SUMMARY
Antibodies to the hemagglutinin (HA) and neuraminidase (NA) glycoproteins are the major mediators of protection against influenza virus infection. Here, we report that current influenza vaccines poorly display key NA epitopes and rarely induce NA-reactive B cells. Conversely, influenza virus infection induces NA-reactive B cells at a frequency that approaches (H1N1) or exceeds (H3N2) that of HA-reactive B cells. NA-reactive antibodies display broad binding activity spanning the entire history of influenza A virus circulation in humans, including the original pandemic strains of both H1N1 and H3N2 subtypes. The antibodies robustly inhibit the enzymatic activity of NA, including oseltamivir-resistant variants, and provide robust prophylactic protection in vivo, including against avian H5N1 viruses. When used therapeutically, NA-reactive antibodies protected mice from lethal influenza virus challenge even 48-hours post-infection. These findings strongly suggest that influenza vaccines should be optimized to improve targeting of NA for durable and broad protection against divergent influenza strains.
In Brief
Current influenza vaccines predominantly produce antibodies targeting the viral hemagglutinin (HA). However, during natural infection the body also produces antibodies targeting the viral neuraminidase (NA). These NA antibodies can provide robust and broad protection and could potentially be elicited prophylactically, via new vaccine strategies, or used therapeutically.
INTRODUCTION
Influenza is an acute respiratory illness that causes up to 5 million cases of influenza virus infection and 250,000 to 640,000 deaths annually around the world (Iuliano et al., 2017). The influenza virus has two main surface glycoproteins, hemagglutinin (HA) and neuraminidase (NA). HA, the more abundant protein, mediates binding to sialic acid receptors and subsequent fusion between the virus and host cell membranes. The less abundant tetrameric NA protein is essential for cleaving terminal sialic acid residues present on host cell surfaces, allowing the release of the newly formed viral particles (Matrosovich et al., 2004; Palese and Compans, 1976).
Currently, the seasonal influenza virus vaccine is the most widely available method to reduce the annual impact of influenza infection (Nichol, 2008). HA-reactive antibodies are typically considered the de facto mediators of protection from influenza infection; indeed, inhibition of HA activity has been the primary measure of influenza vaccine efficacy for decades. Therefore, most of the current approaches for vaccine design focus on inducing an antibody response to influenza virus HA. Influenza vaccine effectiveness can vary widely from season to season such that protection is always limited. For example, vaccine effectiveness ranged from only 19% to 48% during the past three influenza seasons (Flannery, 2017). Studies have shown that HA antigenic drift (viral genome point mutations) is the primary reason for the limited effectiveness of the seasonal influenza vaccine (Karron and Collins, 2013). Due to viral mutations, preexisting antibodies often show limited neutralization against currently circulating viruses (Wohlbold and Krammer, 2014). Although point mutations also occur in the NA protein, among influenza A viruses the rate of antigenic drift around the active site of NA in the head domain is slower than that for HA (Abed et al., 2002; Air, 2012).
Historically, NA has served as an important target for antivirals or therapeutics, due to its critical role in the influenza virus replication cycle (Wohlbold and Krammer, 2014). Inhibition of NA activity is the basis of commonly used influenza therapeutics including oseltamivir (Tamiflu), zanamivir (Relenza), laninamivir (Inavir), and peramivir (Rapivab). Oseltamivir can reduce the median duration of influenza illness by 1.3 days and markedly reduces symptoms compared to placebo if given within 48 hours of symptom onset. In a prophylactic study, oseltamivir decreased rates of influenza infection five-fold from 5% (25/519) for the placebo group to 1% (6/520) for the oseltamivir-treated group (Genentech, 2016). Thus, inhibition of NA activity has become a standard of care for the treatment of influenza virus infections. The limitations of neuraminidase inhibitors such as oseltamivir are that resistant strains of influenza virus have readily emerged (Dharan et al., 2009) and the window for efficacy is limited to the first 48 hours of symptom onset.
There are several mechanisms of NA-reactive antibody inhibition of influenza virus infection (Krammer and Palese, 2015). NA-reactive antibodies can prevent virus budding and egress from infected cells. These antibodies similarly inhibit viral escape from the natural defense proteins that trap the virus via HA-sialic acid interactions on mucosal surfaces. Moreover, NA-reactive antibody bound to NA at the surface of infected cells might aid in the clearance of the virus through antibody-dependent cell-mediated cytotoxicity (ADCC) and complement-dependent cytotoxicity (CDC) (Wan et al., 2013; Wohlbold et al., 2017).
There is a long history of literature suggesting that immunity to NA could protect from influenza infection. The polyclonal antibody response to NA was broadly reactive and conferred protection against heterologous viruses in mice (Schulman et al., 1968). This cross-reactivity is evident even when there is substantial change within strain specific NA epitopes, resulting in a phenomenon of one-way drift (Sandbulte et al., 2011). NA-reactive monoclonal antibodies (mAbs) isolated from mice and rabbits protected against both homologous and heterologous influenza infection in vivo (Doyle et al., 2013; Wan et al., 2013; Wan et al., 2015; Wilson et al., 2016; Wohlbold et al., 2017). Several conserved amino acids were identified in these studies as the basis for the broad reactivity of NA-reactive mAbs (Wan et al., 2013; Wohlbold et al., 2017). Previous studies in humans have also shown that preexisting NA-reactive antibodies can reduce the number of cases of infection and decrease disease severity (Monto and Kendal, 1973; Murphy et al., 1972). However, on the whole little is known about human antibody responses to NA, and most influenza vaccine development efforts both past and present are focused on targeting HA.
Here, we report that, unlike vaccination, natural influenza virus infection readily induces a high proportion of NA-reactive B cells. Thus, from infected patients, we were able to isolate and characterize protective antibodies binding NA epitopes, informing on the design of an NA-based component for influenza vaccination. The NA-reactive antibodies can be induced in human or mouse by infection or immunization with whole virions, but bind epitopes not efficiently detected in the Fluarix® or Fluzone® influenza vaccines. Importantly, these NA-reactive mAbs bind a broad spectrum of influenza virus strains, often spanning the entire circulation history in humans for that NA group. Moreover, these antibodies have robust NA inhibition (NI) activity and provide prophylactic as well as therapeutic protection in vivo. Our data suggest that the next-generation of influenza vaccines should be optimized to improve the NA humoral immune response to induce broadly cross-reactive and protective NA-reactive antibody responses.
RESULTS
NA is frequently targeted during natural influenza virus infection but not after vaccination
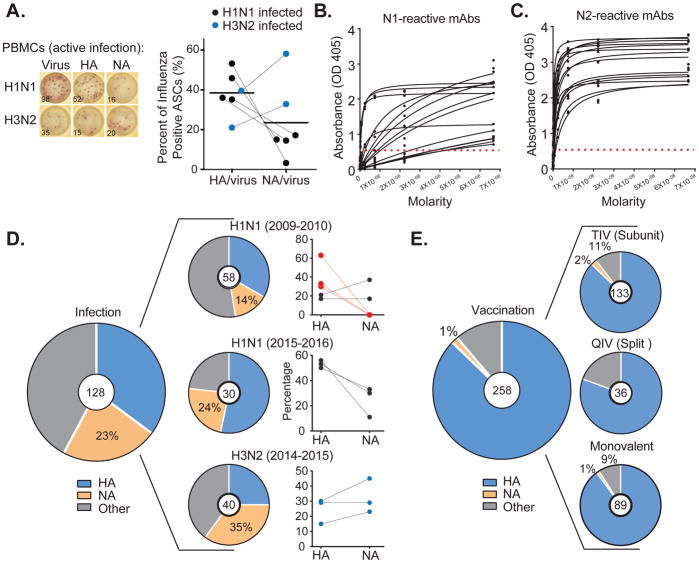
While characterizing the specificity of plasmablasts induced by influenza virus infection, we noted a surprisingly high proportion of NA-reactive cells. The specificity of plasmablasts was evaluated by ELISPOT or mAb characterization from a total of sixteen confirmed influenza-infected patients. Notably, as previously reported (Andrews et al., 2015), plasmablasts are highly representative of the memory compartment, accounting for almost half of the memory cells activated in future responses to similar influenza strains. These patients included eleven subjects infected with the H1N1 pandemic strain (five from 2009, and six from 2016), plus five infected with H3N2 virus strains (three from 2014 and two from 2017) (Table S1). First, we analyzed large numbers of activated plasmablasts in six influenza virus infected patients (four infected with H1N1 in 2016 and two infected with H3N2 in 2017). Scoring of thousands of activated plasmablasts by ELISPOT assay detected an average of 24% that were reactive to NA and 38% to HA (Figure 1A). Surprisingly, plasmablasts from H3N2 infected patients predominantly targeted NA. To more rigorously assess the frequency of NA-reactive B cells activated during infection, we characterized mAbs obtained from patients. For this, the isolated variable region genes from single plasmablasts activated by infection were used to express 128 influenza-binding mAb proteins from 12 of the patients using methods as previously described (Smith et al., 2009; Wardemann et al., 2003; Wrammert et al., 2008). The NA-reactive mAbs were more often encoded by VH3 family genes, but used variable genes that were otherwise similar to HA antibodies (Figure S1). Consistent with the ELISPOT assays, 22.6% (29/128), and on average 24% by year and strain, of plasmablast mAbs activated by influenza virus infection were reactive to recombinant NA (rNA) (Figures 1B, 1C, and 1D). These mAbs included 15/88 that were N1-reactive and 14/40 that were N2-reactive (mAbs by subject indicated in Table S1). Notably, as with the ELISPOT analysis, H3N2 virus infections consistently induced a higher proportion of NA-reactive B cells compared to HA-reactive B cells (Figures 1A and 1D, blue dots). By comparison, activation of NA-reactive B cells was quite rare 7 days after vaccination, accounting for only 1.2% (2 of 258) of vaccine induced plasmablasts relative to 87% that targeted HA and 12% binding other influenza antigens (Figure 1E). This observation was consistent for several influenza virus vaccine compositions, including 1.5% (2 of 133) of NA-reactive cells after immunization with a subunit vaccine (from 2006–2008 and in 2010), 1.1% (1 of 89) after the 2009 H1N1 monovalent vaccine, and none (0 of 36) induced by split vaccines (2008–2010) (Figure 1E). We conclude from this analysis that a quarter of plasmablasts induced by natural influenza virus infection target NA – a percentage that nearly equals that of HA-specific plasmablasts – compared to only 1–2% from influenza vaccination.
Figure 1. Influenza virus infection induces a greater prevalence of NA-reactive antibodies than vaccination.
(A) The proportions of HA- and NA-reactive ASCs out of the total virus-reactive cells were determined by ELISPOT assay. Individuals infected with an H1N1 influenza virus (in black) were compared to individuals infected with an H3N2 influenza virus (in blue). Each dot represents a subject (n=6).
(B–C) Binding of NA-reactive mAbs to rNA proteins by ELISA. Represented are ELISA binding curves. The assays were performed in duplicate at least 3 times for each antibody. (B) Binding to A/California/7/2009 (H1N1) rN1 protein or (C) A/Texas/50/2012 (H3N2) rN2 protein. Numbers of antibodies per subject are indicated in Table S1.
(D–E) Proportion of influenza virus-reactive mAbs that bind to HA, NA or other antigens (D). Pie charts show the percentages of mAbs that bind a given antigen (HA, NA, or other) of the total, indicated in the center circle. Graphed on the right are the percentages of HA- and NA-reactive antibodies per individual. Each dot represents one individual (n=11). Red indicates patients with no NA B cells detected on first exposure to the pandemic H1N1 strain in 2009 (E) The frequency of NA-reactive mAbs induced by vaccination (the vaccinated cohorts are detailed in the methods). As in (D), pie charts show the percentages of mAbs that bind a given antigen (HA, NA, or other) in individuals vaccinated with influenza virus subunit vaccine (seasons 2006–2008 and 2010–2011), influenza virus split vaccine (2008–2010), or monovalent pandemic H1N1 vaccine (2009–2010). For the panels (A) and (D), the blue dots indicate patients infected with an H3N2 virus.
Infection-induced anti-NA antibodies bind epitopes that are not preserved in current influenza vaccines
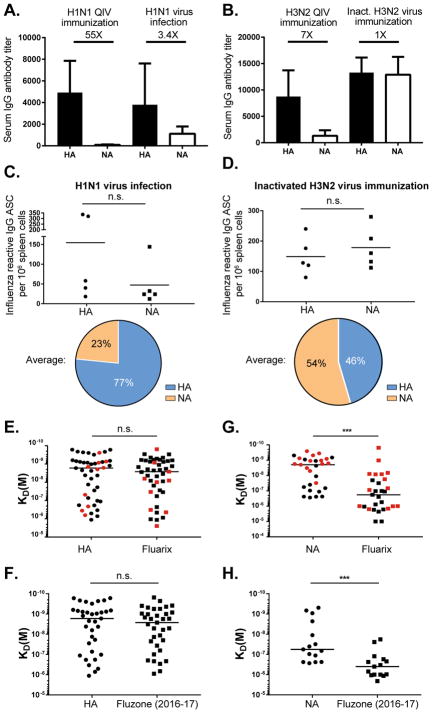
The greater induction of NA-reactive plasmablasts during natural infection may be because the live intact virus displays epitopes not present in the inactivated vaccines. Memory to conserved epitopes appears to play a role in the observed bias, as serological studies have shown an induction of NA-reactive antibodies to past strains (Rajendran et al., 2017). Both HA and NA antibodies were encoded from highly mutated variable genes, supporting a memory cell recall origin (Figure S1). Furthermore, first exposure to the 2009 pandemic influenza virus strain induced NA-reactive plasmablasts at detectable frequencies in only two of the five infected patients that we characterized (black versus red lines in the top row of Figure 1D). Conversely, exposure to that strain seven years later in 2016 or to H3N2 strains that have circulated since 1968 readily induced NA-reactive plasmablasts (Figures 1A and 1D). To determine if infection or exposure to whole virus particles could account for the increased NA targeting, we infected mice with live H1N1 or killed H3N2 whole virions as opposed to split/subunit vaccine. For this, mice were infected intranasally with a sublethal dose of live 2009 pandemic H1N1 virus or immunized intranasally with intact virions of inactivated H3N2 influenza virus, followed by an intranasal boost with the respective virus strains 30 days later. Similar to what we observed in infected humans, serum responses in mice to NA were dramatically increased after immunization with whole influenza virions versus the 2015–2016 Fluarix quadrivalent influenza vaccine (Figures 2A and 2B). To verify the preferred targeting of NA by whole virions at the cellular level, ELISPOT assays on whole splenocytes eight days after secondary infection or immunization were used to measure the proportions of HA- and NA-reactive IgG-secreting cells that were activated. The frequency of NA-reactive cells was common after exposure to whole virions for both the H1N1 and H3N2 strains (Figures 2C and 2D). This observation was not dependent on viral replication as the H3N2 influenza strain was inactivated. As with human infections with an H3N2 virus, more plasmablasts were specific to N2 than to H3 (Figure 2D). Although mice are not an ideal model of human immunity to influenza, it was surprising how accurately the response to whole virions reflected that of humans to both N1 and N2. Together these experiments suggest that NA epitopes present on whole virions are not efficiently targeted by current influenza vaccines. To address this possibility directly, we tested the NA- and HA-reactive mAbs generated from infection-induced plasmablasts for binding to inactivated influenza virus vaccines. While HA-reactive mAbs bound rHA protein with equal avidity to the vaccine, the NA-reactive mAbs had only negligible binding to the Fluarix® (Figures 2E and 2G) or Fluzone® (Figures 2F and 2H) vaccines. There was no notable difference in binding avidity by subtype (H1N1 vs H3N2) or by vaccine batch. Five mAbs that bound poorly to the vaccines maintained full avidity against inactivated H3N2 virus used for the mouse immunization experiments (Figure 2A–D), demonstrating that gentle inactivation protocols can maintain key NA epitopes (Figure S2). We conclude from these studies that current influenza virus vaccines have insufficient NA content or NA protein structural integrity to induce NA-reactive antibody responses efficiently.
Figure 2. Epitopes on NA are not efficiently presented in current commercially available inactivated influenza virus vaccines.
(A–D) Mice were infected with live H1N1 virus or immunized with inactivated H3N2 virus (as detailed in the methods). (A–B) Serum responses in immunized mice were determined by ELISA. (A) HA1 and N1 serum endpoint titers (n=5) were tested by A/California/07/2009 rHA and rNA, respectively. (B) HA3 and N2 serum endpoint titers (n=5) were tested by A/Switzerland/9715293/2013 rHA and A/Texas/50/2012 rNA, respectively. Data are represented as mean ± SD.
(C–D) The proportion of HA and NA-reactive IgG secreting cells (ASCs) in mice after infection (H1N1) or immunization (H3N2). Pie charts show the average frequency of HA versus NA-reactive B cells.
(E–H) HA and NA-reactive mAbs were tested for binding by ELISA to HA, NA and two influenza virus vaccine preparations. Binding avidities (KD) were estimated by Scatchard plot analyses of ELISA data for 35 anti-H1, 15 anti-N1, 10 anti-H3, and 14 anti-N2 mAbs. (E) H1-mAb binding was compared between A/California/7/2009 (H1N1) rHA and Fluarix vaccine (2015–2016), and for H3-mAbs to A/Texas/50/2012 (H3N2) rHA and Fluarix vaccine (2014–2015). (F) Binding of H1-mAbs to A/California/7/2009 (H1N1) rHA compared to the Fluzone vaccine (2016–2017). (G) Binding N1-mAbs to A/California/7/2009 (H1N1) rNA compared to Fluarix vaccine (2015–2016) and N2-mAbs binding to A/Texas/50/2012 (H3N2) rNA was compared to Fluarix vaccine (2014–2015). (H) Binding of N1-mAbs to A/California/7/2009 (H1N1) rNA was compared to Fluzone vaccine (2016–2017). The red points indicate H3- and N2-reactive mAbs. Data are representative of three independent experiments. Statistical significance was determined using the paired nonparametric Wilcoxon test. The line represents the median. n.s., not significant. *p<0.05; **p<0.001; ***p<0.0001.
See also Figure S2.
Human NA-reactive mAbs are broadly cross-reactive
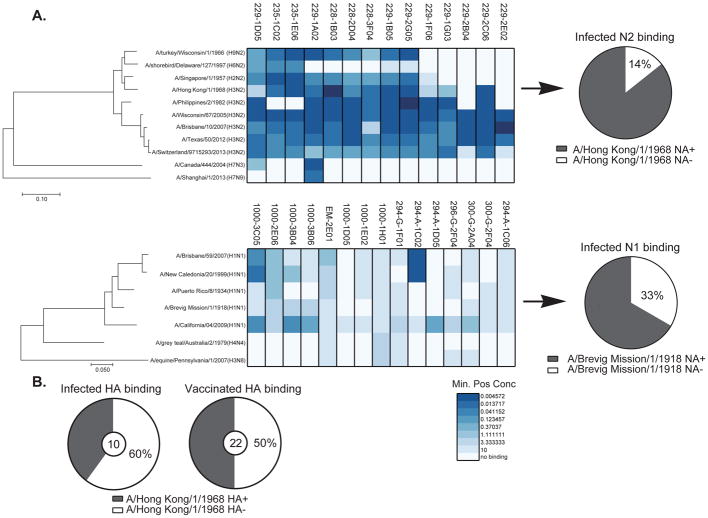
To determine the breadth of binding of the NA-reactive mAbs induced by infection, ELISA was used to test binding against a diverse panel of rNA proteins (Figure 3A). All of the N2-reactive mAbs cross-reacted to all contemporary H3N2 influenza strains, and also a surprising 86% (12 of 14) reacted to the first pandemic H3N2 virus strain known to infect humans (A/Hong Kong/1/1968). Considering that the 1968 H3N2 pandemic virus expressed the NA protein from a reassortant strain with the 1957 H2N2 strain (Scholtissek et al., 1978), it is not surprising that 71% (10 of 14) of the antibodies reacted to the H2N2 influenza strain that circulated since 1957, eleven years prior. By comparison, only 40% of infection-induced and half of the vaccine-induced H3-reactive mAbs were cross-reactive to this 1968 H3N2 strain (Figure 3B). Moreover, 64% (9 of 14) of the N2-reactive mAbs induced by infection were able to bind to avian N2 proteins, including two mAbs with cross-reactivity to heterosubtypic subtypes (N3 and N9) (Figure 3A). Of the N1-reactive mAbs, 67% of cross-reacted to the 1918 pandemic H1N1 strain, 33% reacted to various human H1N1 strains spanning the entire century, plus 20% bound to heterosubtypic strains (Figure 3A). Additionally, demonstrating durability of the NA-epitopes, incubating H3N2 (A/Switzerland/9715293/2013) virus with mAb concentrations up to 250μg did not induce viral escape mutants even after 8 (229-1A02, 229-1D05 and 229-1G03) or 13 passages (228-2D04, 229-2B04 and 229-2C06), an approach that does induce escape to arise from highly conserved HA-stalk mAbs (Anderson et al., 2017). On the whole, the NA-reactive mAbs induced during influenza virus infections are significantly more broadly reactive than antibodies against HA.
Figure 3. NA-reactive mAbs are broadly cross-reactive.
(A) Binding of NA-reactive mAbs to rNA proteins was measured by ELISA. Representative minimum positive concentrations (μg/ml) from three independent experiments are plotted as a heatmap. The different NAs were clustered by amino acid sequence phylogeny. The top panel shows N2-reactive mAbs binding to a panel of NA proteins except for strain A/Switzerland/9715293/2013 (H3N2) which was whole virus. The bottom panel shows N1-reactive mAbs binding to a panel of NA proteins. Pie charts represent the frequency of NA-reactive mAbs binding to historic strains (A/Hong Kong/1/1968 rN2 and A/Brevig Mission/1/1918 rN1).
(B) Binding of 32 HA reactive mAbs isolated from infected or vaccinated subjects to historical past H3N2 strain (A/Hong Kong/1/1968) rH3 were measured by ELISA. Pie charts represent the comparative frequency of HA-reactive mAbs against A/Hong Kong/1/1968 rH3 protein between the infected and vaccinated individuals.
NA-reactive mAbs have broad enzymatic inhibition activity in vitro
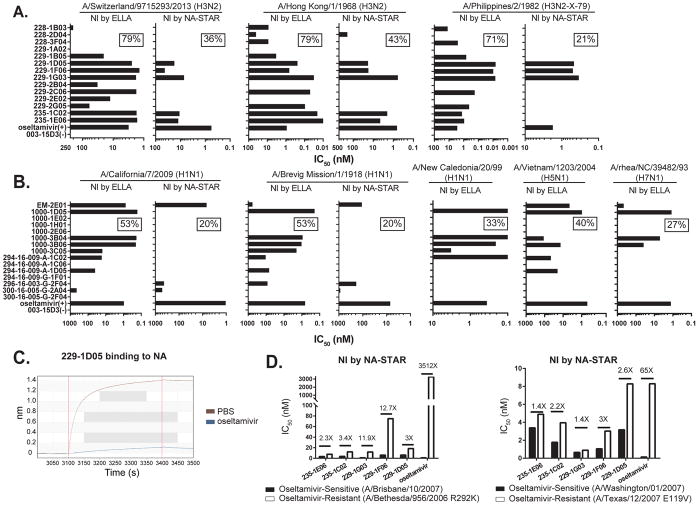
The enzymatic function of NA is to cleave the terminal sialic acid residues allowing viral egress from infected cells. To better access the protective capacity of the NA-reactive mAbs, inhibition of sialic acid cleavage was evaluated using ELLA and NA-STAR assays. For ELLA the sialic acid substrate is found on bulky glycans on the fetuin glycoprotein. If an antibody binds to NA, even outside of the enzymatic site, it sterically blocks access of the enzymatic site to sialic acid, making this assay more sensitive to multiple mechanisms of NI activity. Conversely, the NA-Star assay uses a small, soluble chemiluminescent substrate that easily accesses the enzymatic site unless an antibody binds in close proximity to the enzymatic site. Using ELLA, 79% (11 of 14) of the N2-reactive mAbs inhibited NA activity against A/Switzerland/9715293/2013 (H3N2), of which about half (5 of 14) were also positive in the NA-STAR assay, likely demonstrating activity through blockage or allosteric mechanisms of inhibition. Importantly, by either assay, all of these mAbs inhibited the first pandemic H3N2 strain A/Hong Kong/1/1968 as well as A/Philippines/2/1982 (X-79) (Figure 4A). The antibody 229-1D05 also had broad NI activity against swine influenza virus strain A/swine/Missouri/4296424/2006 (H6N3, H6 is from A/mallard/Sweden/81/2002, PR8 backbone) (Figure S3). Therefore, these mAbs have broad NI activity spanning five decades of H3N2 virus evolution. For mAbs reactive to the 2009 pandemic H1N1 strain, 53% (8 of 15) had NI activity by any means as detected by ELLA, and 20% blocked the enzymatic domain, showing inhibition via the NA-STAR assay. As with the N2-reactive mAbs, N1-reactive mAbs had broad activity against the 1918 pandemic strain A/Brevig Mission/1/1918, against A/New Caledonia/20/99, and against the avian A/Vietnam/1203/2004 (H5N1) and A/rhea/NC/39482/93 (H7N1) strains (Figure 4B).
Figure 4. NA-reactive mAbs exhibit broadly cross-reactive NA–inhibition activity in vitro.
(A) N2-reactive mAbs were tested for inhibiting NA enzymatic activity via ELLA assays and NA-STAR assays against A/Switzerland/9715293/2013 (H3N2), A/Hong Kong/1/1968 (H3N2) and A/Philippines/2/1982 (H3N2-X-79) viruses. Data are represented as half-maximum inhibitory concentration IC50 (nM). Oseltamivir was used as a positive control.
(B) N1-reactive mAbs were tested for inhibiting NA enzymatic activity in ELLA assays and NA-STAR assays against A/California/7/2009 (H1N1) virus, A/Brevig Mission/1/1918 (H1N1) rNA protein, A/New Caledonia/20/99 (H1N1) virus, avian A/Vietnam/1203/2004 (H5N1) virus and A/rhea/NC/39482/93 (H7N1) virus. Data are represented as IC50 (nM). Oseltamivir was used as a positive control.
(C) Binding competition between the N2-reactive mAb 229-1D05 and oseltamivir to A/Texas/50/2012 rNA was measured by bio-layer interferometry.
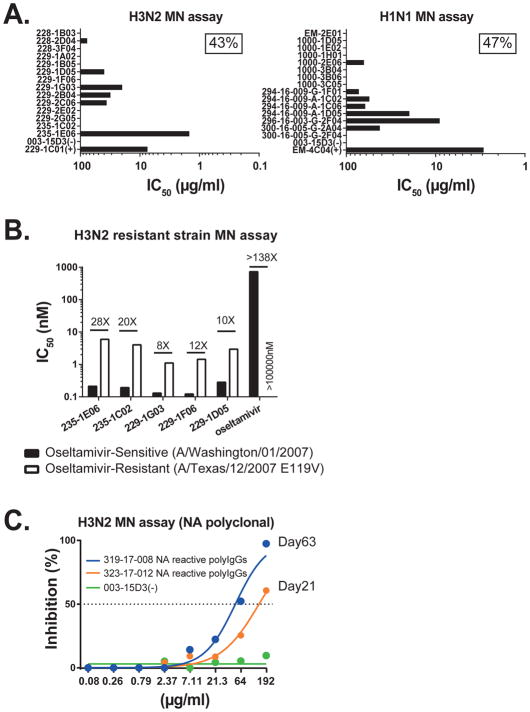
(D) The N2-reactive mAbs were tested for inhibiting NA enzymatic activity by NA-STAR assay against oseltamivir-sensitive strains A/Brisbane/10/2007 (H1N1) and A/Washington/01/2007 (H3N2) and oseltamivir-resistant strains A/Bethesda/956/2006 R292K (H1N1) and A/Texas/12/2007 E119V (H3N2).
Influenza-non-reactive human mAb 003-15D3 is specific for anthrax protective antigen and was used as a negative control in panels A and B and Figure 5.
NA-reactive mAbs may be improved alternatives as therapeutic NA-inhibitor drugs such as oseltamivir, or even more efficacious when efficiently elicited by vaccination. Using bio-layer interferometry, we devised an assay to competitively measure the binding of oseltamivir versus NA-reactive mAbs to the NA protein. Binding of three of the enzymatic domain-targeting mAbs (NA-STAR assay positive, 229-1D05, 229-1F06, and 229-1G03) can be inhibited by prior saturation of NA of an oseltamivir-sensitive strain with oseltamivir (Figures 4C and S4), demonstrating overlap binding of the mAb binding footprints with the pocket occupied by oseltamivir. Because oseltamivir acts by blocking the enzymatic domain, its activity against influenza strains can be assessed by the NA-STAR assay. While oseltamivir had virtually no NI activity, all five of the enzymatic domain-binding mAbs on hand, which is 36% of the N2-reactive mAbs isolated, inhibited the NA activity of typical oseltamivir-resistant strains (A/Bethesda/956/2006 R292K and A/Texas/12/2007 E119V). For 229-1G03 and 235-1E06, the IC50 is nearly identical against the sensitive and resistant A/Texas/12/2007 E119V strains (Figure 4D). These studies demonstrate that the majority of human antibodies against NA analyzed herein can inhibit the enzymatic activity of this protein on highly divergent influenza strains.
NA-reactive human monoclonal and long-term polyclonal antibodies exhibit neutralization activity in vitro
Microneutralization (MN) measures the inhibition of influenza virus replication in vitro, providing another measure of protective activity. Unlike for HA mAbs, this assay underestimates the potency of anti-NA mAbs because the mechanism of action of these antibodies inhibits viral egress, not the initial infection of individual cells. However, MN assays do allow a measure of the frequency of NA antibodies that have protective activity. In total, 45% of the NA reactive mAbs were able to neutralize viruses related to the infecting strain, including; 43% (6 of 14) of the N2-reactive mAbs and 47% (7 of 15) of the N1-reactive mAbs (Figure 5A). The five N2-reactive antibodies that bound the enzymatic site of NA and had NI-activity against oseltamivir-resistant strains were also able to neutralize the resistant A/Texas/12/2007 E119V (H3N2) influenza strain in vitro (Figure 5B). Conversely, oseltamivir had virtually no neutralization activity against this strain. To ensure that the anti-NA antibody response was contributing to long-term serum immunity, we isolated NA-reactive polyclonal antibodies by affinity purification from day 21 and day 63 post-infection serum samples and tested them by using MN assays (Lee et al., 2016). The isolated NA-reactive polyclonal antibodies also readily protected MDCK cells from infection in vitro (Figure 5C). These data show that nearly half of the NA-reactive antibodies characterized herein exhibit neutralization activity, inhibiting virus replication, and suggest that they contribute to long-term serum immunity.
Figure 5. NA-reactive mAbs exhibit neutralization activity in vitro.
(A) NA-reactive mAbs were tested for neutralization by microneutralization (MN) assay using A/Switzerland/9715293/2013 (H3N2) and A/California/7/2009 (H1N1) viruses. Data are represented as IC50 (μg/ml). Positive control mAbs 229-1C01 (anti-H3N2) and EM-4C04 (anti-H1N1) bind HA and neutralize these influenza virus strains.
(B) The N2-reactive mAbs were tested for neutralization by MN assay using A/Washington/01/2007 (oseltamivir-sensitive strain) and A/Texas/12/2007 E119V (oseltamivir-resistant strain) H3N2 viruses. Data are represented as IC50 (nM).
(C) Purified N2 polyclonal antibodies from infected subjects were tested by MN assay against A/Hong Kong/4801/2014 (H3N2) virus.
Data are representative of three independent experiments.
Identification of NA residues crucial for mAb binding and that are disrupted in the inactivated vaccine compositions
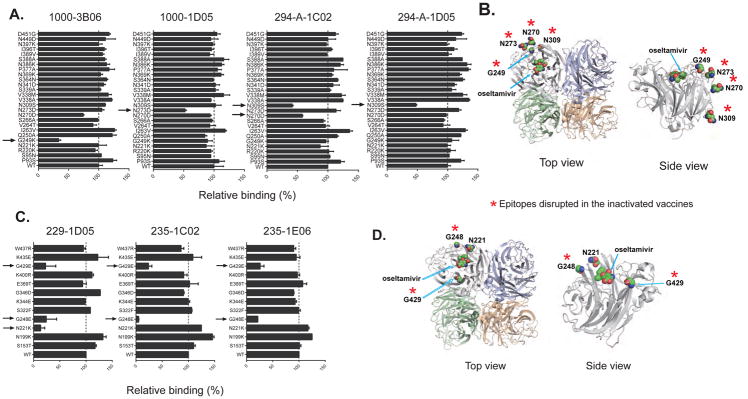
In a previous study, 26 single amino acid escape mutations were identified for common binding sites on N1 (Wan et al., 2013; Wan et al., 2015). To map the epitopes recognized by the N1-reactive mAbs identified herein, these 26 single amino acid mutant A/California/7/2009 N1 proteins were expressed in HEK293. Cell-based ELISAs were carried out to test the binding of all of the NI-active mAbs to the mutant proteins, identifying the binding sites of four antibodies. A G249K mutation significantly affected the binding of 1000-3B06 (70% decrease compared to the wild-type N1). The N273D mutation reduced the binding of 1000-1D05 compared to the wild-type N1 protein. Furthermore, the N309S mutation affected both 294-A-1C02 and 294-A-1D05 binding (Figure 6A). Amino acids N273 and N309 are 99.7% (6835 of 6855 H1 influenza strains) conserved in H1N1 viruses isolated from 1918 until now in the United States. The G249 site is also conserved in H1N1 viruses (90.3%, 6196 of 6855 H1 influenza strains). These residues are all located on the NA head (Figure 6B). To map the epitope(s) targeted by the N2-reactive mAbs, ELISA was used to test the binding avidity of all of the N2-reactive NI-active mAbs against 12 single amino acid mutants of N2 expressed on an A/Minnesota/11/2010 (H6N2-PR8 backbone) purified virus. The 12 amino acids mutated were chosen based on a solved N2-mAb crystal structure and are important for the N2 enzymatic site (Gulati et al., 2002). Three amino acids (N221, G248, and G429) on the NA enzymatic conserved domain are critical for the binding of three of the NI positive N2-reactive mAbs (Figures 6C and 6D). Consistently, all three of these mAbs were also positive in the NA-STAR assay (Figure 4A). Notably, as indicated by the red asterisks in Figures 6B and 6D, these key epitopes on N1 and N2 are disrupted in the inactivated vaccine compositions because the mAbs that bind these sites have substantially reduced or no reactivity to the vaccine.
Figure 6. Identification of critical epitopes targeted by NA-reactive mAbs.
(A) Binding of four N1-reactive mAbs (1000-3B06, 1000-1D05, 294-A-1C02 and 294-A-1D05) to A/California/7/2009 (H1N1) NA mutant proteins transiently expressed on the surface of 293T cells. Hyper-immune mouse serum against A/California/7/2009 (H1N1)-X179A virus was used to verify the expression of NA. Binding to A/California/7/2009 wild type NA is shown in the last bar labeled ‘WT’. Data are represented as mean ± SD. Data are representative of two independent experiments performed in duplicate.
(B) Modeling of N1 was done using PyMOL to show the 4 critical amino acids involved in the binding of the N1-reactive mAbs (PDB: 3TI6) (Vavricka et al., 2011).
(C) Binding of three N2-reactive mAbs (229-1D05, 235-1C02 and 235-1E06) to 12 A/Minnesota/11/2010 (H6N2-PR8 backbone) NA mutant viruses. Data are represented as mean ± SD. Data are representative of two independent experiments performed in duplicate.
(D) Modeling of N2 protein was done using PyMOL to show the three critical amino acid involved in the binding of the N2-reactive mAbs (PDB:4K1J) (Wu et al., 2013).
The mutated sites within epitopes that are also disrupted in the inactivated vaccines (Figure 1), disrupting mAb binding are indicated with red asterisks (mAbs 1000-1D05, 294-A-1C02, 294-A-1D05 did not bind to either Fluarix or Fluzone, and 1000-3B06, 235-1C02 bound poorly).
NA-reactive mAbs protect mice against divergent influenza viruses
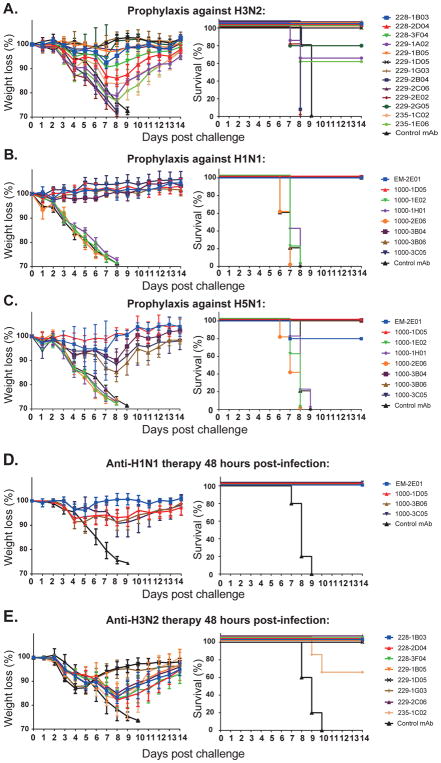
The broad cross-reactivity, as well as widespread in vitro NI activity of NA-reactive mAbs, suggest that they will be broadly protective in vivo. To test this, we measured the prophylactic protection against challenge with divergent strains in vivo. Half-maximal lethal dosages (LD50) of the influenza virus were determined. Mice received 5 mg/kg of NA-reactive mAb or the same dose of a non-binding control mAb by intraperitoneal injection (i.p.). Two hours later, the mice were lethally challenged with 10 LD50 of influenza virus by intranasal inoculation. Recent H3N2 isolates do not replicate well in the mouse model but historical strains like A/Philippines/2/1982 (H3N2, X-79) infect mice readily. This virus is phylogenetically distant from those that caused the human infections from which the mAbs are derived, providing insight into the breadth of protection as well. A selection of N2-reactive mAbs representing all overlapping epitopes were tested. Surprisingly, 84% (11 of 13) of the N2-reactive mAbs showed partial or full protection in the prophylactic challenge experiment against this H3N2 influenza strain that was isolated 35 years ago (Figure 7A). The protection conferred was consistent with the breadth of binding and NI activity of these mAbs. Moreover, non-neutralizing NA-reactive mAbs also provided in vivo prophylactic protection. These data show that neutralizing and non-neutralizing N2-reactive mAbs provide broad prophylactic protection against H3N2 influenza strains in vivo.
Figure 7. NA-reactive mAbs are protective in a prophylactic and therapeutic setting in vivo.
Six week-old female BALB/c mice (5 per experimental condition) were injected intraperitoneally (i.p.) with 5 mg/kg of each NA-reactive mAb individually or with an irrelevant negative control human mAb either 2 h prior to challenge (A–C) or 48 hours after challenge (D–E) with a lethal dose (10 LD50) of virus. The percentage of initial body weight and survival were plotted for each antibody and compared to untreated mice. Data are represented as mean ± SD. Influenza-non-reactive human mAb 003-15D3 (anti-anthrax PA) was used as a negative control in all experiments.
(A) N2-reactive mAb prophylactic protection against A/Philippines/2/1982 (H3N2 - X-79) virus.
(B) N1-reactive mAb prophylactic protection against A/Netherlands/602/2009 virus (pandemic H1N1).
(C) N1-reactive mAbs prophylactic protection against A/Vietnam/1203/2004 (H5N1 - PR8 reassortant) avian influenza virus.
(D) N1-reactive mAbs therapeutic protection from A/Netherlands/602/2009 virus (pandemic H1N1). (E) N2-reactive mAbs therapeutic protection from A/Philippines/2/1982 (H3N2 - X-79) virus.
Variants of the 2009 pandemic H1N1 influenza strains and viruses expressing H5 and N1 from highly pathogenic avian strains are infectious to mice. First, mice treated with N1-reactive mAbs were challenged with a 2009 pandemic H1N1 isolate (A/Netherlands/602/2009). Five out of eight of the mAbs from the 2009–2010 cohort completely protected mice against weight loss and mortality after challenge, whereas mice treated with control mAb lost weight rapidly and were euthanized by day eight post-infection (Figure 7B). Four out of five of the mAbs that prophylactically protected against H1N1 infection (4 of 8 in total) also provided 100% protection from a reverse genetic PR8 variant virus expressing the H5 and N1 of a highly divergent avian influenza virus strain (A/Vietnam/1203/2004, H5N1) (Figure 7C). Thus, half of all mAbs induced against N1 in individuals infected with the 2009 pandemic H1N1 strain provided broad protection against an H5N1 strain. This frequency far exceeded the 10% of HA-reactive mAbs that arose against this H1N1 strain that even bound to H5 (Li et al., 2012; Wrammert et al., 2011). Together, these results suggest that when induced against common infectious influenza virus strains, NA-reactive mAbs are outstanding mediators of broadly protective immunity, even to a virus expressing the H5 and N1 glycoproteins from a divergent avian influenza virus strain with pandemic potential.
The therapeutic efficacy of the NA-reactive mAbs that were protective as prophylactics were also analyzed. Mice that were lethally infected with 10 LD50 of influenza virus were treated with 10 mg/kg of NA-reactive mAbs 48 hours post-infection. All four of the N1-reactive mAbs fully rescued infected mice from severe weight loss and mortality after 2009 pandemic H1N1 influenza virus challenge (Figure 7D). Similarly, 88% (7 of 8) of the N2-reactive mAbs proffered full recovery to the mice challenged with an H3N2 virus (Figure 7E). In sharp contrast, all mice in the control mAb group had to be euthanized around day nine post-infection because of severe weight loss. These results show that the NA-reactive mAbs can be used therapeutically, even after 48 hours of influenza virus infection, suggesting they could be capable alternatives to NA inhibitors such as oseltamivir. With improved vaccine formulations to induce NA antibodies the same benefits as NA-inhibiting drugs could be prophylactically elicited without the need for early administration. Further, unlike NA-inhibiting medications, which lose effectiveness due to the emergence of resistant strains, administration of booster vaccines would control viral resistance.
DISCUSSION
The results presented herein demonstrate that with the right immunogen, NA is capable of inducing a potent, broadly cross-reactive, and protective humoral immune response. In fact, the NA-reactive mAbs were more broadly reactive, the potency of protection and neutralization rivaled that of HA-reactive mAbs. For H3N2 infections there were more NA-reactive than HA-reactive B cells activated. This response is consistent with a recent report that by molar composition, NA is the most immunogenic influenza protein (Angeletti and Yewdell, 2017). The relative conservation of NA epitopes (Sandbulte et al., 2011) also drives a back-boost effect against NAs of historical isolates (Rajendran et al., 2017). In contrast, after vaccination, we find that there is only a 1:87 ratio of NA to HA plasmablasts activated. The NA-reactive mAbs induced by infection reported here have substantially reduced binding to the inactivated vaccines tested, suggesting that the vaccines do not efficiently present important conserved and protective NA epitopes. This observation can be explained by several factors. Firstly, the inactivated influenza vaccines are optimized only for the HA antigen, as the FDA requires that licensed influenza virus vaccines contain at least 15 μg of each HA subtype (Air, 2012). Secondly, antigenic competition between HA and NA may affect the NA humoral immune response (Johansson et al., 1987). However, this mechanism did not appear to preclude the response to NA during infection or to whole virions in mice as reported herein. Thirdly, although influenza vaccine compositions contain varying amounts of NA (Wohlbold et al., 2015), it is unclear if the NA antigen retains its natural tetramer structure, which is important to maintain immunogenicity (Johansson and Cox, 2011). Conversely, during an influenza virus infection, NA replicates along with the virus so that B cells can respond to intact NA on whole virions and infected cells.
The rate of NA antigenic drift is slower than that of HA, which helps to explain the high frequency of broadly cross-reactive antibodies (Sandbulte et al., 2011). The NA-reactive mAbs isolated herein typically cross-bind to heterologous NA proteins from most human influenza A virus strains and a subset also bound to avian H5N1, H7N9 and had some reactivity to H7N3, H4N4, and H3N8 strains. This breadth was evident for the antibodies that were used to map the epitopes. On N1, two of the primary amino acids targeted (N309 and N273) are 99.7% conserved (present in 6835 of 6855 strains) in H1N1 virus from 1918 to 2017 H1N1 strain in the United States (https://www.fludb.org/). Also, N1-reactive mAbs that selected changes at two conserved epitopes (G249 and N273) shared between the human and avian strains were able to mediate prophylactic protection against H5N1 challenge in vivo in mice. Five of the N2-reactive mAbs bind to the conserved enzymatic active site on the head of the NA. The broad reactivity and conservation of the targeted epitopes suggest that NA may be an essential component of universal influenza virus vaccine compositions.
Both NA-inhibiting and non-inhibiting mAbs to N2 protected from influenza virus challenge in vivo. While inhibition of viral release is the typical mechanism of protection, anti-NA antibodies may also mediate protection by other means. For example, several antibodies exhibited high NA-inhibition activity but had low or no microneutralization activity in vitro. However, most that protected in vivo had some degree of NA-inhibiting activity. Thus, the NA-reactive mAbs may alter the functional balance of opposing actions between HA and NA to disrupt efficient viral replication (Benton et al., 2015; Wagner et al., 2002). For non-NI mAbs, there are several mechanisms that likely account for protection. Fc−FcR interactions have been shown to be required for full protection by some NA-reactive mAbs (DiLillo et al., 2016; Henry Dunand et al., 2016; Wohlbold et al., 2017).
Previous studies have shown that infection with influenza virus can induce broader and longer lasting protection than vaccination (Margine et al., 2013a; Nachbagauer et al., 2017; Wrammert et al., 2011). NA inhibiting antibody titers are recognized as a correlate of protection (Clements et al., 1986). Adult influenza virus challenge studies showed that antibodies inhibiting NA but not HA are associated with reduced severity and duration of illness (Memoli et al., 2016). This observation explains why HA and NA inhibiting antibodies are independent correlates of vaccine effectiveness (Monto et al., 2015). We show here that part of such protection is likely mediated by polyclonal NA-reactive antibodies that are not efficiently induced by vaccination. There are some obstacles to exploiting the broadly cross-reactive and protective response to NA for improving influenza virus vaccines. The immunogenicity of NA is strain-dependent (Sultana et al., 2014) and the stability of NAs of each of the vaccine strains differ when subjected to various destabilizing agents. Using recombinant NA that maintains tetrameric form for optimal immunogenicity to induce an NA-based immune response is one solution (Krammer and Palese, 2015). The recombinant proteins used in the work herein were expressed fused to a tetramerization domain to obtain an enzymatically active tetramer (Wohlbold et al., 2015; Wohlbold et al., 2017). This is an important modification since only tetramers are enzymatically active and will display important conformational epitopes (Bucher and Kilbourne, 1972; Paterson and Lamb, 1990). It is challenging to keep the native structure of NA within vaccine formulations (Brett and Johansson, 2006; Eichelberger and Wan, 2015). Another obvious solution is the use of live-attenuated vaccines that express NA on their surface and the surface of infected cells. The findings herein provide a strong impetus to optimize NA content and structural integrity in influenza vaccines.
In conclusion, NA-reactive antibodies can be readily or even dominantly induced, protecting at levels comparable to HA-reactive antibodies. While particular antibodies to HA have been isolated with immense breadth of activity, on a per antibody basis and based on the sampling herein, those to NA appear to have greater breadth than antibodies to HA. The data presented suggest that inclusion of an improved NA component to future influenza vaccine compositions would reduce the severity of infections. With a robust response to NA the degree of protection conferred might protect from any influenza infection occurring at all, and possibly provide broad-ranging protection against potential pandemic strains that express N1 or N2 NAs.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to the Lead Contact, Dr. Patrick C. Wilson (wilsonp@uchicago.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse Models
All mouse experiments were performed in accordance with and with approval of The University of Chicago and the Icahn School of Medicine at Mount Sinai Institutional Animal Care and Use Committees. Six to eight weeks old naive healthy female wild type BALB/c (Strain Code: JAX 000651) mice were purchased from the Jackson Laboratory for these experiments only. Five mice were housed by group in standardized cages with unrestricted access to food and water.
Tissue culture
Madin-Darby canine kidney (MDCK) cells was obtained from the American Type Culture Collection (ATCC), which originally derived from dog kidney. MDCK cells were maintained in culture at 37°C with 5% CO2 in Dulbecco’s minimal essential medium (DMEM, GIBCO) supplemented with 10% fetal bovine serum (GIBCO), 2 mM GlutaMAX (GIBCO), penicillin and streptomycin (100 μg/ml; GIBCO). Human embryo kidney (HEK) 293T cell line was obtained from Thermo Scientific. HEK293T cells originally derived from female fetal cells. The 293T cells were maintained at 37°C with 5% CO2 in Advanced DMEM culture with 2% Ultra-Low IgG FBS (GIBCO), 2 mM GlutaMAX (GIBCO), plus penicillin and streptomycin (100 μg/ml; GIBCO). The cell lines purchased from ATCC and Thermo Scientific are accompanied by authentication documents verifying the identity according to their short tandem repeat profiles and that they are mycoplasma free.
Study approvals and cohorts
All studies were performed with the approval of Emory University, Rochester University, and The University of Chicago institutional review boards. Patients were only included if they had no other concurrent infections and were not being treated with immunosuppressive therapies. The Emory cohort was recruited for a previous study in 2009 for which we published on the HA response (Wrammert et al., 2011). Patient clinical information is detailed in Table S1. Confirmed PCR influenza virus infected patients were recruited. For the human vaccination study, two hundred and fifty-eight mAbs were isolated from 37 influenza vaccinated individuals, including from 19 subjects that received the subunit vaccine (mAbs per subject n=13, 10, 18, 5, 5, 10, 11, 4, 7, 4, 1, 4, 1, 26, 1, 2, 4, 6, 1), 8 subjects that received the split vaccine (mAbs per subject n=4, 11, 8, 4, 2, 1, 2, 4) and 10 subjects received the monovalent vaccine (mAbs per subject n= 7, 4, 8, 5, 11, 4, 11, 14, 11, 14). These cohorts were from previously published studies in our laboratory (Andrews et al., 2015; Wrammert et al., 2008).
METHOD DETAILS
Viruses and recombinant proteins
All influenza virus stocks used for the assays were freshly grown in specific pathogen free (SPF) eggs, harvested, purified and titered. Egg grown virus can accumulate alterations due to egg-selected mutations and different glycosylation. As all mAbs to HA and NA were also verified to bind to recombinant proteins without loss of activity we feel that egg-specific alterations were not a compounding factor for the studies herein. A reassortant H6N2 virus with the backbone from A/Puerto Rico/8/34 (PR8) containing the HA gene of A/turkey/Massachusetts/3740/76 and the NA from A/Minnesota/11/2010 was used to generate the mutant viruses (S153T, N199K, N221K, G248E, S322F, K344E, G346D, E369T, K400R, G429E, K435E and W437R single mutation in the NA gene). A/Switzerland/9715293/2013 (H3N2) was treated with 0.02% methanol free formaldehyde for 48h at 4 °C to generate the inactivated virus particles. The inactivation was verified by injecting treated virus into eggs followed by HA measurements. Recombinant NA proteins derived from A/Puerto Rico/8/1934 (H1N1), A/New Caledonia/20/1999 (H1N1), A/Brisbane/59/2007 (H1N1), A/California/7/2009 (H1N1), A/grey teal/Australia/2/1979 (H4N4), A/Shanghai/1/2013 (H7N9), A/equine/Pennsylvania/1/2007 (H3N8), A/turkey/Wisconsin/1/1966 (H9N2), A/Wisconsin/67/2005 (H3N2) and A/Brisbane/10/2007 (H3N2) were obtained from BEI resources and A/Canada/444/2004 (H7N3) N3 NA was obtained from the Influenza Reagent Resource (IRR). The other recombinant NA and HA proteins were expressed in house in the baculovirus expression system as previously described (Margine et al., 2013b).
Monoclonal antibodies
Antibodies were generated as previously described (Smith et al., 2009; Wrammert et al., 2008). Briefly, peripheral blood was obtained from each subject at or near diagnosis of infection or 7 days after vaccination. Lymphocytes were isolated and enriched for B cells using RosetteSep. Plasmablasts (CD3− CD19+ CD20low CD27hi CD38hi) were single cell-sorted into 96-well plates. Immunoglobulin variable genes from plasmablasts were amplified by reverse transcriptase polymerase chain reaction (RT-PCR) and sequenced, then cloned into human IgG1 expression vectors and co-transfected into HEK293 cells. Secreted mAbs were purified from the supernatant using protein A beads.
Enzyme linked immunosorbent assay (ELISA)
High-protein binding microtiter plates (Costar) were coated with 8 hemagglutinating units (HAU) of whole virus per well or recombinant NAs or HAs at 1μg/ml in phosphate buffered saline (PBS) overnight at 4°C. After blocking, serially diluted antibodies 1:3 starting at 10μg/ml were incubated for 1 h at 37°C. Horse radish peroxidase (HRP)-conjugated goat anti-human IgG antibody diluted 1:1000 (Jackson Immuno Research) was used to detect binding of mAbs, and was developed with Super Aquablue ELISA substrate (eBiosciences). Absorbance was measured at 405nm on a microplate spectrophotometer (BioRad). To standardize the assays, antibodies with known binding characteristics were included on each plate and the plates were developed when the absorbance of the control reached 3.0 OD units. To ensure the NA and HA components were identical and maintained integrity, analyses of mAb binding to vaccines was done on vaccines not expired and with matching influenza virus strains to those causing infection. Competition ELISAs were performed by inhibiting binding of each biotinylated antibody of interest at the half-maximal binding concentration with a 10-fold molar excess of competitor antibody. HRP conjugated streptavidin diluted 1:1000 (Southern Biotech) was used for detection. Plates were developed until samples in the absence of competitor antibody reached an OD of 1 (Henry Dunand et al., 2015).
ELISPOT assay
Filter plates (96-well; Millipore) were incubated overnight at 4°C with anti-human IgG, IgA, and IgM (KPL), 2μg/ml recombinant HAs, 2μg/ml recombinant NAs or 32 HAU/well of whole virus. The plates were washed and blocked by incubation with RPMI containing 10% FBS at 37°C for 2 hours. Freshly isolated human PBMCs or mouse spleen cells were washed three times and were re-suspended in medium (RPMI medium supplemented with 4 mM L-glutamine, 10 mM HEPES, 100 U/ml Pen/Step, 10% FBS). The cells (0.5–1-10E6) underwent 2-fold serial dilutions before being transferred to the ELISPOT plates, which were incubated overnight at 37°C. The plates were washed extensively with PBS and PBS–0.05% Tween before antibody from the antibody-secreting cells (ASC) were detected with either 1:1000 biotinylated anti-human IgG (Mabtech) and biotinylated anti-human IgA (Southern Biotech) or a 1:10000 biotinylated anti-mouse IgG. After a 2 hour incubation at room temperature (RT), a 1:500 dilution of streptavidin-alkaline phosphatase (Southern Biotech) was added to each well. The incubation was repeated before ASC spots were revealed with nitroblue tetrazolium/5-bromo-4-chloro-3-indolylphosphate (NBT/BCIP; Thermo Scientific). The developed plates were scanned using an automated ELISPOT counter (Cellular Technologies, Ltd.).
NA enzyme linked lectin assay (ELLA)
ELLAs were performed as described (Westgeest et al., 2015). Flat-bottom nonsterile 96-well plates (Thermo Scientific) were coated with 100 μl of fetuin (Sigma) at 25 μg/ml at 4°C overnight. 50 μl antibodies were serially diluted (twofold) in Dulbecco’s phosphate-buffered saline (DPBS) containing 0.133 g/L CaCl2 and 0.1 g/L MgCl2 with 0.05% Tween 20 and 1% BSA (DPBSTBSA), then incubated in duplicate fetuin-coated plates with an equal volume of the selected antigen dilution in DPBSTBSA. These plates were subsequently sealed and incubated for 18 h at 37°C. The plates were subsequently washed six times with PBS with 0.05% Tween 20, and 100 μl/well of HRP-conjugated peanut agglutinin lectin (PNA-HRPO, Sigma–Aldrich) in DPBSTBSA was added for 2h at RT in the dark. The plates were washed six times and were developed with Super Aquablue ELISA substrate (eBiosciences). Absorbance was read at 405nm on a microplate spectrophotometer (BioRad). Data points were analyzed using Prism software and the 50% inhibition concentration (IC50) was defined as concentration at which 50% of the NA activity was inhibited compared to the negative control.
NA-STAR assay
The NA-STAR assay was performed according to the Resistance Detection Kit manufacturer’s instructions (Applied Biosystems, Darmstadt, Germany) (Nguyen et al., 2010). Briefly, 25 μl test mAbs in serial two fold dilutions in NA-Star assay buffer (26 mM 2-(N-morpholino) ethanesulfonic acid ; 4 mM calcium chloride; pH 6.0) were mixed with 25 μl of NA protein or 4 X IC50 of virus and incubated at 37°C for 20 min. After adding 10 μl of 1000× diluted NA-Star substrate, the plates were incubated at room temperature for 30 min. The reaction was stopped by adding 60 μl of NA Star accelerator. The chemiluminescent was determined by using the DTX 880 plate reader (Beckman Coulter). Data points were analyzed using Prism software and the 50% inhibition concentration (IC50) was defined as concentration at which 50% of the NA activity was inhibited compared to the negative control.
Competition studies using bio-layer interferometry
A fortéBio Octet K2 instrument was used to measure the competition between the N2-reactive mAbs and oseltamivir. A/Texas/50/2012 rNA (5 μg/ml) in PBS was used to load anti-His probes for 300 s, then the probes were moved to oseltamivir (25 μg/ml) and control PBS for another 300 s, and following by binding of the complex to the N2-reactive mAbs (50 μg/ml) for 300 s to 500 s. The final volume for all the solutions was 200 μl/well. All of the assays were performed with agitation set to 1,000 r.p.m. in PBS buffer supplemented with 1% BSA to minimize nonspecific interactions at 30 °C.
Microneutralization assay (MN)
MN assay for antibody characterization was carried out as previously described (Henry Dunand et al., 2015). Briefly, MDCK cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) at 37°C with 5% CO2. On the day before the experiment, confluent MDCK cells in a 96-well format were washed twice with PBS and incubated in minimal essential medium (MEM) supplemented with 1 μg/ml trypsin-ethylenediamine tetraacetic acid (EDTA). Serial 2-fold dilutions (starting concentration 128 μg/ml) of mAb were mixed with an equal volume of 100 50% tissue culture infectious doses (TCID50) virus and incubated for 1 h at 37°C. The mixture was removed and cells were cultured for 20 h at 37°C with 1X MEM supplemented with 1 μg/ml trypsin-TPCK and appropriate mAb concentration. Cells were washed twice with PBS, fixed with 80% ice cold acetone at −20°C for 1 h, washed 3 times with PBS, blocked for 30 min with 10% FBS and then treated for 30 min with 2% H2O2. An anti-NP-biotinylated antibody (1:3000) in 3% BSA-PBS was incubated for 1h at room temperature. The plates were developed with Super Aquablue ELISA substrate at 405 nm. The signal from uninfected wells were averaged to represent 100% inhibition. Virus infected wells without mAb were averaged to represent 0% inhibition. Duplication wells were used to calculate the mean and SD of neutralization, and inhibitory concentration 50 (IC50) was determined by a sigmoidal dose response curve. The inhibition ratio (%) was calculated as below:
The final concentration of antibody that reduced infection to 50% (IC50) was determined using Prism software (GraphPad).
Purification of NA-reactive IgG from serum
Each serum sample analyzed in this study was passed through a 5 ml Protein G Plus agarose (Pierce) affinity column in gravity mode. Serum flow-through was collected and passed through the column three times. The column was then washed with 15 column volume (CV) of PBS prior to elution with 5 CV of 100 mM glycine-HCl, pH 2.7. The eluate, containing total IgG from serum, was immediately neutralized with 5 ml of 1 M Tris-HCl, pH 8.0. The flow-through was subjected to the same purification process one more time to capture all IgG from serum, and the two eluates were combined. To isolate the NA-reactive IgG, recombinant N2 neuraminidase (rNA) from A/Hong Kong/4801/2014 was first biotinylated using the EZ-link Sulfo-NHS-Biotin (Thermo Scientific) according to the methods provided by the manufacturers. Biotinylated rNA was then bound to NeutrAvidin agarose resins (Pierce) packed into a 0.5 ml chromatography column (Clontech). The resins were equilibrated with 10 CV of PBS. Total IgG was applied to a column packed with Neutravidin agarose resins only, and flow-through was collected in order to remove any resin-binding IgGs. The collected samples were then subjected to the affinity column with rNA in gravity mode, and flow-through was collected and reapplied to the column three times. The column was washed with 10 CV of PBS and eluted with 5 CV of 100 mM glycine-HCl, pH 2.7 and immediately neutralized with 1 M Tris-HCl, pH 8.0. The flow-through from each pull-down was subjected to the same purification process until all of NA-reactive IgGs were isolated. All eluate samples from each donor were combined, then buffer-exchanged into PBS and concentrated using a 30kDa Vivaspin 15 centrifuge tube (Sartorius).
Cell-based ELISA
A/California/7/2009 NA and its mutants were expressed on 293T cells by transfecting with wild type or mutant pCAGGS-CA/09NA plasmids using Lipofectamine 2000 reagent (Invitrogen). ELISA was performed with the transfected cells as described previously (Wan et al., 2013). For all other NAs (mutant and wild type), the signals generated by mAb binding to each NA were normalized to those generated by hyper-immune mouse serum against A/California/7/2009(H1N1)-X179A virus (the background signals generated with mock-transfected cells were subtracted from both the mAb and mouse serum signals) and therefore expressed as relative binding.
Mouse challenge and immunization experiments
To analyze targeting of NA epitopes in a response unbiased by pre-existing immunity in mice, mice were infected with H1N1 or immunized with whole killed H3N2. As only the H1N1 strain infects mice, it was used to examine NA frequency after live virus challenge and the H3N2 strain that does not infect mice was used to assay the inactivated whole virus response. For the immunization assays, mice were infected by 0.25 LD50 of A/Netherlands/602/2009 (H1N1) or immunized with 2 μg of inactivated A/Switzerland/9715293/2013 (H3N2) influenza virus intranasally and boosted on day 30 using the same immunogens/doses. Mice serum and spleen cells were collected on day 38 and analyzed for the HA and NA humoral immune response by ELISA and ELISPOT, respectively. Mice serum were diluted and antibodies were detected by HRP-conjugated goat anti-mouse IgG (1:1000). End point titers were extrapolated from sigmoidal 4PL (where X is log concentration) standard curve. The threshold for end point titers is the mean plus 4 S.D. of naïve mouse sera (Graphpad). In prophylactic studies, five female BALB/c mice (The Jackson Laboratory) per group aged 6 to 8 weeks received a 5 mg/kg dose of mAbs intraperitoneally (i.p.). After 2 h treatment, the mice were anesthetized using a ketamine-xylazine mixture and intranasally infected with 10X the 50% lethal dose (LD50) of A/Netherlands/602/2009 (H1N1), A/Philippines/2/1982 (H3N2, X-79 - surface glycoproteins from A/Philippines/2/1982 and backbone from A/PR/8/34) or A/Vietnam/1203/2004 (H5N1 - surface glycoproteins from A/Vietnam/1203/2004 and backbone from A/PR/8/34, polybasic cleavage site replaced with a regular cleavage site). In a therapeutic setting, mice received a 10 mg/kg dose of each mAbs i.p. 48 h after 10 LD50 virus intranasal inoculation (in a 30 μl inoculum). In all groups, mice were monitored daily for survival and weight loss until day 14 post-infection. Mice that lost 25% or more of their initial body weights were euthanized.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis
All statistical analysis was performed using Prism software (Graphpad Version 7.0). Sample sizes (n) of animals, number of biological repeats of experiments and specific tests for statistical significance used are indicated in the corresponding figure legends. P values equal to or less than 0.05 were considered significant.
Supplementary Material
(A–B) The usage of VH immunoglobulin genes by (A) NA-reactive B cells and (B) HA-reactive B cells
(C) CDR3 length of NA and HA-reactive mAbs, data are represented as mean ± SD.
(D) Total heavy chain mutation number of NA and HA-reactive mAbs based on analysis using the NCBI IgBlast tool (https://www.ncbi.nlm.nih.gov/igblast/), data are represented as mean ± SD.
Several representative HA-reactive or NA-reactive mAbs were tested for binding by ELISA to A/Switzerland/9715293/2013 virus or inactivated A/Switzerland/9715293/2013 virus. (A) HA-reactive mAbs (spots color in red are the HA-stalk reactive mAbs). (B) NA-reactive mAbs (spots colored in red are the NA-star positive mAbs binding around the enzymatic site in Figure 6C). Data are representative of two independent experiments, data are represented as median.
(A) Cross N2-reactive mAbs were tested for inhibiting NA enzymatic activity via ELLA assay against A/swine/Missouri/4296424/2006 (H6N3, H6 is from A/mallard/Sweden/81/2002, PR8 backbone) (B) Cross N1-reactive mAbs were tested for inhibiting NA enzymatic activity via ELLA assay against A/mallard/Sweden/24/2002 (H9N4, H9 is from A/guinea fowl/Hong Kong/WF10/99, PR8 backbone) (C) Cross N1-reactive mAbs were tested for inhibiting NA enzymatic activity via ELLA assay against A/Northern shoveler/Alaska/7MP1708/07 (H3N8). Bar graphs represent IC50 (nM) values. Oseltamivir was used as a positive control and influenza-non-reactive mAb 003-15D3 was used as a negative control in the assays. Data are representative of two independent experiments.
(A) 229-1F06 (B) 229-1G03 (C) 235-1C02 (D) 235-1E06 (E) 229-2C06. Data are representative of three independent experiments.
Highlights.
Flu virus infection induces many neuraminidase (NA)-reactive B cells, which make important antibodies against viral NA
NA antibodies have broad cross-reactivity and inhibit neuraminidase enzyme activity
Current flu vaccines poorly display key NA epitopes, and do not produce NA antibodies
NA-reactive antibodies offer protection against lethal flu virus infection
Acknowledgments
This project was funded in parts from the National Institute of Allergy and Infectious Disease, National Institutes of Health grant numbers U19AI082724 (PCW), U19AI109946 (PCW), U19AI057266 (PCW), U19AI109946 (FK) and the NIAID Centers of Excellence for Influenza Research and Surveillance (CEIRS, HHSN272201400005C, PCW, DJT and JT, and HHSN272201400008C, FK), a CEIRS training grant to (TJW), a by the FDA (JW and MCE). We especially thank all study participants. We thank Jenna Guthmiller for critical comments. We thank Daniel Kaplan and Paul Bunduc for technical assistance. We thank St. Jude Children’s Research Hospital for providing plasmids that were used to generate the reassortant viruses.
Footnotes
AUTHOR CONTRIBUTIONS
Y.C. designed and performed experiments, analyzed data, and wrote the manuscript. T.J.W. expressed NA proteins, performed prophylactic mice experiments and ELLA assays. N.Y.Z purified influenza viruses, provided materials and experimental assistance. M.H. performed mAb cloning. Y.H. expressed mAbs, K.E.N. assisted with PBs sorting and PCR cloning. J.L. and G.G. purified the NA reactive polyIgGs. H.W. and M.C.E. provided the mutant NA systems. K.T.R. patient recruitment and sample collection. E.K. ELLA and escape mutant assays. C.H. provided samples and experimental assistance. A.E.P. and C.T.S. assisted with mice experiments. D.J.T., J. T., J.W. and R.A. provided samples. T.J.W. K.E.N., C.H., A.E.P., D.J.T., M.C.E. and F.K. manuscript editing. F.K. provided rHA and rNA proteins, designed, supervised the project, analyzed data and edited the manuscript. P.C.W. conceived of the project, supervised the work, analyzed data, and wrote the manuscript.
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abed Y, Hardy I, Li Y, Boivin G. Divergent evolution of hemagglutinin and neuraminidase genes in recent influenza A:H3N2 viruses isolated in Canada. J Med Virol. 2002;67:589–595. doi: 10.1002/jmv.10143. [DOI] [PubMed] [Google Scholar]
- Air GM. Influenza neuraminidase. Influenza Other Respir Viruses. 2012;6:245–256. doi: 10.1111/j.1750-2659.2011.00304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson CS, Ortega S, Chaves FA, Clark AM, Yang H, Topham DJ, DeDiego ML. Natural and directed antigenic drift of the H1 influenza virus hemagglutinin stalk domain. Sci Rep. 2017;7:14614. doi: 10.1038/s41598-017-14931-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews SF, Huang Y, Kaur K, Popova LI, Ho IY, Pauli NT, Henry Dunand CJ, Taylor WM, Lim S, Huang M, et al. Immune history profoundly affects broadly protective B cell responses to influenza. Sci Transl Med. 2015;7:316ra192. doi: 10.1126/scitranslmed.aad0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeletti D, Yewdell JW. Is It Possible to Develop a “Universal” Influenza Virus Vaccine? Outflanking Antibody Immunodominance on the Road to Universal Influenza Vaccination. Cold Spring Harb Perspect Biol. 2017 doi: 10.1101/cshperspect.a028852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton DJ, Martin SR, Wharton SA, McCauley JW. Biophysical measurement of the balance of influenza a hemagglutinin and neuraminidase activities. J Biol Chem. 2015;290:6516–6521. doi: 10.1074/jbc.M114.622308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett IC, Johansson BE. Variation in the divalent cation requirements of influenza A virus N1 neuraminidases. J Biochem. 2006;139:439–447. doi: 10.1093/jb/mvj051. [DOI] [PubMed] [Google Scholar]
- Bucher DJ, Kilbourne ED. A 2 (N2) neuraminidase of the X-7 influenza virus recombinant: determination of molecular size and subunit composition of the active unit. J Virol. 1972;10:60–66. doi: 10.1128/jvi.10.1.60-66.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements ML, Betts RF, Tierney EL, Murphy BR. Serum and nasal wash antibodies associated with resistance to experimental challenge with influenza A wild-type virus. J Clin Microbiol. 1986;24:157–160. doi: 10.1128/jcm.24.1.157-160.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharan NJ, Gubareva LV, Meyer JJ, Okomo-Adhiambo M, McClinton RC, Marshall SA, St George K, Epperson S, Brammer L, Klimov AI, et al. Infections with oseltamivir461 resistant influenza A(H1N1) virus in the United States. JAMA. 2009;301:1034–1041. doi: 10.1001/jama.2009.294. [DOI] [PubMed] [Google Scholar]
- DiLillo DJ, Palese P, Wilson PC, Ravetch JV. Broadly neutralizing anti-influenza antibodies require Fc receptor engagement for in vivo protection. The Journal of clinical investigation. 2016;126:605–610. doi: 10.1172/JCI84428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle TM, Hashem AM, Li C, Van Domselaar G, Larocque L, Wang J, Smith D, Cyr T, Farnsworth A, He R, et al. Universal anti-neuraminidase antibody inhibiting all influenza A subtypes. Antiviral Res. 2013;100:567–574. doi: 10.1016/j.antiviral.2013.09.018. [DOI] [PubMed] [Google Scholar]
- Eichelberger MC, Wan H. Influenza neuraminidase as a vaccine antigen. Curr Top Microbiol Immunol. 2015;386:275–299. doi: 10.1007/82_2014_398. [DOI] [PubMed] [Google Scholar]
- Flannery B. Interim estimates of 2016–17 seasonal influenza vaccine effectiveness— United States, February 2017. MMWR Morbidity and Mortality Weekly Report. 2017:66. doi: 10.15585/mmwr.mm6606a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genentech, I. TAMIFLU (R) (oseltamivir phosphate) prescribing. San Francisco, CA: 2016. https://www.gene.com/download/pdf/tamiflu_prescribing.pdf. [Google Scholar]
- Gulati U, Hwang CC, Venkatramani L, Gulati S, Stray SJ, Lee JT, Laver WG, Bochkarev A, Zlotnick A, Air GM. Antibody epitopes on the neuraminidase of a recent H3N2 influenza virus (A/Memphis/31/98) J Virol. 2002;76:12274–12280. doi: 10.1128/JVI.76.23.12274-12280.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry Dunand Carole J, Leon Paul E, Huang M, Choi A, Chromikova V, Ho Irvin Y, Tan Gene S, Cruz J, Hirsh A, Zheng NY, et al. Both Neutralizing and Non-Neutralizing Human H7N9 Influenza Vaccine-Induced Monoclonal Antibodies Confer Protection. Cell Host & Microbe. 2016;19:800–813. doi: 10.1016/j.chom.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry Dunand CJ, Leon PE, Kaur K, Tan GS, Zheng NY, Andrews S, Huang M, Qu X, Huang Y, Salgado-Ferrer M, et al. Preexisting human antibodies neutralize recently emerged H7N9 influenza strains. The Journal of clinical investigation. 2015;125:1255–1268. doi: 10.1172/JCI74374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuliano AD, Roguski KM, Chang HH, Muscatello DJ, Palekar R, Tempia S, Cohen C, Gran JM, Schanzer D, Cowling BJ, et al. Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet. 2017 doi: 10.1016/S0140-6736(17)33293-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson BE, Cox MM. Influenza viral neuraminidase: the forgotten antigen. Expert Rev Vaccines. 2011;10:1683–1695. doi: 10.1586/erv.11.130. [DOI] [PubMed] [Google Scholar]
- Johansson BE, Moran TM, Kilbourne ED. Antigen-presenting B cells and helper T cells cooperatively mediate intravirionic antigenic competition between influenza A virus surface glycoproteins. Proc Natl Acad Sci U S A. 1987;84:6869–6873. doi: 10.1073/pnas.84.19.6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karron RA, Collins PL. Fields Virology: Sixth Edition. Wolters Kluwer Health Adis (ESP); 2013. Parainfluenza viruses. [Google Scholar]
- Krammer F, Palese P. Advances in the development of influenza virus vaccines. Nat Rev Drug Discov. 2015;14:167–182. doi: 10.1038/nrd4529. [DOI] [PubMed] [Google Scholar]
- Lee J, Boutz DR, Chromikova V, Joyce MG, Vollmers C, Leung K, Horton AP, DeKosky BJ, Lee CH, Lavinder JJ, et al. Molecular-level analysis of the serum antibody repertoire in young adults before and after seasonal influenza vaccination. Nat Med. 2016;22:1456–1464. doi: 10.1038/nm.4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GM, Chiu C, Wrammert J, McCausland M, Andrews SF, Zheng NY, Lee JH, Huang M, Qu X, Edupuganti S, et al. Pandemic H1N1 influenza vaccine induces a recall response in humans that favors broadly cross-reactive memory B cells. Proc Natl Acad Sci U S A. 2012;109:9047–9052. doi: 10.1073/pnas.1118979109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margine I, Hai R, Albrecht RA, Obermoser G, Harrod AC, Banchereau J, Palucka K, Garcia-Sastre A, Palese P, Treanor JJ, et al. H3N2 influenza virus infection induces broadly reactive hemagglutinin stalk antibodies in humans and mice. Journal of virology. 2013a;87:4728–4737. doi: 10.1128/JVI.03509-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margine I, Palese P, Krammer F. Expression of functional recombinant hemagglutinin and neuraminidase proteins from the novel H7N9 influenza virus using the baculovirus expression system. J Vis Exp. 2013b:e51112. doi: 10.3791/51112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrosovich MN, Matrosovich TY, Gray T, Roberts NA, Klenk HD. Neuraminidase is important for the initiation of influenza virus infection in human airway epithelium. Journal of virology. 2004;78:12665–12667. doi: 10.1128/JVI.78.22.12665-12667.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memoli MJ, Shaw PA, Han A, Czajkowski L, Reed S, Athota R, Bristol T, Fargis S, Risos K, Powers JH, et al. Evaluation of Antihemagglutinin and Antineuraminidase Antibodies as Correlates of Protection in an Influenza A/H1N1 Virus Healthy Human Challenge Model. MBio. 2016;7:e00417–00416. doi: 10.1128/mBio.00417-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monto AS, Kendal AP. Effect of neuraminidase antibody on Hong Kong influenza. Lancet. 1973;1:623–625. doi: 10.1016/s0140-6736(73)92196-x. [DOI] [PubMed] [Google Scholar]
- Monto AS, Petrie JG, Cross RT, Johnson E, Liu M, Zhong W, Levine M, Katz JM, Ohmit SE. Antibody to Influenza Virus Neuraminidase: An Independent Correlate of Protection. J Infect Dis. 2015;212:1191–1199. doi: 10.1093/infdis/jiv195. [DOI] [PubMed] [Google Scholar]
- Murphy BR, Kasel JA, Chanock RM. Association of serum anti-neuraminidase antibody with resistance to influenza in man. N Engl J Med. 1972;286:1329–1332. doi: 10.1056/NEJM197206222862502. [DOI] [PubMed] [Google Scholar]
- Nachbagauer R, Choi A, Hirsh A, Margine I, Iida S, Barrera A, Ferres M, Albrecht RA, Garcia-Sastre A, Bouvier NM, et al. Defining the antibody cross-reactome directed against the influenza virus surface glycoproteins. Nat Immunol. 2017;18:464–473. doi: 10.1038/ni.3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HT, Sheu TG, Mishin VP, Klimov AI, Gubareva LV. Assessment of pandemic and seasonal influenza A (H1N1) virus susceptibility to neuraminidase inhibitors in three enzyme activity inhibition assays. Antimicrob Agents Chemother. 2010;54:3671–3677. doi: 10.1128/AAC.00581-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichol KL. Efficacy and effectiveness of influenza vaccination. Vaccine. 2008;26(Suppl 4):D17–22. doi: 10.1016/j.vaccine.2008.07.048. [DOI] [PubMed] [Google Scholar]
- Palese P, Compans R. Inhibition of influenza virus replication in tissue culture by 2-deoxy-2, 3-dehydro-N-trifluoroacetylneuraminic acid (FANA): mechanism of action. Journal of General Virology. 1976;33:159–163. doi: 10.1099/0022-1317-33-1-159. [DOI] [PubMed] [Google Scholar]
- Paterson RG, Lamb RA. Conversion of a class II integral membrane protein into a soluble and efficiently secreted protein: multiple intracellular and extracellular oligomeric and conformational forms. J Cell Biol. 1990;110:999–1011. doi: 10.1083/jcb.110.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran M, Nachbagauer R, Ermler ME, Bunduc P, Amanat F, Izikson R, Cox M, Palese P, Eichelberger M, Krammer F. Analysis of Anti-Influenza Virus Neuraminidase Antibodies in Children, Adults, and the Elderly by ELISA and Enzyme Inhibition: Evidence for Original Antigenic Sin. mBio. 2017;8:e02281–02216. doi: 10.1128/mBio.02281-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandbulte MR, Westgeest KB, Gao J, Xu X, Klimov AI, Russell CA, Burke DF, Smith DJ, Fouchier RA, Eichelberger MC. Discordant antigenic drift of neuraminidase and hemagglutinin in H1N1 and H3N2 influenza viruses. Proc Natl Acad Sci U S A. 2011;108:20748–20753. doi: 10.1073/pnas.1113801108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholtissek C, Koennecke I, Rott R. Host range recombinants of fowl plague (influenza A) virus. Virology. 1978;91:79–85. doi: 10.1016/0042-6822(78)90356-2. [DOI] [PubMed] [Google Scholar]
- Schulman JL, Khakpour M, Kilbourne ED. Protective effects of specific immunity to viral neuraminidase on influenza virus infection of mice. Journal of virology. 1968;2:778–786. doi: 10.1128/jvi.2.8.778-786.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K, Garman L, Wrammert J, Zheng NY, Capra JD, Ahmed R, Wilson PC. Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nature protocols. 2009;4:372–384. doi: 10.1038/nprot.2009.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultana I, Yang K, Getie-Kebtie M, Couzens L, Markoff L, Alterman M, Eichelberger MC. Stability of neuraminidase in inactivated influenza vaccines. Vaccine. 2014;32:2225–2230. doi: 10.1016/j.vaccine.2014.01.078. [DOI] [PubMed] [Google Scholar]
- Vavricka CJ, Li Q, Wu Y, Qi J, Wang M, Liu Y, Gao F, Liu J, Feng E, He J, et al. Structural and functional analysis of laninamivir and its octanoate prodrug reveals group specific mechanisms for influenza NA inhibition. PLoS pathogens. 2011;7:e1002249. doi: 10.1371/journal.ppat.1002249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R, Matrosovich M, Klenk HD. Functional balance between haemagglutinin and neuraminidase in influenza virus infections. Rev Med Virol. 2002;12:159–166. doi: 10.1002/rmv.352. [DOI] [PubMed] [Google Scholar]
- Wan H, Gao J, Xu K, Chen H, Couzens LK, Rivers KH, Easterbrook JD, Yang K, Zhong L, Rajabi M, et al. Molecular basis for broad neuraminidase immunity: conserved epitopes in seasonal and pandemic H1N1 as well as H5N1 influenza viruses. Journal of virology. 2013;87:9290–9300. doi: 10.1128/JVI.01203-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H, Yang H, Shore DA, Garten RJ, Couzens L, Gao J, Jiang L, Carney PJ, Villanueva J, Stevens J, et al. Structural characterization of a protective epitope spanning A(H1N1)pdm09 influenza virus neuraminidase monomers. Nat Commun. 2015;6:6114. doi: 10.1038/ncomms7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- Westgeest KB, Bestebroer TM, Spronken MI, Gao J, Couzens L, Osterhaus AD, Eichelberger M, Fouchier RA, de Graaf M. Optimization of an enzyme-linked lectin assay suitable for rapid antigenic characterization of the neuraminidase of human influenza A(H3N2) viruses. J Virol Methods. 2015;217:55–63. doi: 10.1016/j.jviromet.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JR, Guo Z, Reber A, Kamal RP, Music N, Gansebom S, Bai Y, Levine M, Carney P, Tzeng WP. An influenza A virus (H7N9) anti-neuraminidase monoclonal antibody with prophylactic and therapeutic activity in vivo. Antiviral research. 2016;135:48–55. doi: 10.1016/j.antiviral.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlbold TJ, Krammer F. In the shadow of hemagglutinin: a growing interest in influenza viral neuraminidase and its role as a vaccine antigen. Viruses. 2014;6:2465–2494. doi: 10.3390/v6062465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlbold TJ, Nachbagauer R, Xu H, Tan GS, Hirsh A, Brokstad KA, Cox RJ, Palese P, Krammer F. Vaccination with adjuvanted recombinant neuraminidase induces broad heterologous, but not heterosubtypic, cross-protection against influenza virus infection in mice. MBio. 2015;6:e02556. doi: 10.1128/mBio.02556-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlbold TJ, Podolsky KA, Chromikova V, Kirkpatrick E, Falconieri V, Meade P, Amanat F, Tan J, tenOever BR, Tan GS, et al. Broadly protective murine monoclonal antibodies against influenza B virus target highly conserved neuraminidase epitopes. Nat Microbiol. 2017;2:1415–1424. doi: 10.1038/s41564-017-0011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrammert J, Koutsonanos D, Li GM, Edupuganti S, Sui J, Morrissey M, McCausland M, Skountzou I, Hornig M, Lipkin WI, et al. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. The Journal of experimental medicine. 2011;208:181–193. doi: 10.1084/jem.20101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrammert J, Smith K, Miller J, Langley WA, Kokko K, Larsen C, Zheng NY, Mays I, Garman L, Helms C, et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453:667–671. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Qin G, Gao F, Liu Y, Vavricka CJ, Qi J, Jiang H, Yu K, Gao GF. Induced opening of influenza virus neuraminidase N2 150-loop suggests an important role in inhibitor binding. Sci Rep. 2013;3:1551. doi: 10.1038/srep01551. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A–B) The usage of VH immunoglobulin genes by (A) NA-reactive B cells and (B) HA-reactive B cells
(C) CDR3 length of NA and HA-reactive mAbs, data are represented as mean ± SD.
(D) Total heavy chain mutation number of NA and HA-reactive mAbs based on analysis using the NCBI IgBlast tool (https://www.ncbi.nlm.nih.gov/igblast/), data are represented as mean ± SD.
Several representative HA-reactive or NA-reactive mAbs were tested for binding by ELISA to A/Switzerland/9715293/2013 virus or inactivated A/Switzerland/9715293/2013 virus. (A) HA-reactive mAbs (spots color in red are the HA-stalk reactive mAbs). (B) NA-reactive mAbs (spots colored in red are the NA-star positive mAbs binding around the enzymatic site in Figure 6C). Data are representative of two independent experiments, data are represented as median.
(A) Cross N2-reactive mAbs were tested for inhibiting NA enzymatic activity via ELLA assay against A/swine/Missouri/4296424/2006 (H6N3, H6 is from A/mallard/Sweden/81/2002, PR8 backbone) (B) Cross N1-reactive mAbs were tested for inhibiting NA enzymatic activity via ELLA assay against A/mallard/Sweden/24/2002 (H9N4, H9 is from A/guinea fowl/Hong Kong/WF10/99, PR8 backbone) (C) Cross N1-reactive mAbs were tested for inhibiting NA enzymatic activity via ELLA assay against A/Northern shoveler/Alaska/7MP1708/07 (H3N8). Bar graphs represent IC50 (nM) values. Oseltamivir was used as a positive control and influenza-non-reactive mAb 003-15D3 was used as a negative control in the assays. Data are representative of two independent experiments.
(A) 229-1F06 (B) 229-1G03 (C) 235-1C02 (D) 235-1E06 (E) 229-2C06. Data are representative of three independent experiments.