Abstract
Purpose of review
A successful HIV-1 vaccine will require immunogens that induce protective immune responses. However, recent studies suggest that the response to human immunodeficiency virus-type 1 (HIV-1) and perhaps other viruses may be altered by immune system exposure to intestinal microbiota (IM)-antigens. This review will discuss select aspects of these studies.
Recent findings
Naïve CD4 T and B cell repertoires can be imprinted by IM-antigens to respond to virus epitopes prior to virus infection. A multiclade Env gp145 DNA prime, recombinant adenovirus type 5 boost vaccine tested in a HIV Vaccine Trials Network (HVTN) phase IIb human vaccine efficacy trial (HVTN 505) induced a dominant gp41-reactive antibody response that was non-neutralizing and cross-reactive with IM. This vaccine regimen also induced a dominant gp41-reactive, IM-cross-reactive gp41 antibody response in neonatal and adult Rhesus macaques. Studies of naïve CD4 T cells have demonstrated cross-reactivity to both HIV-1 and influenza peptides.
Summary
HIV-1 Env vaccine-induced CD4 T and B cell responses can originate from a pool of IM-cross-reactive immune cells. Moreover, IM-cross-reactive HIV-1 Env antibodies are ineffective in protection against HIV-1 infection. Thus, IM-imprinting of the B cell repertoire may be one of several roadblocks to the induction of protective HIV-1 antibodies.
Keywords: Microbiome, HIV vaccines, CD4 T cells, B cells, Cross-reactive antibodies
Introduction
Both B cell receptor (BCR) and T cell receptor (TCR) diversity contributes to the development of an effective humoral immune response that can recognize pathogens and environmental antigens (1). Many B cells responding to pathogens are polyreactive, and thus are capable of responding to multiple antigens (2). In this regard, naïve B cell subsets can be stimulated with environmental antigens and become primed for responding to pathogens and vaccine-immunogens with shared properties, including sequence and structural motifs (2-4). The notion that environmental antigens can prime naïve B cell subsets to respond to pathogens is important for understanding CD4 T and B cell responses to infectious diseases. Here we discuss recent findings of IM cross-reactivity with viruses, focusing on HIV-1 Env cross-reactivity with bacterial IM.
Cross-reactive T cell responses in viral infections
For generating a T cell response, T cells must be present in sufficient numbers to recognize a specific antigen, but given the remarkable number of environmental antigens, the TCR repertoire must be able to recognize a vast array of peptides in the context of major histocompatibility complex (MHC) class I or class II (5). The ability of T cells to bind multiple ligands may be conferred by cross-reactivity with environmental antigens of similarity in epitopes recognized by T cells or the flexibility of TCR recognizing different epitopes presented by the same MHC (6, 7). Intestinal microbiome proteins are candidate environmental antigens with sequence and structural similarities to viruses that may be able to stimulate naïve T cells (7).
Antigen-specific CD4 T helper cells, particularly T follicular helper (Tfh) cells, are required for optimal B cell affinity maturation and class switching (8). Two studies have shown the existence of memory CD4 T cells that cross-reacted with HIV-1 and IM peptides in HIV-1 uninfected individuals (9, 10). Using a human leukocyte antigen (HLA)-restricted, peptide MHC tetramer enrichment technique, Su and colleagues characterized the CD4 T cell repertoire in 26 healthy adults and found T cells that reacted with tetramers derived from HIV-1, cytomegalovirus (CMV) or herpes simplex virus (HSV) epitopes (10). Reactive T cells had surface markers and gene expression profiles of memory T cells and showed evidence of clonal expansion. Su et al. demonstrated that one mechanism for accruing virus-specific CD4 T cells was naïve CD4 T cell cross-reactivity with environmental antigens, including microbiota antigens (10). Specifically, peptides derived from intestinal commensal bacteria Ruminococcus flavefaciens, Lachanospiraceae bacterium, and Bifidobacterium bifidum had sequence homology with HIV-1 peptides and were found to cross-react with CD4 T cells (10). Similarly, influenza-reactive T cells from two individuals vaccinated with an influenza vaccine responded to an HA 391-410 peptide sequence and could be activated by peptides from a human skin bacterium Finegoldia magna (10). Campion and colleagues used an HLA-unbiased T cell library technique to characterize the naïve and memory T cell repertoires of seven healthy HIV-1 seronegative individuals and found clonally-expanded naïve and memory CD4 T cells that reacted with HIV-1 peptides (9). The HIV-1 peptides that cross-reacted with naïve and memory CD4 T cells from HIV-1-negative individuals had epitope-length matches with microbial sequences of human microbiome proteins, suggesting that microbial proteins could have been responsible for T cell priming (9). Therefore, using two independent approaches, both Su et al. and Campion et al. found naïve and memory CD4 T cells that cross-reacted with virus and IM-antigens, and provided evidence that virus-reactive T cells may be induced by cross-reactivity with environmental antigens, including commensal microbial antigens. Thus, CD4 T cell anti-viral repertoires may be shaped by microbial antigens and can influence the immune response to HIV-1 vaccines.
Pre-existing naïve CD8 T cells that can recognize viral antigens have also been described. Schmidt and colleagues found naïve CD8 T cells that cross-reacted with Hepatitis C virus (HCV)-epitopes in seven HCV-uninfected individuals (11). Interestingly, the HCV epitope that most commonly reacted with naïve precursor CD8 T cells in the uninfected individuals was the most frequently targeted epitope in an independent cohort of 26 HCV infected individuals, suggesting that immunodominance in HCV infection was linked to precursor frequency of CD8 T cells cross-reactive with HCV-specific epitopes (11).
Cross-reactive B cell responses in viral infections
Humans have a diverse B cell repertoire capable of generating antibodies that can mediate immune effector functions against viruses and virus-infected cells (1). High affinity anti-pathogen antibodies are generally monospecific since host tolerance mechanisms, including clonal deletion, anergy and receptor editing normally restrict the maturation of high affinity autoreactive B cells during B cell development (12, 13). However, ~20% of mature naïve B cells are low affinity self-reactive or polyreactive, and provide breadth of response to the B cell receptor repertoire (13). Interestingly, HIV-1 Env-reactive antibodies, including bnAbs are frequently polyreactive (14, 15), and have been suggested to be derived from a polyreactive pool of B cells such as marginal zone B cells and to be controlled by immune tolerance mechanisms (14, 16-18). Polyreactivity may be beneficial for HIV-1 Env-reactive antibodies, since neutralizing antibody epitopes on glycosylated HIV-1 Envs mutate extensively during viral evolution, are shielded by glycans, and each virion contains ~7-10 functional viral spikes (15, 19). Polyreactivity of HIV-1 Env-reactive antibodies may be conferred by high levels of somatic mutations (15), thus suggesting that polyreactive B cells may be positively selected in response to HIV-1 infection.
In acute HIV-1 infection (AHI), the initial humoral immune response consists of plasma IgM and IgG antibodies that target the gp41 region of HIV-1 envelope (Env) and are non-neutralizing and ineffective at controlling viremia (20). It was interesting to note that in AHI, both IgM and IgG responses arise at the same time, implying a mixture of IgM and IgG BCR-expressing responding B cells in AHI (20). Two subsequent studies addressed the origin of predominant gp41-reactive antibodies during AHI (3, 4). Ninety-one percent (61/67) of the Env-reactive antibodies isolated from plasma cells of the five AHI subjects were gp41-reactive, consistent with immunodominance of gp41-reactive plasma antibodies (3, 20). Moreover, plasma cell-derived gp41-reactive antibodies from AHI were highly mutated and thus led to the hypothesis that they originated from a pool of preexisting mutated B cells that cross-reacted with HIV-1 Env gp41 (3). Liao and colleagues went on to isolate gp41-reactive antibodies from plasma cells in peripheral blood of two uninfected individuals and demonstrated that the gp41-reactive antibodies from uninfected and AHI subjects were indeed cross-reactive with IM-antigens (3). Trama and colleagues studied the plasma cell and memory B cell repertoires of the terminal ileum in early and chronically HIV-1 infected individuals (4). Mutated gp41-reactive antibodies that cross-reacted with IM-antigens were also dominant in terminal ileum of six early-HIV-1-infected and three HIV-1-uninfected individuals (4). Thus, both of these studies suggested that the initial gp41-reactive antibody response observed in AHI were derived from a pool of preexisting IM-cross-reactive B cells (3, 4). That gp41-IM cross-reactive antibodies were class-switched and mutated, supported the hypothesis that these antibodies were derived from a pool of preexisting B cells primed by IM-antigens.
Recent studies have assessed how changes in the microbiome following HIV-1 infection may impact disease progression towards AIDS. In a cohort of HIV-1-infected Ugandan individuals, post-infection fecal virome DNA was characterized using next generation sequencing, and the bacterial microbiome was characterized using 16S rRNA gene amplification (21). Analysis in HIV-1 infected Ugandans revealed alterations of virome and bacterial microbiome that were associated with low peripheral CD4 T cell counts. HIV-1 infected Ugandan subjects with CD4 T cell count <200 had significantly less bacterial phylogenetic diversity compared to infected subjects with CD4 T cell count >200 and HIV-1 uninfected subjects (21). In a cohort of rhesus macaques that received an Ad26 prime, Env protein vaccine prior to simian immunodeficiency virus (SIV) challenge, the overall bacterial communities remained relatively stable after SIV infection independent of vaccine-mediated protection, but the fecal samples of unprotected animals had a higher frequency of gastrointestinal adeno-associated viruses and picornaviruses compared to the fecal samples from vaccine-protected animals (22). Moreover, detection of adenovirus sequences in SIV-infected macaques was associated with macaque death due to AIDS-related complications, suggesting a possible emergence of pathogens during lentivirus infections (22). Thus, these studies demonstrated that gut viruses and bacteria are impacted by lentivirus infection and SIV vaccination may prevent overgrowth of pathogens that promote disease progression (21, 22).
Microbiota regulation of B cell responses has also been reported to impact Ab responses to influenza. Germ-free mice immunized with trivalent inactivated influenza vaccine (TIV) had impaired TIV-specific antibody responses that were restored upon recolonization of germ-free mice with strains of E. coli microbiota antigen, thus demonstrating a role for gut microbiota in response to influenza vaccines (23).
Cross-reactive B cell responses in HIV-1 Env vaccination
A National Institute of Health (NIH) Vaccine Research Center (VRC) DNA prime, recombinant adenovirus type 5 (rAd5) boost vaccine (VRC vaccine) that was studied in an HVTN phase IIb human vaccine efficacy trial (HVTN 505) in adult participants showed futility for protection against HIV-1 acquisition (24). This VRC vaccine was also studied in HVTN phase 1b (HVTN 082) and phase 2a (HVTN 204) human clinical trials (25, 26). The dominant plasma and memory B cell-derived antibodies induced by this VRC vaccine were neither neutralizing nor mediated FcR-dependent anti-HIV-1 activities (24, 26). The dominant blood-derived vaccine-induced antibody responses targeted the gp41 region of Env, gp41-reactive antibodies cross-reacted with IM-antigens, and gp41-reactive antibodies originated from B cells cross-reactive with both IM and HIV-1 Env that were present prior to vaccination (26). This phenomenon of dominant gp41 antibody response to gp41-containing Env vaccine was similarly observed in another human clinical trial HVTN 205 that studied a DNA prime, Modified Vaccina Ankara (MVA) boost with HIV-1 Env gp140 (27).
Infants live in a relatively sterile environment prior to birth (28), but neonate B cell repertoires may be imprinted soon after birth by microbiota antigens (29, 30). Thus, one hypothesis for induction of HIV-1 protective antibodies is to vaccinate infants early in life. The immune cells of neonates and adults have phenotypic and functional differences (31), but infants can respond robustly to some vaccines (32). We recently immunized neonate macaques with the VRC DNA prime, rAd5 boost vaccine at ~2-6 days after birth and found that vaccine-induced memory B cell-derived antibodies were predominantly gp41-reactive, and 16S rRNA-derived IM taxa profile was similar for neonate and adult macaques immunized with the same VRC vaccine (33). These data confirmed that the imprinting of the B cell repertoire occurs soon after birth. A recent review highlighted studies that suggested that the infant microbiome can in some cases be seeded in utero (34). Additionally, neonate B cells have been shown to express CD5 and CD1c, markers of marginal zone B cells (35, 36), and marginal zone B cells have low affinity IgM, autoreactive and polyreactive BCRs (37, 38). Since HIV-1 bnAbs are frequently autoreactive and subjected to immune tolerance controls during B cell lineage development in adults (14, 18), the polyreactive nature of infant B cells could be exploited for bnAb induction via HIV-1 vaccination. Indeed, human infants that are HIV-1 infected have a higher frequency of bnAbs and generate them sooner than HIV-1 infected adults (39, 40). HIV-1-induced immune dysfunction with disrupted immune tolerance has been associated with bnAb induction (41). Thus, one hypothesis to explain these observations in infants is that infant immune systems have possibly less stringent immune tolerance controls or respond to HIV-1 infection with more HIV-1-induced immune dysregulation.
Macaques are used as an animal model to study human HIV-1 infections and for testing candidate HIV-1 vaccines (42). Thus, evaluating gp41 immunodominance induced by HIV-1 Env gp140 in macaques is important for the HIV-1 vaccine field. Han and colleagues recently showed that, as in humans, the VRC DNA prime, rAd5 boost vaccine induced a dominant gp41-reactive, memory B cell response in both neonate and adult macaques (33). In this study, Han and colleagues also showed IM-cross-reactivity of the VRC vaccine-induced gp41-reactive antibodies. Thus, neonatal non-human primates may be a model to define immunoregulatory controls of bnAb induction in HIV-1 infection, and to elucidate roles the microbiome might have in imprinting the B cell repertoire to respond to pathogens.
HIV-1 Env and IM crossreactive epitopes
Many cross-reactive epitopes between HIV-1 proteins and human proteins have been described (43, 44). Studies of HIV-1 Env immunization in humans (24-27) and macaques (33) raised the hypothesis that IM-primed B cells can respond to the gp41 component of a HIV-1 Env vaccine. Two IM antigens have been identified, bacterial E. coli RNA polymerase and pyruvate flavodoxin oxidoreductase, that shared sequence and structural motifs with a region in the heptad-repeat 1 (HR1) region of gp41 containing the LLRAIE amino acid residues (4, 26). Interestingly, Han et al. demonstrated that human gp41-IM cross-reactive antibodies bound macaque IM and provided evidence of neonate and adult macaque IM bacterial proteins that encode the gp41-IM cross-reactive epitope found on E. coli RNA polymerase (33). Whether the LLRAIE amino acid sequence motif in Env gp41 HR1 region (26) is the principal candidate IM cross-reactive epitope on gp41 remains unknown. Interestingly, it is likely that additional IM proteins have Env-cross-reactive epitopes, since gp120-reactive antibodies induced by the VRC DNA prime, rAd5 boost vaccine (26, 33) and HIV-1 infection (45) have also been isolated that cross-react with IM. Identifying IM-antigens and epitopes for Env-IM cross-reactivity will provide a toolbox of IM proteins that can be used to evaluate the IM-cross-reactivity of candidate HIV-1 Env vaccine immunogens. The new generation of stabilized trimers (SOSIPs) has been designed to present only bnAb epitopes that can be recognized by the immune system during vaccination (46-49). The membrane proximal external region (MPER) region targeted by gp41-reactive bnAbs was not included in the SOSIP trimers, but a portion of the gp41 ectodomain region remained at the base of these proteins. While most studies have found SOSIP trimers to induce primarily autologous tier 2 nAbs (50-52), a recent study in mice have shown that BG505 SOSIP.664 trimers induced non-neutralizing antibodies that targeted the base of the SOSIP (53). It will be key to determine if the new generation of HIV-1 Env immunogens induce gp41-dominant responses in non-human primates or in humans.
Conclusions
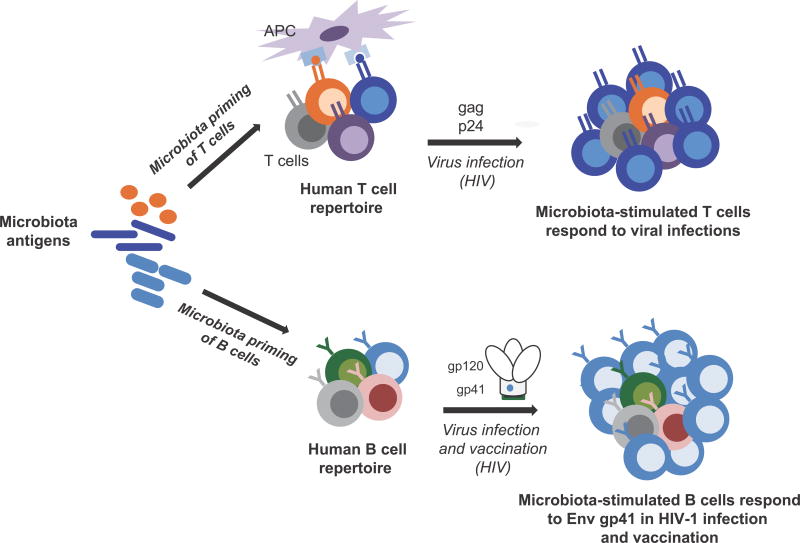
The microbiome can shape the repertoire of immune cells to respond to viruses, and influence the response to vaccine immunogens (Figure 1). Analysis of individuals vaccinated with Env-immunogens has demonstrated that IM-antigen cross-reactivity with HIV-1 Env can possibly divert HIV-1 Env vaccine-induced antibody responses away from protective immunity. Here we have reviewed evidence for three hypotheses: (i) the microbiome can influence the specificity of B and CD4 T cell responses to infections; (ii) the microbiome can modulate the response to HIV-1 Env immunization; and (iii) modification of the microbiome may enhance vaccine responses.
Figure 1.
Microbiota priming of CD4 T and B cell repertoires. For T cells (upper path); HIV-1-reactive T cells can be activated by antigens with sequence homology between HIV and microbiota proteins. For B cells (lower path); a dominant B cell response to HIV-1 infection and vaccination can be shaped by microbiota stimulation of a pre-existing pool of polyreactive B cells. Thus, microbiota-stimulated CD4 T and B cells can respond to viral antigens in the setting of infection or vaccination.
Key bullet points.
The microbiome can influence immune responses to infections.
Cross-reactivity of naïve and memory CD4 T and B cells occurs with intestinal microbiota antigens
The microbiome can modulate antibody responses to HIV-1 Env vaccines.
Modification of the microbiome and HIV-1 Env-Intestinal microbiota cross-reactive epitopes may enhance HIV-1 Env vaccine responses.
Acknowledgments
Financial support and sponsorship
This work has been funded with grants provided by the National Institute of Health (NIH), National Institute of Allergy and Infectious Diseases (NIAID), Division of AIDS; UM-1 grant for the Duke Center for HIV/AIDS Vaccine Immunology-Immunogen Discovery (CHAVI ID; UM1 AI100645), Duke University Center for AIDS Research (CFAR; P30-Al-64518), and HVTN Laboratory Center UM1 AI068618.
Footnotes
Conflicts of interest
The authors report no conflicts of interest.
References and recommended reading
Papers of particular interest, published within the annual period of review, (the last 2 years) have been highlighted as:
* of special interest
- 1.Jackson KJ, Kidd MJ, Wang Y, Collins AM. The shape of the lymphocyte receptor repertoire: lessons from the B cell receptor. Front Immunol. 2013;4:263. doi: 10.3389/fimmu.2013.00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mouquet H, Nussenzweig MC. Polyreactive antibodies in adaptive immune responses to viruses. Cell Mol Life Sci. 2012;69(9):1435–45. doi: 10.1007/s00018-011-0872-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liao HX, Chen X, Munshaw S, Zhang R, Marshall DJ, Vandergrift N, et al. Initial antibodies binding to HIV-1 gp41 in acutely infected subjects are polyreactive and highly mutated. J Exp Med. 2011;208(11):2237–49. doi: 10.1084/jem.20110363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trama AM, Moody MA, Alam SM, Jaeger FH, Lockwood B, Parks R, et al. HIV-1 envelope gp41 antibodies can originate from terminal ileum B cells that share cross-reactivity with commensal bacteria. Cell Host Microbe. 2014;16(2):215–26. doi: 10.1016/j.chom.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mason D. A very high level of crossreactivity is an essential feature of the T-cell receptor. Immunol Today. 1998;19(9):395–404. doi: 10.1016/s0167-5699(98)01299-7. [DOI] [PubMed] [Google Scholar]
- 6.Reinherz EL, Tan K, Tang L, Kern P, Liu J, Xiong Y, et al. The crystal structure of a T cell receptor in complex with peptide and MHC class II. Science. 1999;286(5446):1913–21. doi: 10.1126/science.286.5446.1913. [DOI] [PubMed] [Google Scholar]
- 7.Su LF, Davis MM. Antiviral memory phenotype T cells in unexposed adults. Immunol Rev. 2013;255(1):95–109. doi: 10.1111/imr.12095. [DOI] [PubMed] [Google Scholar]
- 8.Havenar-Daughton C, Lee JH, Crotty S. Tfh cells and HIV bnAbs, an immunodominance model of the HIV neutralizing antibody generation problem. Immunol Rev. 2017;275(1):49–61. doi: 10.1111/imr.12512. [DOI] [PubMed] [Google Scholar]
- 9.Campion SL, Brodie TM, Fischer W, Korber BT, Rossetti A, Goonetilleke N, et al. Proteome-wide analysis of HIV-specific naive and memory CD4(+) T cells in unexposed blood donors. J Exp Med. 2014;211(7):1273–80. doi: 10.1084/jem.20130555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Su LF, Kidd BA, Han A, Kotzin JJ, Davis MM. Virus-specific CD4(+) memory-phenotype T cells are abundant in unexposed adults. Immunity. 2013;38(2):373–83. doi: 10.1016/j.immuni.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidt J, Neumann-Haefelin C, Altay T, Gostick E, Price DA, Lohmann V, et al. Immunodominance of HLA-A2-restricted hepatitis C virus-specific CD8+ T cell responses is linked to naive-precursor frequency. J Virol. 2011;85(10):5232–6. doi: 10.1128/JVI.00093-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelsoe G, Verkoczy L, Haynes BF. Immune System Regulation in the Induction of Broadly Neutralizing HIV-1 Antibodies. Vaccines (Basel) 2014;2(1):1–14. doi: 10.3390/vaccines2010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science. 2003;301(5638):1374–7. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- 14.Haynes BF, Fleming J, St Clair EW, Katinger H, Stiegler G, Kunert R, et al. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science. 2005;308(5730):1906–8. doi: 10.1126/science.1111781. [DOI] [PubMed] [Google Scholar]
- 15.Mouquet H, Scheid JF, Zoller MJ, Krogsgaard M, Ott RG, Shukair S, et al. Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature. 2010;467(7315):591–5. doi: 10.1038/nature09385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haynes BF, Moody MA, Verkoczy L, Kelsoe G, Alam SM. Antibody polyspecificity and neutralization of HIV-1: a hypothesis. Hum Antibodies. 2005;14(3–4):59–67. [PMC free article] [PubMed] [Google Scholar]
- 17.Bonsignori M, Wiehe K, Grimm SK, Lynch R, Yang G, Kozink DM, et al. An autoreactive antibody from an SLE/HIV-1 individual broadly neutralizes HIV-1. J Clin Invest. 2014;124(4):1835–43. doi: 10.1172/JCI73441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haynes BF, Verkoczy L. AIDS/HIV. Host controls of HIV neutralizing antibodies. Science. 2014;344(6184):588–9. doi: 10.1126/science.1254990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422(6929):307–12. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 20.Tomaras GD, Yates NL, Liu P, Qin L, Fouda GG, Chavez LL, et al. Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J Virol. 2008;82(24):12449–63. doi: 10.1128/JVI.01708-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21*.Monaco CL, Gootenberg DB, Zhao G, Handley SA, Ghebremichael MS, Lim ES, et al. Altered Virome and Bacterial Microbiome in Human Immunodeficiency Virus-Associated Acquired Immunodeficiency Syndrome. Cell Host Microbe. 2016;19(3):311–22. doi: 10.1016/j.chom.2016.02.011. Defined how changes in the microbiome following HIV-1 infection impact disease progression towards AIDS. Alterations in the enteric virome and bacterial microbiome of HIV-1 infected individuals were found to be associated with poor clinical biomarkers (low CD4 T cell counts) during HIV-1 pathogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22*.Handley SA, Desai C, Zhao G, Droit L, Monaco CL, Schroeder AC, et al. SIV Infection-Mediated Changes in Gastrointestinal Bacterial Microbiome and Virome Are Associated with Immunodeficiency and Prevented by Vaccination. Cell Host Microbe. 2016;19(3):323–35. doi: 10.1016/j.chom.2016.02.010. Defined how changes in the microbiome following macaque SIV-infection impact disease progression. Enteric viruses and bacteria were found to impacted by lentivirus infection, including the outgrowth of pathogens that promoted disease progression. SIV vaccination was found to prevent the outgrowth of pathogenic viruses and bacteria. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oh JZ, Ravindran R, Chassaing B, Carvalho FA, Maddur MS, Bower M, et al. TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity. 2014;41(3):478–92. doi: 10.1016/j.immuni.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hammer SM, Sobieszczyk ME, Janes H, Karuna ST, Mulligan MJ, Grove D, et al. Efficacy trial of a DNA/rAd5 HIV-1 preventive vaccine. N Engl J Med. 2013;369(22):2083–92. doi: 10.1056/NEJMoa1310566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Churchyard GJ, Morgan C, Adams E, Hural J, Graham BS, Moodie Z, et al. A phase IIA randomized clinical trial of a multiclade HIV-1 DNA prime followed by a multiclade rAd5 HIV-1 vaccine boost in healthy adults (HVTN204) PLoS One. 2011;6(8):e21225. doi: 10.1371/journal.pone.0021225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26*.Williams WB, Liao HX, Moody MA, Kepler TB, Alam SM, Gao F, et al. HIV-1 VACCINES. Diversion of HIV-1 vaccine-induced immunity by gp41-microbiota cross-reactive antibodies. Science. 2015;349(6249) doi: 10.1126/science.aab1253. aab1253. Demonstrated the origin of the gp41-IM cross-reactive Ab responses induced by a VRC HIV-1 Env gp41-containing vaccine, and suggested that IM cross-reactivity of HIV-1 Env possibly diverting the vaccine-induced protective antibody respones was one reason this VRC vaccine was not efficacious. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goepfert PA, Elizaga ML, Seaton K, Tomaras GD, Montefiori DC, Sato A, et al. Specificity and 6-month durability of immune responses induced by DNA and recombinant modified vaccinia Ankara vaccines expressing HIV-1 virus-like particles. J Infect Dis. 2014;210(1):99–110. doi: 10.1093/infdis/jiu003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neu J. The microbiome during pregnancy and early postnatal life. Semin Fetal Neonatal Med. 2016;21(6):373–9. doi: 10.1016/j.siny.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 29.Nguyen QN, Himes JE, Martinez DR, Permar SR. The Impact of the Gut Microbiota on Humoral Immunity to Pathogens and Vaccination in Early Infancy. PLoS Pathog. 2016;12(12):e1005997. doi: 10.1371/journal.ppat.1005997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wesemann DR, Portuguese AJ, Meyers RM, Gallagher MP, Cluff-Jones K, Magee JM, et al. Microbial colonization influences early B-lineage development in the gut lamina propria. Nature. 2013;501(7465):112–5. doi: 10.1038/nature12496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martinez DR, Permar SR, Fouda GG. Contrasting Adult and Infant Immune Responses to HIV Infection and Vaccination. Clin Vaccine Immunol. 2015;23(2):84–94. doi: 10.1128/CVI.00565-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ota MO, Vekemans J, Schlegel-Haueter SE, Fielding K, Whittle H, Lambert PH, et al. Hepatitis B immunisation induces higher antibody and memory Th2 responses in new-borns than in adults. Vaccine. 2004;22(3–4):511–9. doi: 10.1016/j.vaccine.2003.07.020. [DOI] [PubMed] [Google Scholar]
- 33*.Han Q, Williams WB, Saunders KO, Seaton KE, Wiehe KJ, Vandergrift N, et al. HIV DNA-Adenovirus Multiclade Envelope Vaccine Induces Gp41 Antibody Immunodominance in Rhesus Macaques. J Virol. 2017 doi: 10.1128/JVI.00923-17. Demonstrated that the dominant gp41-intestinal microbiota (IM) cross-reactive antibody response observed in individuals immunized with a VRC HIV-1 Env vaccine also occurred in macaques who received the same vaccine, and suggested that macaques may be an appropriate animal model to test candidate Env immunogens for gp41-IM cross-reactivity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perez-Muñoz ME, Arrieta MC, Ramer-Tait AE, Walter J. A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: implications for research on the pioneer infant microbiome. Microbiome. 2017;5(1):48. doi: 10.1186/s40168-017-0268-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Durandy A, Thuillier L, Forveille M, Fischer A. Phenotypic and functional characteristics of human newborns’ B lymphocytes. J Immunol. 1990;144(1):60–5. [PubMed] [Google Scholar]
- 36.Plebani A, Proserpio AR, Guarneri D, Buscaglia M, Cattoretti G. B and T lymphocyte subsets in fetal and cord blood: age-related modulation of CD1c expression. Biol Neonate. 1993;63(1):1–7. doi: 10.1159/000243901. [DOI] [PubMed] [Google Scholar]
- 37.Mackenzie LE, Youinou PY, Hicks R, Yuksel B, Mageed RA, Lydyard PM. Auto- and polyreactivity of IgM from CD5+ and CD5- cord blood B cells. Scand J Immunol. 1991;33(3):329–35. doi: 10.1111/j.1365-3083.1991.tb01778.x. [DOI] [PubMed] [Google Scholar]
- 38.Cerutti A, Cols M, Puga I. Marginal zone B cells: virtues of innate-like antibody-producing lymphocytes. Nat Rev Immunol. 2013;13(2):118–32. doi: 10.1038/nri3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goo L, Chohan V, Nduati R, Overbaugh J. Early development of broadly neutralizing antibodies in HIV-1-infected infants. Nat Med. 2014;20(6):655–8. doi: 10.1038/nm.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40*.Muenchhoff M, Adland E, Karimanzira O, Crowther C, Pace M, Csala A, et al. Nonprogressing HIV-infected children share fundamental immunological features of nonpathogenic SIV infection. Sci Transl Med. 2016;8(358):358ra125. doi: 10.1126/scitranslmed.aag1048. Compared to HIV-1-infected adults, a higher frequency of HIV-1-infected children generated HIV-1 envelope (Env)-reactive broadly neutralizing antibodies (bnAbs). These data suggest that the infant immune system may be more permissible for the development of polyreactive HIV-1 Env bnAbs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41*.Moody MA, Pedroza-Pacheco I, Vandergrift NA, Chui C, Lloyd KE, Parks R, et al. Immune perturbations in HIV-1-infected individuals who make broadly neutralizing antibodies. Sci Immunol. 2016;1(1) doi: 10.1126/sciimmunol.aag0851. aag0851. HIV-1-infected individuals who make HIV-1 envelope (Env)-reactive, broadly neutralizing antibodies (bnAbs) had immune dysfunction with disrupted immune tolerance mechanisms Thus, HIV-1 vaccines may require transient immune system perturbations for induction of polyreactive HIV-1 Env bnAbs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hatziioannou T, Evans DT. Animal models for HIV/AIDS research. Nat Rev Microbiol. 2012;10(12):852–67. doi: 10.1038/nrmicro2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Süsal C, Kröpelin M, Daniel V, Opelz G. Molecular mimicry between HIV-1 and antigen receptor molecules: a clue to the pathogenesis of AIDS. Vox Sang. 1993;65(1):10–7. doi: 10.1111/j.1423-0410.1993.tb04518.x. [DOI] [PubMed] [Google Scholar]
- 44.Silvestris F, Williams RC, Dammacco F. Autoreactivity in HIV-1 infection: the role of molecular mimicry. Clin Immunol Immunopathol. 1995;75(3):197–205. doi: 10.1006/clin.1995.1072. [DOI] [PubMed] [Google Scholar]
- 45*.Jeffries TL, Sacha CR, Pollara J, Himes J, Jaeger FH, Dennison SM, et al. The function and affinity maturation of HIV-1 gp120-specific monoclonal antibodies derived from colostral B cells. Mucosal Immunol. 2016;9(2):414–27. doi: 10.1038/mi.2015.70. Identification of HIV-1 envelope (Env) gp120-reactive antibodies from colostrum B cells of HIV-1-infected mothers that cross-reacted with intestinal microbiota (IM) antigens. Thus, IM-antigens with molecular mimicry to HIV-1 Env gp120 may also shape the B cell response to HIV-1 antigens. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013;9(9):e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Julien JP, Lee JH, Ozorowski G, Hua Y, Torrents de la Peña A, de Taeye SW, et al. Design and structure of two HIV-1 clade C SOSIP.664 trimers that increase the arsenal of native-like Env immunogens. Proc Natl Acad Sci U S A. 2015;112(38):11947–52. doi: 10.1073/pnas.1507793112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chuang GY, Geng H, Pancera M, Xu K, Cheng C, Acharya P, et al. Structure-Based Design of a Soluble Prefusion-Closed HIV-1 Env Trimer with Reduced CD4 Affinity and Improved Immunogenicity. J Virol. 2017;91(10) doi: 10.1128/JVI.02268-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ringe RP, Ozorowski G, Rantalainen K, Struwe WB, Matthews K, Torres JL, et al. Reducing V3 Antigenicity and Immunogenicity on Soluble, Native-Like HIV-1 Env SOSIP Trimers. J Virol. 2017;91(15) doi: 10.1128/JVI.00677-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, Burger JA, et al. HIV-1 VACCINES. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science. 2015;349(6244) doi: 10.1126/science.aac4223. aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Havenar-Daughton C, Carnathan DG, Torrents de la Peña A, Pauthner M, Briney B, Reiss SM, et al. Direct Probing of Germinal Center Responses Reveals Immunological Features and Bottlenecks for Neutralizing Antibody Responses to HIV Env Trimer. Cell Rep. 2016;17(9):2195–209. doi: 10.1016/j.celrep.2016.10.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pauthner M, Havenar-Daughton C, Sok D, Nkolola JP, Bastidas R, Boopathy AV, et al. Elicitation of Robust Tier 2 Neutralizing Antibody Responses in Nonhuman Primates by HIV Envelope Trimer Immunization Using Optimized Approaches. Immunity. 2017;46(6):1073–88.e6. doi: 10.1016/j.immuni.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu JK, Crampton JC, Cupo A, Ketas T, van Gils MJ, Sliepen K, et al. Murine Antibody Responses to Cleaved Soluble HIV-1 Envelope Trimers Are Highly Restricted in Specificity. J Virol. 2015;89(20):10383–98. doi: 10.1128/JVI.01653-15. [DOI] [PMC free article] [PubMed] [Google Scholar]