. 2009;10:992–1000. doi: 10.1016/S1470-2045(09)70240-2. [DOI] [PubMed] [Google Scholar]
- 9.Klatte T, Rao PN, de Martino M, LaRochelle J, Shuch B, Zomorodian N, et al. Cytogenetic profile predicts prognosis of patients with clear cell renal cell carcinoma. J Clin Oncol. 2009;27:746–53. doi: 10.1200/JCO.2007.15.8345. [DOI] [PubMed] [Google Scholar]
- 10.Pei J, Feder MM, Al-Saleem T, Liu Z, Liu A, Hudes GR, et al. Combined classical cytogenetics and microarray-based genomic copy number analysis reveal frequent 3;5 rearrangements in clear cell renal cell carcinoma. Genes Chromosomes Cancer. 2010;49:610–9. doi: 10.1002/gcc.20771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dondeti VR, Wubbenhorst B, Lal P, Gordan JD, D'Andrea K, Attiyeh EF, et al. Integrative genomic analyses of sporadic clear cell renal cell carcinoma define disease subtypes and potential new therapeutic targets. Cancer Res. 2012;72:112–21. doi: 10.1158/0008-5472.CAN-11-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liotta LA, Kohn EC. The microenvironment of the tumour-host interface. Nature. 2001;411:375–9. doi: 10.1038/35077241. [DOI] [PubMed] [Google Scholar]
- 13.Edwards IJ. Proteoglycans in prostate cancer. Nat Rev Urol. 2012;9:196–206. doi: 10.1038/nrurol.2012.19. [DOI] [PubMed] [Google Scholar]
- 14.Du WW, Yang W, Yee AJ. Roles of versican in cancer biology--tumorigenesis, progression and metastasis. Histol Histopathol. 2013;28:701–13. doi: 10.14670/HH-28.701. [DOI] [PubMed] [Google Scholar]
- 15.Gao R, Cao C, Zhang M, Lopez MC, Yan Y, Chen Z, et al. A unifying gene signature for adenoid cystic cancer identifies parallel MYB-dependent and MYB-independent therapeutic targets. Oncotarget. 2014;5:12528–42. doi: 10.18632/oncotarget.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang W, Yee AJ. Versican V2 isoform enhances angiogenesis by regulating endothelial cell activities and fibronectin expression. FEBS Lett. 2013;587:185–92. doi: 10.1016/j.febslet.2012.11.023. [DOI] [PubMed] [Google Scholar]
- 17.Fujii K, Karpova MB, Asagoe K, Georgiev O, Dummer R, Urosevic-Maiwald M. Versican upregulation in Sezary cells alters growth, motility and resistance to chemotherapy. Leukemia. 2015;29:2024–32. doi: 10.1038/leu.2015.103. [DOI] [PubMed] [Google Scholar]
- 18.Suwiwat S, Ricciardelli C, Tammi R, Tammi M, Auvinen P, Kosma VM, et al. Expression of extracellular matrix components versican, chondroitin sulfate, tenascin, and hyaluronan, and their association with disease outcome in node-negative breast cancer. Clin Cancer Res. 2004;10:2491–8. doi: 10.1158/1078-0432.ccr-03-0146. [DOI] [PubMed] [Google Scholar]
- 19.Ricciardelli C, Mayne K, Sykes PJ, Raymond WA, McCaul K, Marshall VR, et al. Elevated levels of versican but not decorin predict disease progression in early-stage prostate cancer. Clin Cancer Res. 1998;4:963–71. [PubMed] [Google Scholar]
- 20.Suhovskih AV, Aidagulova SV, Kashuba VI, Grigorieva EV. Proteoglycans as potential microenvironmental biomarkers for colon cancer. Cell Tissue Res. 2015;361:833–44. doi: 10.1007/s00441-015-2141-8. [DOI] [PubMed] [Google Scholar]
- 21.Ghosh S, Albitar L, LeBaron R, Welch WR, Samimi G, Birrer MJ, et al. Up-regulation of stromal versican expression in advanced stage serous ovarian cancer. Gynecol Oncol. 2010;119:114–20. doi: 10.1016/j.ygyno.2010.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arichi N, Mitsui Y, Hiraki M, Nakamura S, Hiraoka T, Sumura M, et al. Versican is a potential therapeutic target in docetaxel-resistant prostate cancer. Oncoscience. 2015;2:193–204. doi: 10.18632/oncoscience.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitsui Y, Hirata H, Arichi N, Hiraki M, Yasumoto H, Chang I, et al. Inactivation of bone morphogenetic protein 2 may predict clinical outcome and poor overall survival for renal cell carcinoma through epigenetic pathways. Oncotarget. 2015;6:9577–91. doi: 10.18632/oncotarget.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wight TN, Merrilees MJ. Proteoglycans in atherosclerosis and restenosis: key roles for versican. Circ Res. 2004;94:1158–67. doi: 10.1161/01.RES.0000126921.29919.51. [DOI] [PubMed] [Google Scholar]
- 25.Cattaruzza S, Schiappacassi M, Kimata K, Colombatti A, Perris R. The globular domains of PG-M/versican modulate the proliferation-apoptosis equilibrium and invasive capabilities of tumor cells. FASEB J. 2004;18:779–81. doi: 10.1096/fj.03-0660fje. [DOI] [PubMed] [Google Scholar]
- 26.Du WW, Fang L, Yang W, Sheng W, Zhang Y, Seth A, et al. The role of versican G3 domain in regulating breast cancer cell motility including effects on osteoblast cell growth and differentiation in vitro - evaluation towards understanding breast cancer cell bone metastasis. BMC Cancer. 2012;12:341. doi: 10.1186/1471-2407-12-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fanhchaksai K, Okada F, Nagai N, Pothacharoen P, Kongtawelert P, Hatano S, et al. Host stromal versican is essential for cancer-associated fibroblast function to inhibit cancer growth. Int J Cancer. 2016;138:630–41. doi: 10.1002/ijc.29804. [DOI] [PubMed] [Google Scholar]
- 28.Mocellin S, Rossi CR, Pilati P, Nitti D. Tumor necrosis factor, cancer and anticancer therapy. Cytokine Growth Factor Rev. 2005;16:35–53. doi: 10.1016/j.cytogfr.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 29.Al-Lamki RS, Sadler TJ, Wang J, Reid MJ, Warren AY, Movassagh M, et al. Tumor necrosis factor receptor expression and signaling in renal cell carcinoma. Am J Pathol. 2010;177:943–54. doi: 10.2353/ajpath.2010.091218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caldwell MC, Hough C, Furer S, Linehan WM, Morin PJ, Gorospe M. Serial analysis of gene expression in renal carcinoma cells reveals VHL-dependent sensitivity to TNFalpha cytotoxicity. Oncogene. 2002;21:929–36. doi: 10.1038/sj.onc.1205140. [DOI] [PubMed] [Google Scholar]
- 31.Qi H, Ohh M. The von Hippel-Lindau tumor suppressor protein sensitizes renal cell carcinoma cells to tumor necrosis factor-induced cytotoxicity by suppressing the nuclear factor-kappaB-dependent antiapoptotic pathway. Cancer Res. 2003;63:7076–80. [PubMed] [Google Scholar]
- 32.Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 33.Gross A, McDonnell JM, Korsmeyer SJ. BCL-2 family members and the mitochondria in apoptosis. Genes Dev. 1999;13:1899–911. doi: 10.1101/gad.13.15.1899. [DOI] [PubMed] [Google Scholar]
- 34.Ricciardelli C, Russell DL, Ween MP, Mayne K, Suwiwat S, Byers S, et al. Formation of hyaluronan- and versican-rich pericellular matrix by prostate cancer cells promotes cell motility. J Biol Chem. 2007;282:10814–25. doi: 10.1074/jbc.M606991200. [DOI] [PubMed] [Google Scholar]
- 35.Ween MP, Oehler MK, Ricciardelli C. Role of versican, hyaluronan and CD44 in ovarian cancer metastasis. Int J Mol Sci. 2011;12:1009–29. doi: 10.3390/ijms12021009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Halpert I, Sires UI, Roby JD, Potter-Perigo S, Wight TN, Shapiro SD, et al. Matrilysin is expressed by lipid-laden macrophages at sites of potential rupture in atherosclerotic lesions and localizes to areas of versican deposition, a proteoglycan substrate for the enzyme. Proc Natl Acad Sci U S A. 1996;93:9748–53. doi: 10.1073/pnas.93.18.9748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2:161–74. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 38.Miyata Y, Iwata T, Ohba K, Kanda S, Nishikido M, Kanetake H. Expression of matrix metalloproteinase-7 on cancer cells and tissue endothelial cells in renal cell carcinoma: prognostic implications and clinical significance for invasion and metastasis. Clin Cancer Res. 2006;12:6998–7003. doi: 10.1158/1078-0432.CCR-06-1626. [DOI] [PubMed] [Google Scholar]
- 39.Wang L, Huang T, Chen W, Gao X, Zhou T, Wu Z, et al. Silencing of CXCR4 by RNA interference inhibits cell growth and metastasis in human renal cancer cells. Oncol Rep. 2012;28:2043–8. doi: 10.3892/or.2012.2028. [DOI] [PubMed] [Google Scholar]
- 40.Tang B, Tang F, Li Y, Yuan S, Li B, Wang Z, et al. Clinicopathological significance of CXCR4 expression in renal cell carcinoma: a meta-analysis. Ann Surg Oncol. 2015;22:1026–31. doi: 10.1245/s10434-014-4019-5. [DOI] [PubMed] [Google Scholar]
- 41.Du Y, Long Q, Guan B, Mu L. Prognostic Value of High CXCR4 Expression in Renal Cell Carcinoma: A System Review and Meta-Analysis. Dis Markers. 2015;2015:568980. doi: 10.1155/2015/568980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gassenmaier M, Chen D, Buchner A, Henkel L, Schiemann M, Mack B, et al. CXC chemokine receptor 4 is essential for maintenance of renal cell carcinoma-initiating cells and predicts metastasis. Stem Cells. 2013;31:1467–76. doi: 10.1002/stem.1407. [DOI] [PubMed] [Google Scholar]
- 43.Ierano C, Santagata S, Napolitano M, Guardia F, Grimaldi A, Antignani E, et al. CXCR4 and CXCR7 transduce through mTOR in human renal cancer cells. Cell Death Dis. 2014;5:e1310. doi: 10.1038/cddis.2014.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Micucci C, Matacchione G, Valli D, Orciari S, Catalano A. HIF2alpha is involved in the expansion of CXCR4-positive cancer stem-like cells in renal cell carcinoma. Br J Cancer. 2015;113:1178–85. doi: 10.1038/bjc.2015.338. [DOI] [PMC free article] [PubMed] [Google Scholar]