Abstract
Osteoarthritis affects over 250 million individuals worldwide. Currently, there are no options for early diagnosis of osteoarthritis, demonstrating the need for biomarker discovery. To find biomarkers of osteoarthritis in human synovial fluid, we used high performance liquid-chromatography mass spectrometry for global metabolomic profiling. Metabolites were extracted from human osteoarthritic (n=5), rheumatoid arthritic (n=3), and healthy (n=5) synovial fluid, and a total of 1233 metabolites were detected. Principal components analysis clearly distinguished the metabolomic profiles of diseased from healthy synovial fluid. Synovial fluid from rheumatoid arthritis patients contained expected metabolites consistent with the inflammatory nature of the disease. Similarly, unsupervised clustering analysis found that each disease state was associated with distinct metabolomic profiles and clusters of co-regulated metabolites. For osteoarthritis, co-regulated metabolites that were upregulated compared to healthy synovial fluid mapped to known disease processes including chondroitin sulfate degradation, arginine and proline metabolism, and nitric oxide metabolism. We utilized receiver operating characteristic analysis to determine the diagnostic value of each metabolite and identified 35 metabolites as potential biomarkers of osteoarthritis, with an area under the receiver operating characteristic curve > 0.9. These metabolites included phosphatidylcholine, lysophosphatidylcholine, ceramides, myristate derivatives, and carnitine derivatives. This pilot study provides strong justification for a larger cohort-based study of human osteoarthritic synovial fluid using global metabolomics. The significance of these data is the demonstration that metabolomic profiling of synovial fluid can identify relevant biomarkers of joint disease.
Keywords: osteoarthritis, metabolomics, mass spectrometry, biomarkers, joint disease
Introduction
Osteoarthritis (OA) is the most common degenerative joint disease involving the breakdown of the extracellular matrix in cartilage. OA affects over 250 million individuals, with the majority being adults over the age of 65[1]. Currently, symptoms include joint pain and loss of function. Unfortunately, diagnosis at the onset of symptoms typically occurs after irreversible structural degeneration. Joint replacement is commonly utilized for advanced disease[1]. There are currently no available tests for diagnosing early OA or tracking its progression, demonstrating the need for biomarkers of OA. Previous studies have attempted identifying biochemical biomarkers in urine[2, 3], blood serum[4, 5], or joint synovial fluid (SF)[6–8] samples using proteomics, genomics, and most recently, metabolomics[2–7, 9, 10].
Among the studies utilizing metabolomics for OA biomarker identification, many use urine and blood serum samples[2, 3, 5]. While obtaining a blood sample is less invasive than SF, metabolites from the diseased joint must move across the synovium prior to entering the serum for detection. This dilutes potential biomarkers and leaves them susceptible to potential degradation in the circulatory compartment. One study showed that the composition of metabolites and their corresponding concentrations were altered in osteoarthritic SF[7]. The SF is also in direct contact with the diseased joint tissue, making it a promising candidate for biomarker discovery in OA.
Many studies have used 1H NMR for metabolite biomarker discovery[11]. While 1H NMR has very little sample preparation and high reproducibility, it is mainly used for metabolites that exist in high concentrations due to sensitivity limitations[7]. Alternatively, mass spectrometry has a much higher sensitivity than 1H NMR and is inherently capable of detecting larger numbers of metabolites. Liquid chromatography coupled to mass spectrometry (LC-MS) shows great promise for analyzing complex biofluids such as SF because it provides chromatographic separation of these heterogeneous samples prior to reaching the mass analyzer.
Here, we characterized global metabolomic profiles from SF of patients with various joint diseases using mass spectrometry. Metabolomic profiles of rheumatoid arthritis (RA) synovial fluid were used to validate the method by confirming established findings in RA. To our knowledge, this is the first study to perform LC-MS-based global metabolomic profiling of osteoarthritic SF for biomarker discovery. The long-term goal of this research is to identify biomarkers of OA phenotypes for earlier diagnosis and quantification of disease progression. The objective of this study was to evaluate global LC-MS-based metabolomic profiles as a tool for quantifying biomarkers within SF. We hypothesized that (1) RA profiles would be distinct from OA with RA showing increases in inflammatory metabolites and (2) OA patients would exhibit diverse metabolomics profiles suggesting distinct disease phenotypes that correlate with clinical symptoms.
Methods
Patients and Synovial Fluid Samples
OA (n=5) and RA (n=3) SF samples were obtained under IRB approval. Healthy SF samples (n=5) were purchased from Articular Engineering (Northbrook, IL) post mortem with partial clinical data including age, gender, race, and cause of death. All samples were frozen at −80°C after harvest until analysis.
Metabolite Extraction
Metabolites were extracted from 100 μL of SF prior to analysis by LC-MS. Samples were thawed on ice for 3–5 minutes prior to centrifuging at 4°C at 500 × g for 10 minutes to remove cells and debris. The supernatant was collected and evaporated, and dried samples were re-suspended in 100 μL of 50:50 water:acetonitrile. 500 μL of acetone was added and the sample was mechanically shaken for 3 min to precipitate polymers. The samples were then refrigerated for 10 minutes prior to centrifuging at 16100 × g for 5 min. The supernatant was transferred to a new tube and further evaporated to remove acetone. The remaining sample volume was doubled with acetonitrile for a final 50:50 water:acetonitrile LC-MS injection buffer.
Metabolomic Profiling and Data Processing
Metabolites extracted from SF samples were analyzed using an Agilent 1290 UPLC system connected to an Agilent 6538 Q-TOF mass spectrometer (Agilent, Santa Clara, CA). The samples were run in normal phase, using a Cogent Diamond Hydride HILIC 150 × 2.1 mm column (MicroSolv, Eatontown, NJ). The HPLC solvents used were 0.1% formic acid in water (solvent A) and 0.1% formic acid in acetonitrile (solvent B). The elution gradient consisted of 100 to 25% solvent B over 13 minutes, and each run began with 2 minutes wash (15 minutes total run time).
Metabolite detection was performed in positive mode with a resolution of ~20,000 and accuracy of ~5 ppm. Agilent MSConvert converted data from Agilent’s proprietary file format, .d files, to mzXML files to filter data in MZMine 2.14 for retention time, noise level, and peak detection[12]. An intensity threshold of 1000 was applied to detect peaks above the noise. The processed chromatograms were then normalized and aligned for metabolite (m/z value) detection. Metabolites with median intensity values of zero across the OA, RA, and healthy datasets were removed from the analysis. To identify the metabolites, untargeted metabolite mass to charge (m/z) values were loaded into METLIN, a database containing over 80,000 metabolites[13]. The batch search used a mass tolerance of 15 parts per million (ppm), including positively charged molecules with +1H+ or +1Na+ adducts but excluding peptides, toxins, and drugs. All m/z values detected were crosschecked against our library of metabolite standards. To confirm metabolite identities, both m/z value and retention time had to match within a m/z value tolerance of 0.01 or 30 ppm and retention time tolerance of 0.25 minutes[14].
Statistical analyses
Statistical analysis was performed in Matlab (Mathworks, Natick, MA USA). To identify metabolites differentially expressed between OA, RA, and healthy SF, we used Student’s T-tests and one-way analysis of variance (ANOVA) with false-discovery rate (FDR) corrections at a significance level of 0.05. Additionally, we used volcano plots to visualize metabolites significantly upregulated and downregulated in OA or RA in comparison to healthy SF[15]. The negative log10 of the p-value (y-axis) for each m/z value and the fold change (log2(ratio)) of the median intensity between disease state and healthy SF (x-axis) were plotted against one another to yield both significance and magnitude of change. M/z values with zero intensity for either group were excluded from the volcano plot analysis. Principal component analysis (PCA) was applied to fold change normalized, log-transformed metabolites to examine sources of underlying variation in the dataset. To visualize the unique metabolites in each dataset, we created a scatterplot of median metabolite intensities from each group against one another (+/− standard deviation). Where applicable, error bars show standard error.
Hierarchical cluster analysis (HCA) was used to provide a visual representation of global metabolomic profiles and co-regulated metabolites. Co-regulated metabolites within clusters were identified using METLIN and subsequently analyzed for pathway enrichment using IMPaLA[16]. Based on the metabolites in those clusters, IMPaLA identifies potential pathways implicated by over-representation analysis using established biochemical pathway databases (i.e. KEGG, Reactome, HumanCyc, etc.).
Potential biomarkers were determined using receiver operating characteristic (ROC) analysis. By analyzing the area under the receiver operating curve (AUC), we identified metabolites that could accurately classify between two cases, such as OA and healthy. We focused on metabolites with an AUC>0.9 as potential biomarkers.
Results
Metabolomic Profiling of Osteoarthritis, Rheumatoid Arthritis, and Healthy Synovial Fluid
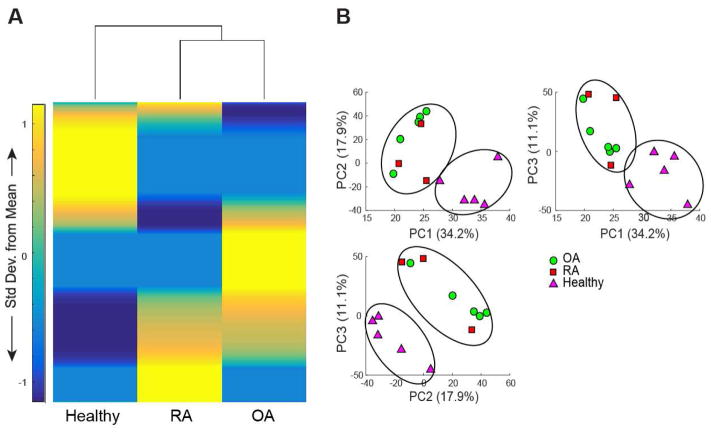
We detected a total of 1233 metabolite features across all OA, RA, and healthy SF samples using LC-MS analysis. HCA of all metabolites in OA, RA, and healthy SF demonstrated that OA and RA have more similar metabolomic profiles than healthy SF, as indicated by the dendrogram (Fig. 1A). PCA of all three datasets revealed clear separation between diseased and healthy SF metabolomic profiles, with the first two principle components associated with 52.1% of the total variation (PC1=34.2%, PC2=17.9%, Fig. 1B). 26 metabolites were significantly different between OA, RA, and healthy SF as determined by one-way ANOVA (pfdr <0.05). 15 metabolites were confidently identified by both m/z value and retention time using our library of standards, with 6 of these being significantly different (pfdr<0.05). Citric acid, D-lactic acid methyl ester, hydroxyl-L-proline, L-isoleucine, and L-methionine were significantly upregulated in healthy SF in comparison to diseased. In contrast, L-citrulline was significantly upregulated in OA over both RA and healthy SF.
Figure 1.
Global metabolomic profiles of diseased (OA and RA) SF are distinct from healthy SF. (A) Clustered heatmap of median metabolite intensities from healthy (n=5), OA (n=5), and RA (n=3) SF. 1233 metabolites, represented by the rows in this heatmap, were detected in human SF. Dendrogram of HCA demonstrates that OA and RA metabolomic profiles are more similar to one another than healthy SF. (B) PCA of metabolite intensities in OA, RA, and healthy SF, comparing PC1 (34.2%), PC2 (17.9%), and PC3 (11.1%). PCA shows clear discrimination between diseased and healthy metabolomic profiles.
Metabolomic Profiles of OA Synovial Fluid
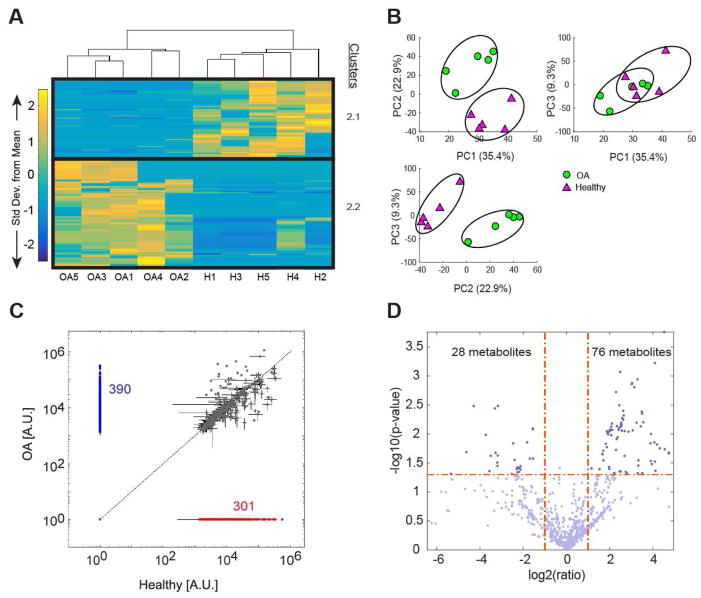
1098 metabolites were detected in OA and healthy SF. 58 metabolites were significantly different between OA and healthy SF (pfdr<0.05). HCA was performed on all 318 significantly different metabolites without FDR corrections to remain consistent with the RA and healthy analysis (Supplemental Figure 1). HCA showed 2 distinct clusters of co-regulated metabolites, with cluster 2.1 containing 132 metabolites downregulated in OA and cluster 2.2 containing 186 metabolites upregulated in OA (Fig. 2A). Enrichment of cluster 2.1 identified similar pathways as cluster S1.1 (metabolites downregulated in healthy compared to RA) in Supplemental Figure 1A, including purine metabolism, tRNA charging, and gene expression. Enrichment of cluster 2.2, however, yielded arginine and proline metabolism, chondroitin sulfate degradation, amino oxidase reactions, COX reactions, and creatine biosynthesis (Supplemental Table 1, Supplemental Table 6).
Figure 2.
OA and healthy SF exhibit distinct metabolomic features. (A) Clustered heatmap of OA (n=5) and healthy (n=5) SF median metabolite intensities (H=healthy). 1098 metabolites were detected in OA and healthy SF, with 318 being significantly different as represented in this heatmap. Clusters of co-regulated metabolites are outlined in black and labeled as 2.1 and 2.2. Cluster 2.1 contains 132 metabolites downregulated in OA and cluster 2.2 contains 186 metabolites upregulated in OA. (B) PCA of OA and healthy SF metabolite intensities. PC1 (35.4%), PC2 (22.9%), and PC3 (9.3%) were associated with 67.6% of the variation within the dataset. Comparison of PC1, PC2, and PC3 show clear separation between OA and healthy SF metabolomic profiles. (C) Scatterplot comparing OA and healthy metabolite intensity levels (A.U.=Arbitrary Units). 301 metabolites were exclusively detected in OA SF (blue) and 290 metabolites were exclusively detected in healthy SF (red). (D) Volcano plot of OA and healthy SF, plotting the −log(p-value) against the fold change (log2(ratio)) of individual metabolites. Vertical dashed lines mark the twofold change and the horizontal line marks the cutoff p-value of 0.05. Metabolites in the upper right and left quadrants represent metabolites with a p-value less than 0.05 and a greater than twofold change. 76 metabolites were significantly upregulated in OA (upper right quadrant) and 28 metabolites were significantly downregulated in OA (upper left quadrant) in comparison to healthy (p-value<0.05; fold change>2).
PCA showed clear separation between OA and healthy SF, with the first two principle components (PC1=35.4%, PC2=22.9%) associated with 58.3% of the variation (Fig. 2B). Of the 1098 metabolites detected in OA and healthy SF, 390 metabolites were exclusively detected in OA SF and 301 metabolites were exclusively detected in healthy SF (Fig. 2C). Volcano plot analysis identified 76 metabolites upregulated and 28 metabolites downregulated in OA, with a p-value<0.05 and fold change>2 (Fig 2D). Enrichment of these metabolites yielded similar pathways implicated in clusters 2.1 and 2.2 (Supplemental Table 2). Heatmap clustering, PCA discrimination, scatterplot visualization, and volcano plot analysis clearly demonstrate that OA and healthy SF represent distinct populations with phenotypic differences.
Differences between Pathological and Healthy Synovial Fluid
OA and RA metabolite intensities were normalized to healthy SF to determine how metabolomic profiles of SF change with disease. HCA of the normalized metabolites exhibited distinct clusters of co-regulated metabolites (Fig. 3A). Cluster 3.1 included 276 co-regulated metabolites downregulated in diseased compared to healthy (Fig. 3A). These metabolites mapped to SLC-mediated transport, gamma-glutamyl cycle, tRNA aminoacylation, and various amino acid transporters (Supplemental Table 1, Supplemental Table 7). Cluster 3.2 included 258 co-regulated metabolites upregulated in OA in comparison to healthy and RA (Fig. 3A). These metabolites mapped to creatine biosynthesis, alpha-linolenic acid metabolism, galactose metabolism, vitamin B6 metabolism, chondroitin sulfate degradation, arginine biosynthesis, urea cycle, and nitric oxide metabolism (Supplemental Table 1, Supplemental Table 7). Cluster 3.3 included 305 co-regulated metabolites upregulated in diseased compared to healthy (Fig. 3A). These metabolites mapped to phospholipid biosynthesis, mTOR signaling, arginine and proline metabolism, organic cation transporters, synthesis of PS, acyl chain remodeling, glycerophospholipid metabolism, and phosphatidylcholine biosynthesis (Supplemental Table 1, Supplemental Table 7). Cluster 3.4 included 152 co-regulated metabolites upregulated in RA over healthy and OA, mapping to histidine, lysine, phenylalanine, tyrosine, glycine, proline, and tryptophan metabolism, ibuprofen metabolism, steroid biosynthesis, glucocorticoid and mineralocorticoid metabolism, alpha-linolenic acid metabolism, and leukotriene biosynthesis (Supplemental Table 1, Supplemental Table 7). Select metabolites implicated in each cluster are displayed in Figure 3B.
Figure 3.
Metabolomic profiling reveals features of each disease state with co-regulated metabolites upregulated or downregulated in comparison to healthy SF. (A) Clustered heatmap of 1233 median metabolite intensities normalized to healthy. Four clusters of interest were identified as clusters 3.1–3.4, with co-regulated metabolites upregulated or downregulated in OA and RA. (B) Bar graphs of median metabolite intensities +/− standard deviation (A.U. = arbitrary units, N.D. = not detected) for 2 individual metabolites implicated in each of the 4 clusters. Row 1 corresponds to cluster 3.1, row 2 corresponds to cluster 3.2, and so on. Each bar graph contains the m/z value, the metabolite identification, and its suspected adduct.
Biomarker Candidates for Osteoarthritis
We used ROC analysis to determine the potential diagnostic value of each detected metabolite as an OA biomarker. ROC analysis identified 65 m/z values as potential biomarkers of OA, all having an AUC>0.9. 30 of these 65 m/z values were also identified as potential biomarkers of RA and were consequently removed from the analysis. To identify the remaining 35 m/z values as metabolites, we searched METLIN for a full list of potential metabolites that match these m/z values. Of these 35 m/z values, 16 did not have any METLIN database matches, thus classifying them as potentially novel metabolites for future analysis. The remaining 19 metabolites with an AUC>0.9 mapped to 91 putative metabolites identities (Supplemental Table 3). Of those 91 identified metabolites identities, over half were phospholipids (PC, PAF, PG, PE, PS, and ceramides). Other potential biomarkers for OA included 2,3,6-trihydroxypyridine, myristate derivatives, arachidonyl carnitine, N-methyl arachidonyl amine, oleanolic acid acetate, vitamin D3 derivatives, sulfonic acid derivatives, and ursolic acid derivatives (Supplemental Table 3). Additional studies are needed to validate these metabolites as OA biomarkers and determine the implications of their involvement in OA pathogenesis.
Discussion
This is the first study to perform global LC-MS-based metabolomic profiling on human OA synovial fluid. We used LC-MS for both (1) biomarker discovery and (2) understanding disease pathophysiology. PCA and HCA of metabolite data distinguished between OA and healthy SF, creating distinct metabolomic profiles with identifiable features. We detected a total of 1098 metabolites in OA and healthy SF, more than any previous study we know of[7, 17–20], with 58 significantly different metabolites between OA and healthy SF. Enrichment analysis of metabolites that were upregulated in OA identified nitric oxide production and chondroitin sulfate degradation both of which have previously been linked to OA pathogenesis[5, 21, 22]. Chondroitin sulfate is a glycosaminoglycan that plays an important structural role in the cartilage matrix. Altered levels of chondroitin sulfate have previously been found in OA cartilage[21, 22]. Nitric oxide aids in OA disease progression by promoting expression of proinflammatory cytokines, inhibiting proteoglycan and collagen synthesis, mediating apoptosis, and activating metalloproteinases[5].
ROC analysis is valuable for determining the diagnostic value of each metabolite as a potential biomarker. ROC analysis identified 35 potential biomarkers of OA, including phosphatidylcholines (PC), lysophoshatidylcholines (lysoPC), ceramides (Cer), myristate derivatives, and carnitine derivatives. Phospholipids are important components of the SF that contribute to the mechanical function of the SF to lubricate articular cartilage surfaces[6]. Altered composition and concentration of lubricating agents in the SF are associated with damaged articular cartilage surfaces in both OA and RA[23–25]. However, the small sample size of this pilot study prevents broader interpretation about these metabolites as potential biomarkers and their implications in OA pathogenesis.
Global metabolic profiles of RA patient SF found anti-inflammatory drug pathways, amino acid metabolism, leukotriene biosynthesis, alpha-linolenic acid metabolism, glucocorticoid and mineralocorticoid metabolism, and steroid biosynthesis implicated in RA (Supplemental Table 1, Supplemental Table 2). Given that rheumatoid arthritis is highly inflammatory, the identification and upregulation of anti-inflammatory drug pathways is expected in RA SF and validates the use of our LC-MS global metabolomic profiling approach.
While this pilot study holds great promise for biomarker discovery, this is discovery phase research with important limitations. First, healthy SF samples were harvested post mortem. In contrast, the SF from OA and RA joints was harvested from living patients. While post mortem SF may differ from that of living patients, previous studies found that post mortem SF maintained normal lubricant components, composition, and function[26, 27]. Second, patient information was limited. Thus, the ability to decipher effects of age, BMI, or gender on metabolomic profiles was not possible. The third, and main limitation of this study is the small sample size. This was a pilot study meant to assess global metabolomic profiling of human SF for biomarker discovery. Future studies will increase the sample sizes of all patient groups as well as age and gender match samples to identify potential biomarkers of OA.
In summary, this study demonstrates the advantages of global metabolomic profiling for both biomarker discovery and further understanding of OA pathogenesis. Despite extremely small sample sizes, we were able to detect a number of potential metabolites for future validation studies. Global metabolomic profiling via LC-MS may provide physicians with a high sensitivity method for tracking treatment effectiveness, disease progression, and biomarker discovery for OA and other diseases.
Supplementary Material
Supplemental Figure 1. RA and healthy SF metabolomic profiles are distinct from one another. (A) Clustered heatmap of RA (n=3) and healthy (n=5) median metabolite intensities (H=healthy). 1018 metabolites were detected in RA and healthy SF, with 162 metabolites being significantly different (p<0.05). Clusters of co-regulated metabolites are outlined in black and labeled as S1.1 and S1.2. Cluster S1.1 contains 55 metabolites downregulated in RA and cluster S1.2 contains 107 metabolites upregulated in RA. (B) PCA of metabolite intensities in RA and healthy SF. PC1 (33.6%), PC2 (17.8%), and PC3 (15.2%) accounted for 62.6% of the variation within the dataset. Comparison of PC1, PC2, and PC3 show clear separation between RA and healthy SF metabolomic profiles. (C) Scatter plot comparing metabolite intensities in RA and healthy SF (A.U.=Arbitrary Units). Of the 1018 metabolites detected, 310 metabolites were exclusively detected in RA SF (blue) and 282 metabolites were exclusively detected in healthy SF (red). (D) Volcano plot of RA and healthy SF, plotting the −log(p-value) against the fold change (log2(ratio)) of individual metabolites. Vertical dashed lines represent the twofold change and the horizontal dashed line represents the cutoff p-value of 0.05. Metabolites in the upper right and left quadrants represent metabolites with a p-value less than 0.05 and a greater than twofold change.12 metabolites were significantly upregulated in RA (upper right quadrant) and 25 metabolites were significantly downregulated in RA (upper left quadrant) in comparison to healthy (p-value<0.05; fold change>2).
Supplemental Table 1: Pathway Enrichment of Metabolite Clusters
Metabolic pathways are altered in OA and RA SF. Metabolites in clusters from Figures 2A, 3A, and Supplemental Figure 1A were identified using METLIN and enriched via IMPaLA for relevant biochemical pathways.
Supplemental Table 2: Volcano Plot Enrichment of Significantly Upregulated and Downregulated Metabolites in Diseased SF
Metabolic profiling revealed metabolites in diseased SF upregulated or downregulated greater than twofold. Significant m/z values with a p-value<0.05 and greater than twofold change in the upper right and upper left quadrants of Figures 2D and Supplemental Figure 1D were identified using METLIN.
Supplemental Table 3: METLIN Identification of Potential OA Biomarkers
ROC analysis identified 35 potential SF biomarkers for OA. M/z values with an AUC>0.9 were matched with known metabolite identities using the METLIN metabolite database. 19 of the 35 m/z values mapped to 91 potential metabolite identities with a 15 ppm tolerance. The remaining 16 m/z values did not map to known metabolite identities and thus further experimentation is needed to determine if they are novel metabolites.
Supplemental Table 4: METLIN Identification of Potential RA Biomarkers
ROC analysis identified 30 potential SF biomarkers for rheumatoid arthritis with an AUC>0.9. Of these 30 m/z values, 15 matched with 119 potential metabolite identities with a 15 ppm tolerance. The remaining 15 did not map to any known metabolite identities, thus may be potentially novel metabolites.
Supplemental Table 5: METLIN Identification and Pathway Enrichment of Supplemental Figure 1 Heatmap
Metabolic profiling revealed pathways altered in RA SF. M/z values in Supplemental Figure 1, clusters S1.1 and S1.2 were identified using METLIN and consequently enriched via IMPaLA for relevant pathways.
Supplemental Table 6: METLIN Identification and Pathway Enrichment of Figure 2 Heatmap
Metabolic profiling revealed pathways altered in OA SF. M/z values in Figure 2 clusters 2.1 and 2.2 were identified using METLIN and consequently enriched via IMPaLA for relevant pathways.
Supplemental Table 7: METLIN Identification and Pathway Enrichment of Figure 3 Heatmap
Pathway enrichment revealed distinct pathways altered in diseased SF compared to healthy. M/z values in Figure 3, clusters 3.1–3.4 were identified using METLIN and consequently enriched via IMPaLA for relevant pathways.
Highlights.
Untargeted metabolomics using liquid chromatography-mass spectrometry clearly discriminates between osteoarthritis, rheumatoid arthritis, and healthy synovial fluid, showing distinct metabolomic profiles with key features of each disease state.
Global metabolomic profiling confirmed previously identified pathways implicated in osteoarthritis including chondroitin sulfate degradation and nitric oxide metabolism.
Of the 1233 metabolite features identified in human synovial fluid using our method, 65 features are potential biomarkers of osteoarthritis based on receiver operating characteristic analysis (AUC>0.9) including various phospholipids, vitamin D3 derivatives, carnitine derivatives, and oleanolic acid acetate.
Acknowledgments
We would like to acknowledge the Mass Spectrometry Core at Montana State University for technical assistance.
Role of the funding source
This work was supported by NSF grant 1554708.
The Proteomics, Metabolomics, and Mass Spectrometry facility at MSU received support from the Murdock Charitable Trust the National Institute of General Medical Sciences of the National Institutes of Health under Award Number P20GM103474.
The funding sources had no role in the design or execution of this study.
Abbreviations
- OA
osteoarthritis
- RA
rheumatoid arthritis
- SF
synovial fluid
- m/z
mass-to-charge ratio
- LC-MS
liquid chromatography mass spectrometry
- ROC
receiver operating characteristic curve
- PCA
principal component analysis
- HCA
hierarchical cluster analysis
Footnotes
Competing interest statement
Dr. June owns stock in Beartooth Biotech. This company was not involved in this study. The remaining authors have no competing interests in this work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hunter DJ, Nevitt M, Losina E, Kraus V. Biomarkers for osteoarthritis: current position and steps towards further validation. Best Pract Res Clin Rheumatol. 2014;28:61–71. doi: 10.1016/j.berh.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lamers RJ, van Nesselrooij JH, Kraus VB, Jordan JM, Renner JB, Dragomir AD, Luta G, van der Greef J, DeGroot J. Identification of an urinary metabolite profile associated with osteoarthritis. Osteoarthritis Cartilage. 2005;13:762–768. doi: 10.1016/j.joca.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 3.Loeser RF, Pathmasiri W, Sumner SJ, McRitchie S, Beavers D, Saxena P, Nicklas BJ, Jordan J, Guermazi A, Hunter DJ, Messier SP. Association of urinary metabolites with radiographic progression of knee osteoarthritis in overweight and obese adults: an exploratory study. Osteoarthritis Cartilage. 2016;24:1479–1486. doi: 10.1016/j.joca.2016.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhai G, Wang-Sattler R, Hart DJ, Arden NK, Hakim AJ, Illig T, Spector TD. Serum branched-chain amino acid to histidine ratio: a novel metabolomic biomarker of knee osteoarthritis. Ann Rheum Dis. 2010;69:1227–1231. doi: 10.1136/ard.2009.120857. [DOI] [PubMed] [Google Scholar]
- 5.Zhang W, Sun G, Likhodii S, Liu M, Aref-Eshghi E, Harper PE, Martin G, Furey A, Green R, Randell E, Rahman P, Zhai G. Metabolomic analysis of human plasma reveals that arginine is depleted in knee osteoarthritis patients. Osteoarthritis Cartilage. 2016;24:827–834. doi: 10.1016/j.joca.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 6.Kosinska MK, Liebisch G, Lochnit G, Wilhelm J, Klein H, Kaesser U, Lasczkowski G, Rickert M, Schmitz G, Steinmeyer J. A lipidomic study of phospholipid classes and species in human synovial fluid. Arthritis Rheum. 2013;65:2323–2333. doi: 10.1002/art.38053. [DOI] [PubMed] [Google Scholar]
- 7.Mickiewicz B, Kelly JJ, Ludwig TE, Weljie AM, Wiley JP, Schmidt TA, Vogel HJ. Metabolic analysis of knee synovial fluid as a potential diagnostic approach for osteoarthritis. J Orthop Res. 2015;33:1631–1638. doi: 10.1002/jor.22949. [DOI] [PubMed] [Google Scholar]
- 8.Zheng K, Shen N, Chen H, Ni S, Zhang T, Hu M, Wang J, Sun L, Yang X. Global and targeted metabolomics of synovial fluid discovers special osteoarthritis metabolites. J Orthop Res. 2017 doi: 10.1002/jor.23482. [DOI] [PubMed] [Google Scholar]
- 9.Liao W, Li Z, Zhang H, Li J, Wang K, Yang Y. Proteomic analysis of synovial fluid as an analytical tool to detect candidate biomarkers for knee osteoarthritis. Int J Clin Exp Pathol. 2015;8:9975–9989. [PMC free article] [PubMed] [Google Scholar]
- 10.van Meurs JB. Osteoarthritis year in review 2016: genetics, genomics and epigenetics. Osteoarthritis Cartilage. 2017;25:181–189. doi: 10.1016/j.joca.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 11.Damyanovich AZ, Staples JR, Marshall KW. 1H NMR investigation of changes in the metabolic profile of synovial fluid in bilateral canine osteoarthritis with unilateral joint denervation. Osteoarthritis Cartilage. 1999;7:165–172. doi: 10.1053/joca.1998.0205. [DOI] [PubMed] [Google Scholar]
- 12.Pluskal T, Castillo S, Villar-Briones A, Oresic M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics. 2010;11:395. doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu ZJ, Schultz AW, Wang J, Johnson CH, Yannone SM, Patti GJ, Siuzdak G. Liquid chromatography quadrupole time-of-flight mass spectrometry characterization of metabolites guided by the METLIN database. Nat Protoc. 2013;8:451–460. doi: 10.1038/nprot.2013.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, Fan TW, Fiehn O, Goodacre R, Griffin JL, Hankemeier T, Hardy N, Harnly J, Higashi R, Kopka J, Lane AN, Lindon JC, Marriott P, Nicholls AW, Reily MD, Thaden JJ, Viant MR. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI) Metabolomics. 2007;3:211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hur M, Campbell AA, Almeida-de-Macedo M, Li L, Ransom N, Jose A, Crispin M, Nikolau BJ, Wurtele ES. A global approach to analysis and interpretation of metabolic data for plant natural product discovery. Nat Prod Rep. 2013;30:565–583. doi: 10.1039/c3np20111b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cavill R, Kamburov A, Ellis JK, Athersuch TJ, Blagrove MS, Herwig R, Ebbels TM, Keun HC. Consensus-phenotype integration of transcriptomic and metabolomic data implies a role for metabolism in the chemosensitivity of tumour cells. PLoS Comput Biol. 2011;7:e1001113. doi: 10.1371/journal.pcbi.1001113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adams SB, Jr, Setton LA, Kensicki E, Bolognesi MP, Toth AP, Nettles DL. Global metabolic profiling of human osteoarthritic synovium. Osteoarthritis Cartilage. 2012;20:64–67. doi: 10.1016/j.joca.2011.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hugle T, Kovacs H, Heijnen IA, Daikeler T, Baisch U, Hicks JM, Valderrabano V. Synovial fluid metabolomics in different forms of arthritis assessed by nuclear magnetic resonance spectroscopy. Clin Exp Rheumatol. 2012;30:240–245. [PubMed] [Google Scholar]
- 19.Kim S, Hwang J, Kim J, Ahn JK, Cha HS, Kim KH. Metabolite profiles of synovial fluid change with the radiographic severity of knee osteoarthritis. Joint Bone Spine. 2016 doi: 10.1016/j.jbspin.2016.05.018. [DOI] [PubMed] [Google Scholar]
- 20.Kim S, Hwang J, Xuan J, Jung YH, Cha HS, Kim KH. Global metabolite profiling of synovial fluid for the specific diagnosis of rheumatoid arthritis from other inflammatory arthritis. PLoS One. 2014;9:e97501. doi: 10.1371/journal.pone.0097501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shinmei M, Miyauchi S, Machida A, Miyazaki K. Quantitation of chondroitin 4-sulfate and chondroitin 6-sulfate in pathologic joint fluid. Arthritis Rheum. 1992;35:1304–1308. doi: 10.1002/art.1780351110. [DOI] [PubMed] [Google Scholar]
- 22.Caterson B, Mahmoodian F, Sorrell JM, Hardingham TE, Bayliss MT, Carney SL, Ratcliffe A, Muir H. Modulation of native chondroitin sulphate structure in tissue development and in disease. J Cell Sci. 1990;97(Pt 3):411–417. doi: 10.1242/jcs.97.3.411. [DOI] [PubMed] [Google Scholar]
- 23.Elsaid KA, Jay GD, Warman ML, Rhee DK, Chichester CO. Association of articular cartilage degradation and loss of boundary-lubricating ability of synovial fluid following injury and inflammatory arthritis. Arthritis Rheum. 2005;52:1746–1755. doi: 10.1002/art.21038. [DOI] [PubMed] [Google Scholar]
- 24.Mazzucco D, Scott R, Spector M. Composition of joint fluid in patients undergoing total knee replacement and revision arthroplasty: correlation with flow properties. Biomaterials. 2004;25:4433–4445. doi: 10.1016/j.biomaterials.2003.11.023. [DOI] [PubMed] [Google Scholar]
- 25.Hills BA, Crawford RW. Normal and prosthetic synovial joints are lubricated by surface-active phospholipid: a hypothesis. J Arthroplasty. 2003;18:499–505. doi: 10.1016/s0883-5403(03)00072-x. [DOI] [PubMed] [Google Scholar]
- 26.Ludwig TE, McAllister JR, Lun V, Wiley JP, Schmidt TA. Diminished cartilage-lubricating ability of human osteoarthritic synovial fluid deficient in proteoglycan 4: Restoration through proteoglycan 4 supplementation. Arthritis Rheum. 2012;64:3963–3971. doi: 10.1002/art.34674. [DOI] [PubMed] [Google Scholar]
- 27.Sarkioja T, Yla-Herttuala S, Solakivi T, Nikkari T, Hirvonen J. Stability of plasma total cholesterol, triglycerides, and apolipoproteins B and A-I during the early postmortem period. J Forensic Sci. 1988;33:1432–1438. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. RA and healthy SF metabolomic profiles are distinct from one another. (A) Clustered heatmap of RA (n=3) and healthy (n=5) median metabolite intensities (H=healthy). 1018 metabolites were detected in RA and healthy SF, with 162 metabolites being significantly different (p<0.05). Clusters of co-regulated metabolites are outlined in black and labeled as S1.1 and S1.2. Cluster S1.1 contains 55 metabolites downregulated in RA and cluster S1.2 contains 107 metabolites upregulated in RA. (B) PCA of metabolite intensities in RA and healthy SF. PC1 (33.6%), PC2 (17.8%), and PC3 (15.2%) accounted for 62.6% of the variation within the dataset. Comparison of PC1, PC2, and PC3 show clear separation between RA and healthy SF metabolomic profiles. (C) Scatter plot comparing metabolite intensities in RA and healthy SF (A.U.=Arbitrary Units). Of the 1018 metabolites detected, 310 metabolites were exclusively detected in RA SF (blue) and 282 metabolites were exclusively detected in healthy SF (red). (D) Volcano plot of RA and healthy SF, plotting the −log(p-value) against the fold change (log2(ratio)) of individual metabolites. Vertical dashed lines represent the twofold change and the horizontal dashed line represents the cutoff p-value of 0.05. Metabolites in the upper right and left quadrants represent metabolites with a p-value less than 0.05 and a greater than twofold change.12 metabolites were significantly upregulated in RA (upper right quadrant) and 25 metabolites were significantly downregulated in RA (upper left quadrant) in comparison to healthy (p-value<0.05; fold change>2).
Supplemental Table 1: Pathway Enrichment of Metabolite Clusters
Metabolic pathways are altered in OA and RA SF. Metabolites in clusters from Figures 2A, 3A, and Supplemental Figure 1A were identified using METLIN and enriched via IMPaLA for relevant biochemical pathways.
Supplemental Table 2: Volcano Plot Enrichment of Significantly Upregulated and Downregulated Metabolites in Diseased SF
Metabolic profiling revealed metabolites in diseased SF upregulated or downregulated greater than twofold. Significant m/z values with a p-value<0.05 and greater than twofold change in the upper right and upper left quadrants of Figures 2D and Supplemental Figure 1D were identified using METLIN.
Supplemental Table 3: METLIN Identification of Potential OA Biomarkers
ROC analysis identified 35 potential SF biomarkers for OA. M/z values with an AUC>0.9 were matched with known metabolite identities using the METLIN metabolite database. 19 of the 35 m/z values mapped to 91 potential metabolite identities with a 15 ppm tolerance. The remaining 16 m/z values did not map to known metabolite identities and thus further experimentation is needed to determine if they are novel metabolites.
Supplemental Table 4: METLIN Identification of Potential RA Biomarkers
ROC analysis identified 30 potential SF biomarkers for rheumatoid arthritis with an AUC>0.9. Of these 30 m/z values, 15 matched with 119 potential metabolite identities with a 15 ppm tolerance. The remaining 15 did not map to any known metabolite identities, thus may be potentially novel metabolites.
Supplemental Table 5: METLIN Identification and Pathway Enrichment of Supplemental Figure 1 Heatmap
Metabolic profiling revealed pathways altered in RA SF. M/z values in Supplemental Figure 1, clusters S1.1 and S1.2 were identified using METLIN and consequently enriched via IMPaLA for relevant pathways.
Supplemental Table 6: METLIN Identification and Pathway Enrichment of Figure 2 Heatmap
Metabolic profiling revealed pathways altered in OA SF. M/z values in Figure 2 clusters 2.1 and 2.2 were identified using METLIN and consequently enriched via IMPaLA for relevant pathways.
Supplemental Table 7: METLIN Identification and Pathway Enrichment of Figure 3 Heatmap
Pathway enrichment revealed distinct pathways altered in diseased SF compared to healthy. M/z values in Figure 3, clusters 3.1–3.4 were identified using METLIN and consequently enriched via IMPaLA for relevant pathways.