Abstract
The discovery of adipose triglyceride lipase (ATGL) and its coactivator comparative gene identification-58 (CGI-58) provided a major paradigm shift in the understanding of intracellular lipolysis in both adipocytes and nonadipocyte cells. The subsequent discovery of G0/G1 switch gene 2 (G0S2) as a potent endogenous inhibitor of ATGL revealed a unique mechanism governing lipolysis and fatty acid (FA) availability. G0S2 is highly conserved in vertebrates, and exhibits cyclical expression pattern between adipose tissue and liver that is critical to lipid flux and energy homeostasis in these two tissues. Biochemical and cell biological studies have demonstrated that a direct interaction with ATGL mediates G0S2’s inhibitory effects on lipolysis and lipid droplet degradation. In this review we examine evidence obtained from recent in vitro and in vivo studies that lends support to the proof-of-principle concept that G0S2 functions as a master regulator of tissue-specific balance of TG storage vs. mobilization, partitioning of metabolic fuels between adipose and liver, and the whole-body adaptive energy response.
Keywords: G0S2, ATGL, lipolysis, lipid droplet, fatty acid, triglyceride
1. Introduction
Metabolic regulation and energy availability are essential for all biological functions. In rodents and humans, adipose tissue and liver function in synchronization to confer adaptive response to fasting and maintain systemic energy homeostasis [1, 2]. During periods of food consumption when dietary glucose is used as the primary source of energy, insulin promotes the deposition of excess glucose in glycogen in the liver and the storage of dietary fatty acids (FAs) as triglycerides (TGs) in cytosolic lipid droplets (LDs) of adipocytes. In response to nutrient deprivation or fasting, changes in both carbohydrate and lipid metabolism are initiated through nutritional and hormonal signaling. Hepatic glycogen stores are depleted through glycogenolysis to produce glucose as a rapid source of energy. When fasting is prolonged, TG stores in adipocytes are mobilized via hydrolytic degradation (lipolysis), resulting in the release of FAs and glycerol into circulation. A large percentage of these adipose-derived FAs and glycerol are destined for uptake by the liver, where they are used as substrates for β-oxidation to drive ketogenesis as well as for synthesis of TGs that are packed in VLDL particles for secretion. Since the amount of incoming FAs often exceeds the net utilization, excess FAs are deposited as TGs within LDs in hepatocytes [3–5]. Glycerol, a second product from adipose lipolysis, is also transported to the liver, where it is used for glucose production through gluconeogenesis or when phosphorylated, serves as the backbone for TG synthesis [6].
The flux of FAs between adipose tissue and liver is a delicate balance of TG synthesis vs. mobilization in these two organs. Adipocytes are the primary site for storage of FAs as neutral TGs in most organisms. Imbalanced TG synthesis and FA mobilization are major contributors to the onset of pathologies including obesity and obesity-associated metabolic abnormalities. Unlike adipocytes, hepatocytes are not designed to effectively store TGs. In a physiological setting, a cycle exists between adipose tissue and liver with regards to the movement of FA and TG across these depots. A marked increase in intrahepatic TG in the fasted state is of no negative consequence and is reversed upon refeeding. However, if a continued dietary assault is present during refeeding, as occurs often in obese individuals or those that consistently consume a high fat or high carbohydrate diet, pathological conditions including non-alcoholic fatty liver disease (NAFLD) and the more severe non-alcoholic steatohepatitis (NASH) can develop [7–10]. Presumably, hepatic steatosis can also stem from defective clearance of intrahepatic TG through the action of neural lipases or autophagic degradation [11, 12].
While obesity, NAFLD and NASH elicit a devastating impact, together these dysfunctions in adipose tissue and liver often lead to further complications. The most characterized and detrimental disease conditions resulting from obesity and liver pathologies are insulin resistance and diabetes. By disrupting the normal flow of FAs and their ectopic deposition as TG and toxic lipids across peripheral tissues, alterations in energy metabolism, glucose utilization, inflammation state and plasma metabolite concentrations occur. As a result, whole-body and tissue-specific insulin sensitivity can be negatively affected [13, 14]. Chronic insulin resistance and sustained hyperlipidemia resulting from exacerbated FA and TG levels globally promote the development of type 2 diabetes. Therefore, the “FA cycle” that occurs between adipose tissue and liver is of vital importance to not only energy homeostasis but overall metabolic health as well.
A key mechanism in regulating intracellular lipid content and mobilization of FAs from TG stores is lipolysis. This is the sequential hydrolytic breakdown of TG into FAs and glycerol. The rate-limiting step is the cleavage of the first ester bond in TGs by adipose triglyceride lipase (ATGL) [15, 16]. Alterations in ATGL expression or/and function ultimately impact a plethora of FA-dependent processes including oxidative activity and lipid accumulation in a variety of tissues. Therefore, refereeing the enzymatic function of ATGL is of immense importance. In 2010, our laboratory reported the discovery of the ATGL inhibitory properties of G0/G1 Switch Gene 2 (G0S2) [17]. In this article, we summarize recent advances in research exploring the function of G0S2 in the regulation of ATGL-mediated lipolysis as well as of adipose-liver lipid flux and whole-body energy metabolism.
2. ATGL-mediated intracellular lipolysis
The biochemical process of intracellular lipolysis is likewise conserved in adipocyte and non-adipocyte cell types. Intracellular lipolysis is mediated by cytosolic lipases that catalyze the hydrolysis of TGs in response to fasting and/or nutrient deprivation. For a long time it was believed that the enzyme hormone sensitive lipase (HSL) was the major TG hydrolase in mammalian cells. However, studies over the past 12 years have identified and confirmed the rate-limiting enzyme of lipolysis as ATGL [16, 18–21], also named desnutrin and patatin-like phospholipase domain-containing protein 2 (PNPLA2). ATGL is responsible for catalyzing the first step in a sequence of three step-wise reactions that define lipolysis [21]. Structurally, the N-terminal portion of ATGL contains a predicted α/β-hydrolase fold and an overlapping patatin-like domain. A nucleophilic serine residue is located within the patatin-like domain at position 47, and is part of an unconventional catalytic dyad (Ser47, Asp166) similar to the one found in human cytosolic phospholipase A2 (cPLA2). A hydrophobic stretch of 45 amino acids is present in the C-terminal region and has been shown to mediate targeting of ATGL to the surface of TG-containing LDs. It is now known that the in vivo enzymatic action of ATGL is decided not only by its own lipase activity, but also by its LD localization and interaction with its co-activator comparative gene identification 58 (CGI-58) [22]. In humans, mutations of CGI-58 and ATGL have been identified as a causative factor for various forms of neutral lipid storage disease (NLSD) characterized by TG deposition in various non-adipose tissues [23].
The phenotypic evaluation of ATGL knockout mice has extended our understanding of ATGL’s role in the control of lipid and energy metabolism in vivo. Whole-body and adipose-specific ATGL knockout mice both exhibited impaired lipolysis in adipose tissue and reduced FA availability in the plasma, leading to compensatory increases in overall glucose utilization and insulin sensitivity [24–27]. In addition, ATGL ablation in brown adipose tissue (BAT) caused severely impaired cold adaptation and an increased expression of white adipose tissue (WAT)-enriched genes. The results suggest that ATGL-mediated lipolysis is critical for generating FAs for oxidation and uncoupling protein-1 (UCP1)-dependent thermogenesis. In nonadipose tissues such as liver and cardiac muscle, ATGL selectively channels hydrolyzed FAs to β-oxidation in mitochondria [28–30]. In fact, massive lipid accumulation in cardiac myocytes caused congestive heart failure and early death of the whole-body knockout mice [24]. Exercise-induced lipolysis was also blunted in these mice. In mouse liver, ATGL ablation led to the impairment in FA oxidation along with the development of hepatic steatosis [26, 29]. When fed with a NASH-inducing diet or challenged by bacterial endotoxin, ATGL knockout mice showed exacerbated hepatic steatosis as well as enhanced hepatic inflammation, increased mortality and torpor [31]. Conversely, adenovirus-mediated overexpression of ATGL in liver promoted FA oxidation, reduced lipotoxicity and ameliorated steatosis in mice with diet-induced obesity (DIO) [32]. Apart from its effects on TG mobilization and FA trafficking, ATGL is also an important regulator of peroxisome proliferator-activated receptor-α (PPARα) in oxidative tissues. Conditional ATGL deficiency decreased mRNA expression of PPARα target genes, resulting in reduced oxidative capacity in BAT, cardiac muscle and liver [25, 27, 29, 30]. In humans, mutations in ATGL cause neutral lipid storage disease characterized by TG deposition in multiple nonadipose tissues and mild myopathy (NLSDM) [23, 33]. Intriguingly, neither NLSDM patients nor adipose-specific ATGL knockout mice were found to be exceedingly obese. These somewhat surprising results are explained, at least in part, by the observations that an adaptive reduction in lipid uptake and synthesis occurs in WAT in response to ATGL deficiency [27, 34].
3. Identification and in vitro characterization of G0S2 as an ATGL inhibitor
G0S2 was originally identified in cultured mononuclear cells in response to drug-induced cell cycle transition from the G0 to G1 phase [35] [36]. In both mouse and human genomes G0S2 is located on chromosome 1 and encodes a small basic protein of 103 amino acids. The G0S2 protein is highly conserved between species with 78% identity between the mouse and human isoforms. Secondary structure prediction suggests the G0S2 protein contains two α-helices separated by a hydrophobic region that has the potential to generate turns and assume a β-sheet conformation.
The expression of G0S2 mRNA has been profiled in a variety of human and mouse cell types. Although results from a limited number of studies have implied that G0S2 is a multifaceted protein involved in a variety of cellular functions including proliferation, apoptosis, inflammation, metabolism, oxidative phosphorylation and possibly carcinogenesis [37–43], recent studies have provided convincing evidence that G0S2 is abundantly expressed in metabolically active tissues such as adipose tissue, liver, heart and skeletal muscle, and acts as a molecular brake on TG catabolism.
A major advance in understanding the function of G0S2 was the discovery that G0S2 is able to directly bind to ATGL and inhibit its TG hydrolase activity [17]. Initial findings showed that G0S2 inhibits ATGL activity and its ability to degrade intracellular LDs even in the presence of its co-activator CGI-58 [44–46]. Subsequent mutagenesis analysis revealed that the interaction and the inhibition require the central hydrophobic domain (HD) of G0S2 and the patatin-like region of ATGL. A more recent study by Cerk et al. demonstrated that a peptide derived from the G0S2 HD covering residues Lys20 to Ala52 is capable of inhibiting ATGL in a dose dependent, non-competitive manner and with an IC50 in the nanomolar range [47]. The sequence of this peptide is highly conserved between human and mouse G0S2. The inhibition is specific for ATGL since the peptide did not inhibit various lipases tested in the same study including HSL, monoacylglycerol lipase (MGL), lipoprotein lipase (LPL), or lysophospholipases from the PNPLA protein family such as PNPLA6 and PNPLA7 [47].
Studies using cultured adipocytes provided first pieces of evidence supporting an inhibitory role for G0S2 in adipocyte lipolysis. In differentiated mouse 3T3-L1 and human Simpson–Golabi–Behmel syndrome (SGBS) adipocytes, overexpression of G0S2 was capable of decreasing both basal and adrenergically stimulated lipolysis, whereas knockdown of endogenous G0S2 enhanced lipolysis and FA efflux under the same conditions [17, 48]. β-adrenergic signals are known to up-regulate the enzymatic action of ATGL in adipocytes. Mechanisms for ATGL activation have been suggested to involve dissociation of CGI-58 from phosphorylated perilipin 1 [48, 49] as well as direct phosphorylation of ATGL by PKA [50]. In addition, our group and others found that ATGL also undergoes HSL-like translocation from cytosol to LDs in a manner dependent on perilipin 1 phosphorylation [45, 51, 52]. Interestingly, G0S2 was observed to translocate to LDs along with ATGL in response to β-adrenergic agonists, and this translocation was abolished when ATGL expression was suppressed [17]. On the other hand, binding of G0S2 enabled LD localization of ATGL mutants that are C-terminally truncated and thus disabled in self-anchoring to LDs [53]. Thus, an interdependent relationship appears to exist between G0S2 and ATGL in their localization to LDs, though the functional relevance of this mutual recruitment remains unclear. Furthermore, TNF-α is known to potently stimulate basal lipolysis in adipocytes, which may contribute to hyperlipidemia and peripheral insulin resistance in obesity [54–56]. Work by our laboratory demonstrated that TNF-α treatment causes a rapid abrogation of G0S2 expression, and ectopic expression of G0S2 is able to significantly decrease TNF-α-stimulate lipolysis mediated by ATGL [57]. The results indicate that the early reduction of G0S2 content is permissive for TNF-α-induced lipolysis in adipocytes.
4. Physiologic role of G0S2 in regulating lipid and energy metabolism
In the fed state, G0S2 protein expression increases in adipose tissue and decreases in the liver; these changes are reversed by subsequent fasting [58]. The generation and phenotypic characterization of multiple G0S2 knockout and overexpression animal models have shed light on the physiologic function of G0S2 in tissue-specific lipolysis and global energy homeostasis [58–66]. In general, the findings obtained from different studies have been remarkably consistent in showing in different tissue settings that G0S2 inhibits lipolysis by direct protein-protein interaction with ATGL.
4.1 G0S2 and adipose lipolysis
In three G0S2 whole-body knockout mouse models [58–60], basal as well as stimulated lipolytic rates were increased in the WAT. Consequently, these mice were lean, cold tolerant and resistant to HFD-induced obesity and insulin resistance. The increased energy expenditure observed in these mice was explained by augmented adipocyte FA oxidation, which promoted thermogenic function of BAT and browning-like changes in WAT. Conversely, transgenic overexpression of G0S2 in mice strongly attenuated adipose lipolysis and FA flux to liver in response to fasting and β-adrenergic stimulation [62]. As a result, these mice experienced difficulty in shifting from carbohydrate to FA oxidation during fasting. Moreover, G0S2 overexpression promoted accumulation of more and larger LDs in brown adipocytes, leading to defective cold adaptation in the transgenic mice. In response to HFD feeding, the transgenic mice displayed a greater gain of body weight and adiposity along with decreases in the fasting plasma levels of free FAs, TGs, and insulin. However, glucose and insulin tolerance was improved in these mice [62]. In a transgenic quail model, overexpression of G0S2 in the adipose tissue resulted in a reduced elevation of plasma free FA levels and smaller reduction in fat mass in response to food restriction and fasting [63]. Overexpression of G0S2 also inhibited adipose lipolysis during early and active laying, leading to decreased supply of lipids for yolk synthesis and delayed onset of egg production [67]. In free-ranging brown bears, G0S2 expression was found to markedly elevate in adipose tissue in late summer prior to hibernation, coinciding with lipolytic suppression and increased bodyweight and fat mass gain, glucose utilization and insulin sensitivity [68]. Together, these studies have demonstrated a key role for G0S2 as a regulator of adipose lipolysis and the overall FA availability. The evidence on a whole has revealed a molecular mechanism that can contribute to explaining the paradox of improved insulin sensitivity and glucose tolerance during weight and fat mass gain.
4.2 Hepatic G0S2 and steatosis
One of the most drastic changes observed in the G0S2 knockout mice was their lack of liver TG. Specifically, these mice exhibited an impaired fasting response in terms of hepatic TG accumulation and were resistant to HFD-induced liver steatosis [58, 60]. Both of these effects were also demonstrated with liver-specific knockdown of G0S2 expression [58]. In normal chow-fed wild type mice, hepatic G0S2 expression peaks early within 6 h of fasting. Loss of G0S2 in the liver led to increased expression of FA oxidation genes and ketogenesis as well as accelerated gluconeogenesis and decelerated glycogen breakdown [58]. Conversely, liver-specific overexpression of G0S2 in mice resulted in steatosis and elicited opposite effects including reduced rates of lipolysis, FA oxidation, and ketogenesis [58, 64]. The data indicate that by limiting FA availability during the early stage of fasting, hepatic G0S2 may act to ensure the usage of glycogen-derived glucose as the primary source of rapid energy output. In HFD-fed mice, we found that hepatic G0S2 levels rise in the fed state. Interestingly, whole-body glucose tolerance and insulin sensitivity were improved in response to both global and hepatic loss of G0S2 [58]. Consistent with the findings obtained from mice, liver-specific G0S2 overexpression in male Wistar rats promoted hepatic insulin resistance by exacerbating HFD-induced hepatic steatosis [65]. Together, the results from these studies imply that while G0S2 plays a critical role in regulating the hepatic adaptive energy response to fasting, sustained G0S2 expression in liver during HFD feeding promotes a detrimental effect on systemic insulin response and energy balance.
4.3 Role of G0S2 in muscle
Although the current studies focus primarily on the function of G0S2 in adipose tissue and liver, emerging evidence has also pointed to an important role of G0S2 in regulating TG and FA metabolism in striated muscles. For example, G0S2 was recently shown to express and function in cardiac muscle, where G0S2 expression pattern resembles that of adipose tissue, i.e. lower in fasted than in ad libitum fed state. G0S2 deficiency resulted in a de-repression of cardiac lipolysis and decreased cardiac TG content [66]. Transgenic overexpression of G0S2, on the other hand, inhibited cardiac lipolysis and caused severe cardiac steatosis [66]. Moreover, a separate study has found that G0S2 protein is highly expressed in mouse soleus muscle (largely made up of slow oxidative fibers), and G0S2 expression in rat and human skeletal muscle is positively associated with the oxidative capacity and lipid content [69, 70]. Overexpression and knockdown experiments using human primary myotubes and mouse skeletal muscle showed that G0S2 controls lipolysis and FA oxidation as well as TG content in an ATGL-dependent manner. Importantly, knockdown of G0S2 also reduced glucose metabolism and enhanced mitochondrial function [69]. Together, these studies have offered insight to the role of G0S2 as an important modulator of lipolysis and substrate utilization in oxidative muscle types.
5. Regulation of G0S2 mRNA and protein expression
5.1 G0S2 expression in adipose tissue
In situ hybridization analysis of mouse embryos provided the first evidence for the predominant expression of G0S2 in adipose tissue [71]. Successive studies confirmed high-level expression of G0S2 in adipose tissue of adult humans and animals including mice, pigs, bears and avian species [68, 72–77]. In cultured human SGBS and mouse 3T3-L1 pre-adipocyte cell lines, both G0S2 mRNA and protein have been shown to exhibit adipogenesis-dependent expression [45, 61, 72, 75]. In humans, G0S2 mRNA expression in subcutaneous adipose tissue was found to be 7 times higher in mature adipocytes than in the cells of corresponding stroma-vascular fraction [77]. Sequence analysis revealed that the G0S2 promoter region encompasses a potential PPAR-responsive element (PPRE), and Zandbergen et al. subsequently provided evidence that G0S2 expression in adipogenesis is increased by PPARγ [72]. A separate study recently showed that knockdown of either PPARγ or G0S2 resulted in apoptotic induction in 3T3-L1 cells before terminal differentiation [61]. The lack of notable defects in the adipose development of G0S2 whole-body knockout mice [58–60], however, argues against a significant role of G0S2 in adipocyte differentiation in vivo. In addition to PPARγ agonism, the adipose expression of G0S2 is subject to regulation under different nutritional conditions. Specifically, it is at very low levels during fasting but increases following feeding [58, 73, 78]. This expression pattern is consistent with the observation derived from cultured adipocytes where G0S2 is upregulated by insulin and downregulated in response to β-adrenergic stimulation [58]. Furthermore, the G0S2 protein has a relatively short half-life of approximately 15 min [79]. While transcriptional regulation of G0S2 expression is no doubt a major contributor to intracellular protein levels, recent evidence has shown G0S2 stabilization at the protein level by the presence of ATGL and in response to FA-induced TG accumulation. The observation that G0S2 protein but not mRNA levels were reduced in the adipose tissue of ATGL-deficient mice corroborates the involvement of ATGL in the stabilization of G0S2 protein [79].
5.2 Control of hepatic G0S2 transcription
Compared to its high expression in adipose tissue in the fed state, levels of G0S2 in liver are low when nutrients are abundant. In response to fasting, hepatic expression of G0S2 mRNA and protein is robustly increased [58, 80]. An early study demonstrated that fasting-induced G0S2 expression was abolished in the liver of mice lacking PPARα, the master transcriptional regulator of FA catabolism. In addition, treatment with Wy14643, a synthetic PPARα ligand, was able to upregulate G0S2 mRNA levels in mouse liver and isolated hepatocytes in a PPARα-dependent manner [72]. These results strongly suggest that G0S2 is a target gene of PPARα in liver. Despite the fact that the G0S2 promoter region contains a PPRE, however, whether hepatic G0S2 expression is directly activated by PPARα remains uncertain. Overexpression of PPARα and treatment with a PPARα agonist in combination failed to stimulate the promoter activity of G0S2 in a cell reporter assay [72]. The PPARα agonist Wy14643 also failed to considerably increase G0S2 expression in liver in the absence of adipose FA influx [80]. Additionally, our most recent study showed that chemical antagonism of PPARα in mice elicited no effect on fasting-induced hepatic G0S2 expression [81]. Collectively, the existing evidence points to a more complex scenario of G0S2 regulation by PPARα. Furthermore, the G0S2 gene contains a carbohydrate response element (ChoRE) within its promoter region, and results from a previous study suggest that ChREBP may regulate hepatic G0S2 in response to glucose [82]. However, this pathway is unlikely to account for increased G0S2 expression in the fasting liver as glucose availability would be limited.
Recent evidence obtained from studies of adipose-specific ATGL and CGI-58 knockout mice has demonstrated that during fasting FAs derived from adipose lipolysis are critical for promoting hepatic G0S2 expression [80, 81]. Our co-culture experiments further revealed that the effect is the consequence of a direct induction of G0S2 gene transcription in hepatocytes by unsaturated FAs such as oleic acid and linoleic acid, which are released from lipolytically stimulated adipose tissue [81]. In the same study, we obtained compelling evidence that G0S2 is a direct target gene of liver X receptor α subtype (LXRα). LXRs are transcription factors essential for cholesterol homeostasis and lipogenesis [83, 84]. LXRα has previously been implicated in regulating hepatic TG accumulation both during fasting [85] and upon stimulation of de novo FA synthesis [86–89]. Using ChIP analysis and cell reporter assays, we demonstrated that transcriptional activation of G0S2 expression is conferred by LXRα binding to a direct repeat 4 (DR4) motif in the G0S2 promoter [81]. The importance of LXRα for G0S2 expression was further recognized in experiments using LXRα knockout mice where induction of both hepatic G0S2 expression by fasting or LXR agonist agonists was abolished [81].
Despite the data strongly supporting a predominant role for LXRα, the mechanisms underlying the specific activation of G0S2 by LXRα during fasting are less clear, given that oxysterols but not FAs are widely considered to be ligands of LXRs. In the fed state, LXRs are known to interact with insulin in the control of hepatic SREBP-1c and genes for de novo FA synthesis [90]. In fasting liver, it is possible that the absence of insulin and the presence of unsaturated FAs alter the composition of cofactors and lipid mediators, thereby decreasing the binding LXRα to the SREBP-1c promoter while enhancing the specificity of LXRα for the G0S2 promoter. Since LXR functions in a heterodimeric complex with the retinoid X receptor (RXR), it is also possible that the FA responsiveness of LXRα:RXR is mediated via FA modification of RXR. In this regard, RXR has long been recognized as an FA receptor that can be activated upon FA binding [91–95]. Further studies are needed to unveil the mechanistic details of LXRα-mediated G0S2 transcription in the presence of FAs. Moreover, several previous studies have demonstrated that extensive crosstalk between LXR and PPARα drives lipid metabolism in response to oxysterols and FAs [96–101]. According to a recent study, simultaneous activation of LXR/PPARα synergistically exacerbates hepatic steatosis via modulating the expression of a set of genes involved in lipid and glucose metabolism as well as FGF21 [102]. Thus, it would be tempting to determine whether G0S2 is also under the dual regulation of LXRα and PPARα. Furthermore, Zhang et al. have recently shown that FoxO1 promotes intrahepatic lipolysis and FA oxidation through concomitantly stimulating ATGL and suppressing G0S2 expression [103]. Interestingly, FoxO1 is known to antagonize de novo lipogenesis via reducing the binding of LXRα to the SREBP-1c promoter [104, 105]. It is possible that a similar mechanism may be employed for FoxO1 to downregulate G0S2 transcription by LXRα.
6. G0S2 and human pathologies
Since the discovery of G0S2 as a lipolytic inhibitor, several studies on the expression pattern of G0S2 have shed light on its potential role in regulating adipose lipolysis and FA mobilization in humans. Previous studies by Nielsen and colleagues have shown in humans that prolonged fasting is associated with a substantial decrease in adipose G0S2 expression [78]. A recent study also showed that G0S2 mRNA expression was substantially higher in subcutaneous compared to omental adipose tissue, and in both depots cell size inversely correlated with G0S2 expression [77]. Intriguingly, obese subjects exhibited decreased mRNA levels of G0S2 in subcutaneous adipose tissue despite decreased lipolysis per unit of fat mass [77]. In patients with poorly controlled type 2 diabetes, reduced expression of G0S2 and perilipin 1 in adipose tissue was observed, which might contribute to increased ATGL lipolysis and circulating free FA levels post insulin withdrawal [106]. In an experimental human model of diabetic ketoacidosis (DKA), lipolytic stimulation resulting from the release of proinflammatory cytokines and stress hormones was found to be associated with decreased G0S2 and increased CGI-58 expression in adipose tissue [107]. Given that G0S2 does inhibit ATGL activity in cultured human adipocytes, results from these studies indicate that this regulatory mechanism is highly conserved in the adipose tissue of humans.
7. Perspectives & discussion
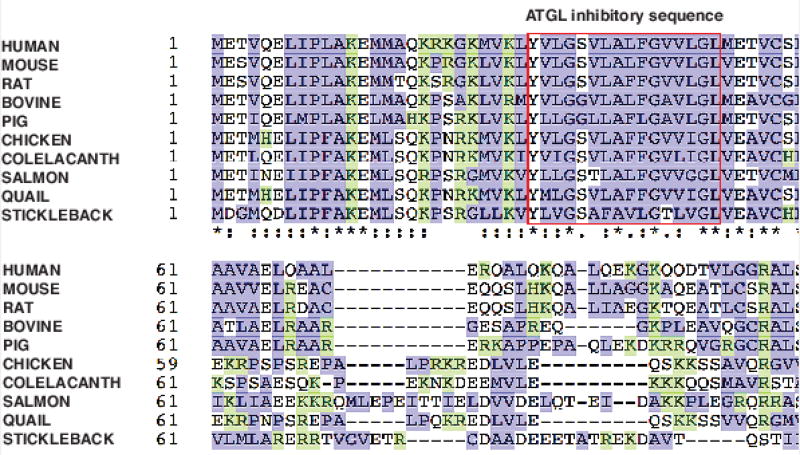
The G0S2 and ATGL functional studies have revealed the significance of the lipolytic mechanism and its regulation. In eukaryotic evolution, TG synthesis and catabolism emerged as one of the preferred pathways for mitigating fluctuations in energy demand and availability. This evolutionary establishment of TG as a primary stored nutrient to buffer energy homeostasis is often referred to as adaptive metabolism and is governed by the thrifty gene hypothesis, whereby genes promoting TG deposition and regulation were highly selected [108–113]. Concurrently, enzymes were needed to promote the mobilization of this energy reserve, hence the appearance of ATGL homologs in the genomes of early eukaryotic species such as yeast and fruit fly [114, 115]. Through genetic refinement and environmental pressures, ATGL became the predominant intracellular TG hydrolase in higher and more complex organisms. To counteract the role of ATGL and to establish a regulatory mechanism for intracellular lipolysis, G0S2 appeared in later evolution as it is only found in vertebrates [37]. As a small protein regulator, G0S2 has remained minimally altered throughout evolution since its emergence (Figure 1). Sequence identities between mammalian species range from 77% to 95% when compared to human G0S2. In particular, the G0S2 hydrophobic sequence responsible for mediating ATGL inhibition is extremely similar in vertebrates ranging from stickleback fish to birds and to rodents and humans (Figure 1). We hypothesize that this high level of conservation reflects the importance of G0S2 to the survival of lower vertebrates by promoting energy preservation. In higher vertebrates such as mammals, G0S2 evolved to become paramount for the acquisition of metabolic adaptations that allow processing of fuel utilization and storage in a cyclical manner.
Figure 1. G0S2 protein sequence alignment.
Amino acid sequences of G0S2 proteins from different species were aligned using Clustal Omega program. Overall, the sequences are well conserved with high content of basic (shown in green) and hydrophobic (shown in blue) residues. Red box contains the hydrophobic sequences required for ATGL inhibition. Identical amino acids in all proteins are marked with an asterisk (*), conservative substitutions with a colon (:), and semi-conservative substitutions with a period (.).
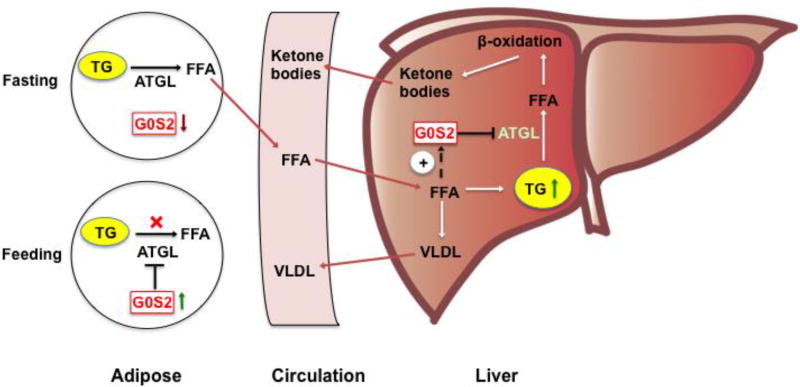
In support of the above-described evolutionary hypothesis, studies using cell and animal models have yielded compelling evidence to prove that the interplay between ATGL and G0S2 is critical for balancing tissue-specific TG storage vs. mobilization as well as for partitioning of metabolic fuels between adipose and liver (Figure 2). During periods of food abundance, increased expression of G0S2 promotes lipolytic suppression and the net TG deposition in WAT. In the periods of food shortage, G0S2 levels decrease in WAT, facilitating the release of free FAs to circulation. In response to the influx of adipose-derived FAs, G0S2 expression increases in liver, where it acts to control hepatic substrate utilization by decelerating the rates of ATGL-mediated TG hydrolysis and FA oxidation (Figure 2). We speculate that early human populations that were food insecure would have benefited from this arrangement. The increased hepatic TG storage and controlled FA mobilization conferred by G0S2 might allow for a longer period of energy availability between meals, and it may represent a metabolic advantage for surviving extended periods of famine.
Figure 2. Control of adipose-liver FA flux by G0S2.
In the fed state, G0S2 is expressed at high levels in WAT when ATGL is mostly inactive and the lipolytic rate is low. In the fasted state, G0S2 is downregulated in WAT and upregulated in the liver, favoring the mobilization of FFAs through increased adipose lipolysis and their deposition as TGs in liver. G0S2 in liver acts to coordinate hepatic substrate utilization by limiting the rates of TG hydrolysis, FA oxidation, gluconeogenesis, glycogen breakdown, and ketogenesis. Upon refeeding, these fasting-induced changes in G0S2 and substrate metabolism are reversed.
The function of G0S2 may have for the most part remained consistent over time, yet nutritional and environmental pressures have changed. In today’s society where there is an almost unlimited access to food, the role of G0S2 and many other TG promoting genes may constitute a detriment to metabolic health. Under physiologic conditions, the fasting-refeeding transition decreases the hepatic G0S2 levels when the WAT is set back to the mode of storing instead of releasing FAs. This in turn would lead to enhanced intrahepatic lipolysis, facilitating the TG clearance in liver. When a high calorie diet is readily accessible, however, the hepatic G0S2 content likely remains high after refeeding due to the influx of dietary FAs or increased de novo FA synthesis. In DIO, decreased expression of G0S2 and impaired insulin sensitivity in WAT may also lead to elevated basal adipose lipolysis and FA flux to liver. Consequently, the presence of high levels of hepatic G0S2 then becomes disadvantageous, predisposing liver to chronic steatosis, insulin resistance and other related problems. Supportive of this postulation are the findings derived from mice that ablation of G0S2 or activation of ATGL reduces adiposity, increases energy expenditure, and alleviates HFD-induced hepatic steatosis and insulin resistance [32, 58, 60]. Although data pertaining to G0S2 expression in the liver of obese and diabetic patients are currently lacking, high hepatic G0S2 levels would advocate for the benefits of down-modulating G0S2 function or expression in addressing metabolic health concerns in humans. Given that it only has a single isoform and bears no structural or sequence similarities to any other known proteins, G0S2 presents a highly attractive target for therapeutic development for treating obesity-associated disorders such as NAFLD and insulin resistance.
Another significant advance was the identification of G0S2 as a direct target gene of LXRα. In humans and rodents, a major function of LXRα is in the regulation of cholesterol metabolism. LXR agonists such as T0901317 have been shown to be extremely potent in inducing rapid decreases in global cholesterol content in human trials [116–122]. LXR agonism leads to increased reverse cholesterol transport (RCT), where low-density lipoprotein (LDL) particles (bad cholesterol) are converted to high-density lipoprotein (HDL) particles (good cholesterol) in macrophage-derived foam cells and trafficked to liver for excretion in bile and feces, thereby decreasing cumulative cholesterol content. Despite the established beneficial effects on cholesterol, pharmacological activation of LXR results in increased plasma TG and the development of hepatic steatosis [116–123]. These side effects render LXR agonists essentially useless in the clinical treatment of high cholesterol. The recent finding that G0S2 promotes TG deposition in response to LXR agonism has led to new insight into our understanding of the mechanism that underlies these TG-associated side effects of LXR agonists [81]. In particular, our data show that when G0S2 is ablated, T0901317 is rendered incapable of inducing plasma TG elevation and of causing hepatic steatosis while still retaining its positive effect on cholesterol/reverse cholesterol trafficking [81]. Thus, inhibition of G0S2 may be a means to alleviate hepatic steatosis and hypertriglyceridemia and allow for the use of LXR agonists in the treatment of hypercholesterolemia and atherosclerosis. Finally, given the key role of LXR in mediating de novo lipogenesis, it is tempting to speculate that G0S2 may also possess an ATGL-independent function in promoting synthesis of newly generated FAs to TGs. Further studies of G0S2 biochemistry and animal models with ATGL/G0S2 double deficiency will be required to provide definitive information on this possibility.
8. Concluding remarks
G0S2 has proven to be an interesting and unique regulator of lipid metabolism and energy homeostasis. The ability of G0S2 to have such a profound impact on TG deposition is intriguing. Moreover, its ability to alter energy utilization simply through restriction of FA substrates illuminates the importance of G0S2 to organism survival during the “feast or famine” cycle. In addition, work discussed in this review has provided a proof-of-principle concept that modulation of G0S2 expression could be a putative treatment avenue for a host of metabolically linked diseases characterized by ectopic TG accumulation. Furthermore, the ongoing studies suggesting of functions outside of metabolic regulation have yielded importance to other physiologic impact of G0S2. Future research is needed to uncover the full spectrum of G0S2 function in different cellular settings.
Highlights.
G0/G1 switch gene 2 (G0S2) is a specific inhibitor of intracellular lipolysis mediated by adipose triglyceride lipase (ATGL).
The review covers expression and function of G0S2 in adipose tissue and liver as a regulator of lipid and energy metabolism.
Targeting G0S2-ATGL interaction should be considered in the future.
Acknowledgments
This work was supported by research grants from the National Institutes of Health (DK089178 and DK109096) to J.L.
Abbreviations
- G0S2
G0/G1 Switch Gene 2
- HSL
hormone-sensitive lipase
- ATGL
adipose triglyceride lipase
- TG
triacylglycerol/triglyceride
- FA
fatty acid
- BAT
brown adipose tissue
- WAT
white adipose tissue
- PPAR
peroxisome proliferator-activated receptor
- PPRE
PPAR-responsive element
- LDs
lipid droplets
- SGBS
Simpson–Golabi–Behmel syndrome
- NAFLD
non-alcoholic fatty liver disease
- NASH
non-alcoholic steatohepatitis
- cPLA2
human cytosolic phospholipase A2
- CGI-58
comparative gene identification-58
- NLSD
neutral lipid storage disease
- ChoRE
carbohydrate response element
- TNF-α
tumor necrosis factor- alpha
- LXR
liver x receptor
- LDL
low density lipoprotein
- HDL
high density lipoprotein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
We have no potential conflicts of interest to report.
References
- 1.van den Berghe G. The role of the liver in metabolic homeostasis: implications for inborn errors of metabolism. Journal of inherited metabolic disease. 1991;14:407–420. doi: 10.1007/BF01797914. [DOI] [PubMed] [Google Scholar]
- 2.Cohen JC, Horton JD, Hobbs HH. Human fatty liver disease: old questions and new insights. Science. 2011;332:1519–1523. doi: 10.1126/science.1204265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin X, Yue P, Chen Z, Schonfeld G. Hepatic triglyceride contents are genetically determined in mice: results of a strain survey. American journal of physiology. Gastrointestinal and liver physiology. 2005;288:G1179–1189. doi: 10.1152/ajpgi.00411.2004. [DOI] [PubMed] [Google Scholar]
- 4.Guan HP, Goldstein JL, Brown MS, Liang G. Accelerated fatty acid oxidation in muscle averts fasting-induced hepatic steatosis in SJL/J mice. The Journal of biological chemistry. 2009;284:24644–24652. doi: 10.1074/jbc.M109.034397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nishikawa S, Doi K, Nakayama H, Uetsuka K. The effect of fasting on hepatic lipid accumulation and transcriptional regulation of lipid metabolism differs between C57BL/6J and BALB/cA mice fed a high-fat diet. Toxicologic pathology. 2008;36:850–857. doi: 10.1177/0192623308323920. [DOI] [PubMed] [Google Scholar]
- 6.Lin EC. Glycerol utilization and its regulation in mammals. Annual review of biochemistry. 1977;46:765–795. doi: 10.1146/annurev.bi.46.070177.004001. [DOI] [PubMed] [Google Scholar]
- 7.Malhi H, Gores GJ. Molecular mechanisms of lipotoxicity in nonalcoholic fatty liver disease. Seminars in liver disease. 2008;28:360–369. doi: 10.1055/s-0028-1091980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jensen MD. Role of body fat distribution and the metabolic complications of obesity. The Journal of clinical endocrinology and metabolism. 2008;93:S57–63. doi: 10.1210/jc.2008-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petta S, Gastaldelli A, Rebelos E, Bugianesi E, Messa P, Miele L, Svegliati-Baroni G, Valenti L, Bonino F. Pathophysiology of Non Alcoholic Fatty Liver Disease. International journal of molecular sciences. 2016;17 doi: 10.3390/ijms17122082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mashek DG, Khan SA, Sathyanarayan A, Ploeger JM, Franklin MP. Hepatic lipid droplet biology: Getting to the root of fatty liver. Hepatology. 2015;62:964–967. doi: 10.1002/hep.27839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ueno T, Komatsu M. Autophagy in the liver: functions in health and disease. Nature reviews. Gastroenterology & hepatology. 2017;14:170–184. doi: 10.1038/nrgastro.2016.185. [DOI] [PubMed] [Google Scholar]
- 12.Cingolani F, Czaja MJ. Regulation and Functions of Autophagic Lipolysis. Trends in endocrinology and metabolism: TEM. 2016;27:696–705. doi: 10.1016/j.tem.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samuel VT, Shulman GI. The pathogenesis of insulin resistance: integrating signaling pathways and substrate flux. The Journal of clinical investigation. 2016;126:12–22. doi: 10.1172/JCI77812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glass CK, Olefsky JM. Inflammation and lipid signaling in the etiology of insulin resistance. Cell metabolism. 2012;15:635–645. doi: 10.1016/j.cmet.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zechner R, Kienesberger PC, Haemmerle G, Zimmermann R, Lass A. Adipose triglyceride lipase and the lipolytic catabolism of cellular fat stores. J Lipid Res. 2009;50:3–21. doi: 10.1194/jlr.R800031-JLR200. [DOI] [PubMed] [Google Scholar]
- 16.Zechner R, Zimmermann R, Eichmann TO, Kohlwein SD, Haemmerle G, Lass A, Madeo F. FAT SIGNALS--lipases and lipolysis in lipid metabolism and signaling. Cell metabolism. 2012;15:279–291. doi: 10.1016/j.cmet.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang X, Lu X, Lombes M, Rha GB, Chi YI, Guerin TM, Smart EJ, Liu J. The G(0)/G(1) switch gene 2 regulates adipose lipolysis through association with adipose triglyceride lipase. Cell Metab. 2010;11:194–205. doi: 10.1016/j.cmet.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Girousse A, Langin D. Adipocyte lipases and lipid droplet-associated proteins: insight from transgenic mouse models. International journal of obesity. 2012;36:581–594. doi: 10.1038/ijo.2011.113. [DOI] [PubMed] [Google Scholar]
- 19.Jenkins CM, Mancuso DJ, Yan W, Sims HF, Gibson B, Gross RW. Identification, cloning, expression, and purification of three novel human calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. J Biol Chem. 2004;279:48968–48975. doi: 10.1074/jbc.M407841200. [DOI] [PubMed] [Google Scholar]
- 20.Villena JA, Roy S, Sarkadi-Nagy E, Kim KH, Sul HS. Desnutrin, an adipocyte gene encoding a novel patatin domain-containing protein, is induced by fasting and glucocorticoids: ectopic expression of desnutrin increases triglyceride hydrolysis. J Biol Chem. 2004;279:47066–47075. doi: 10.1074/jbc.M403855200. [DOI] [PubMed] [Google Scholar]
- 21.Zimmermann R, Strauss JG, Haemmerle G, Schoiswohl G, Birner-Gruenberger R, Riederer M, Lass A, Neuberger G, Eisenhaber F, Hermetter A, Zechner R. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 2004;306:1383–1386. doi: 10.1126/science.1100747. [DOI] [PubMed] [Google Scholar]
- 22.Lass A, Zimmermann R, Haemmerle G, Riederer M, Schoiswohl G, Schweiger M, Kienesberger P, Strauss JG, Gorkiewicz G, Zechner R. Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman Syndrome. Cell Metab. 2006;3:309–319. doi: 10.1016/j.cmet.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 23.Schweiger M, Lass A, Zimmermann R, Eichmann TO, Zechner R. Neutral lipid storage disease: genetic disorders caused by mutations in adipose triglyceride lipase/PNPLA2 or CGI-58/ABHD5. Am J Physiol Endocrinol Metab. 2009;297:E289–296. doi: 10.1152/ajpendo.00099.2009. [DOI] [PubMed] [Google Scholar]
- 24.Haemmerle G, Lass A, Zimmermann R, Gorkiewicz G, Meyer C, Rozman J, Heldmaier G, Maier R, Theussl C, Eder S, Kratky D, Wagner EF, Klingenspor M, Hoefler G, Zechner R. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science. 2006;312:734–737. doi: 10.1126/science.1123965. [DOI] [PubMed] [Google Scholar]
- 25.Ahmadian M, Abbott MJ, Tang T, Hudak CS, Kim Y, Bruss M, Hellerstein MK, Lee HY, Samuel VT, Shulman GI, Wang Y, Duncan RE, Kang C, Sul HS. Desnutrin/ATGL is regulated by AMPK and is required for a brown adipose phenotype. Cell metabolism. 2011;13:739–748. doi: 10.1016/j.cmet.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kienesberger PC, Lee D, Pulinilkunnil T, Brenner DS, Cai L, Magnes C, Koefeler HC, Streith IE, Rechberger GN, Haemmerle G, Flier JS, Zechner R, Kim YB, Kershaw EE. Adipose triglyceride lipase (ATGL) deficiency causes tissue-specific changes in insulin signaling. J Biol Chem. 2009 doi: 10.1074/jbc.M109.047787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schoiswohl G, Stefanovic-Racic M, Menke MN, Wills RC, Surlow BA, Basantani MK, Sitnick MT, Cai L, Yazbeck CF, Stolz DB, Pulinilkunnil T, O'Doherty RM, Kershaw EE. Impact of Reduced ATGL-Mediated Adipocyte Lipolysis on Obesity-Associated Insulin Resistance and Inflammation in Male Mice. Endocrinology. 2015;156:3610–3624. doi: 10.1210/en.2015-1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu JW, Wang SP, Alvarez F, Casavant S, Gauthier N, Abed L, Soni KG, Yang G, Mitchell GA. Deficiency of liver adipose triglyceride lipase in mice causes progressive hepatic steatosis. Hepatology. 2011;54:122–132. doi: 10.1002/hep.24338. [DOI] [PubMed] [Google Scholar]
- 29.Ong KT, Mashek MT, Bu SY, Greenberg AS, Mashek DG. Adipose triglyceride lipase is a major hepatic lipase that regulates triacylglycerol turnover and fatty acid signaling and partitioning. Hepatology. 2011;53:116–126. doi: 10.1002/hep.24006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haemmerle G, Moustafa T, Woelkart G, Buttner S, Schmidt A, van de Weijer T, Hesselink M, Jaeger D, Kienesberger PC, Zierler K, Schreiber R, Eichmann T, Kolb D, Kotzbeck P, Schweiger M, Kumari M, Eder S, Schoiswohl G, Wongsiriroj N, Pollak NM, Radner FP, Preiss-Landl K, Kolbe T, Rulicke T, Pieske B, Trauner M, Lass A, Zimmermann R, Hoefler G, Cinti S, Kershaw EE, Schrauwen P, Madeo F, Mayer B, Zechner R. ATGL-mediated fat catabolism regulates cardiac mitochondrial function via PPAR-alpha and PGC-1. Nature medicine. 2011;17:1076–1085. doi: 10.1038/nm.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jha P, Claudel T, Baghdasaryan A, Mueller M, Halilbasic E, Das SK, Lass A, Zimmermann R, Zechner R, Hoefler G, Trauner M. Role of adipose triglyceride lipase (PNPLA2) in protection from hepatic inflammation in mouse models of steatohepatitis and endotoxemia. Hepatology. 2014;59:858–869. doi: 10.1002/hep.26732. [DOI] [PubMed] [Google Scholar]
- 32.Reid BN, Ables GP, Otlivanchik OA, Schoiswohl G, Zechner R, Blaner WS, Goldberg IJ, Schwabe RF, Chua SC, Jr, Huang LS. Hepatic overexpression of hormone-sensitive lipase and adipose triglyceride lipase promotes fatty acid oxidation, stimulates direct release of free fatty acids, and ameliorates steatosis. J Biol Chem. 2008;283:13087–13099. doi: 10.1074/jbc.M800533200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fischer J, Lefevre C, Morava E, Mussini JM, Laforet P, Negre-Salvayre A, Lathrop M, Salvayre R. The gene encoding adipose triglyceride lipase (PNPLA2) is mutated in neutral lipid storage disease with myopathy. Nat Genet. 2007;39:28–30. doi: 10.1038/ng1951. [DOI] [PubMed] [Google Scholar]
- 34.Schreiber R, Hofer P, Taschler U, Voshol PJ, Rechberger GN, Kotzbeck P, Jaeger D, Preiss-Landl K, Lord CC, Brown JM, Haemmerle G, Zimmermann R, Vidal-Puig A, Zechner R. Hypophagia and metabolic adaptations in mice with defective ATGL-mediated lipolysis cause resistance to HFD-induced obesity. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:13850–13855. doi: 10.1073/pnas.1516004112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Russell L, Forsdyke DR. A human putative lymphocyte G0/G1 switch gene containing a CpG-rich island encodes a small basic protein with the potential to be phosphorylated. DNA Cell Biol. 1991;10:581–591. doi: 10.1089/dna.1991.10.581. [DOI] [PubMed] [Google Scholar]
- 36.Siderovski DP, Blum S, Forsdyke RE, Forsdyke DR. A set of human putative lymphocyte G0/G1 switch genes includes genes homologous to rodent cytokine and zinc finger protein-encoding genes. DNA and cell biology. 1990;9:579–587. doi: 10.1089/dna.1990.9.579. [DOI] [PubMed] [Google Scholar]
- 37.Heckmann BL, Zhang X, Xie X, Liu J. The G0/G1 switch gene 2 (G0S2): regulating metabolism and beyond. Biochimica et biophysica acta. 2013;1831:276–281. doi: 10.1016/j.bbalip.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kioka H, Kato H, Fujikawa M, Tsukamoto O, Suzuki T, Imamura H, Nakano A, Higo S, Yamazaki S, Matsuzaki T, Takafuji K, Asanuma H, Asakura M, Minamino T, Shintani Y, Yoshida M, Noji H, Kitakaze M, Komuro I, Asano Y, Takashima S. Evaluation of intramitochondrial ATP levels identifies G0/G1 switch gene 2 as a positive regulator of oxidative phosphorylation. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:273–278. doi: 10.1073/pnas.1318547111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yim CY, Sekula DJ, Hever-Jardine MP, Liu X, Warzecha JM, Tam J, Freemantle SJ, Dmitrovsky E, Spinella MJ. G0S2 Suppresses Oncogenic Transformation by Repressing a MYC-Regulated Transcriptional Program. Cancer research. 2016;76:1204–1213. doi: 10.1158/0008-5472.CAN-15-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y, Zhang Y, Zhu Y, Zhang P. Lipolytic inhibitor G0/G1 switch gene 2 inhibits reactive oxygen species production and apoptosis in endothelial cells. American journal of physiology. Cell physiology. 2015;308:C496–504. doi: 10.1152/ajpcell.00317.2014. [DOI] [PubMed] [Google Scholar]
- 41.Matsunaga N, Ikeda E, Kakimoto K, Watanabe M, Shindo N, Tsuruta A, Ikeyama H, Hamamura K, Higashi K, Yamashita T, Kondo H, Yoshida Y, Matsuda M, Ogino T, Tokushige K, Itcho K, Furuichi Y, Nakao T, Yasuda K, Doi A, Amamoto T, Aramaki H, Tsuda M, Inoue K, Ojida A, Koyanagi S, Ohdo S. Inhibition of G0/G1 Switch 2 Ameliorates Renal Inflammation in Chronic Kidney Disease. EBioMedicine. 2016;13:262–273. doi: 10.1016/j.ebiom.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zagani R, El-Assaad W, Gamache I, Teodoro JG. Inhibition of adipose triglyceride lipase (ATGL) by the putative tumor suppressor G0S2 or a small molecule inhibitor attenuates the growth of cancer cells. Oncotarget. 2015;6:28282–28295. doi: 10.18632/oncotarget.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamada T, Park CS, Shen Y, Rabin KR, Lacorazza HD. G0S2 inhibits the proliferation of K562 cells by interacting with nucleolin in the cytosol. Leukemia research. 2014;38:210–217. doi: 10.1016/j.leukres.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cornaciu I, Boeszoermenyi A, Lindermuth H, Nagy HM, Cerk IK, Ebner C, Salzburger B, Gruber A, Schweiger M, Zechner R, Lass A, Zimmermann R, Oberer M. The minimal domain of adipose triglyceride lipase (ATGL) ranges until leucine 254 and can be activated and inhibited by CGI-58 and G0S2, respectively. PLoS One. 2011;6:e26349. doi: 10.1371/journal.pone.0026349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang X, Lu X, Lombes M, Rha GB, Chi YI, Guerin TM, Smart EJ, Liu J. The G(0)/G(1) switch gene 2 regulates adipose lipolysis through association with adipose triglyceride lipase. Cell metabolism. 2010;11:194–205. doi: 10.1016/j.cmet.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu X, Yang X, Liu J. Differential control of ATGL-mediated lipid droplet degradation by CGI-58 and G0S2. Cell Cycle. 2010;9:2719–2725. doi: 10.4161/cc.9.14.12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cerk IK, Salzburger B, Boeszoermenyi A, Heier C, Pillip C, Romauch M, Schweiger M, Cornaciu I, Lass A, Zimmermann R, Zechner R, Oberer M. A Peptide Derived from G0/G1 Switch Gene 2 Acts as Non-competitive Inhibitor of Adipose Triglyceride Lipase. The Journal of biological chemistry. 2014 doi: 10.1074/jbc.M114.602599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Granneman JG, Moore HP, Granneman RL, Greenberg AS, Obin MS, Zhu Z. Analysis of lipolytic protein trafficking and interactions in adipocytes. J Biol Chem. 2007;282:5726–5735. doi: 10.1074/jbc.M610580200. [DOI] [PubMed] [Google Scholar]
- 49.Granneman JG, Moore HP, Krishnamoorthy R, Rathod M. Perilipin controls lipolysis by regulating the interactions of AB-hydrolase containing 5 (Abhd5) and adipose triglyceride lipase (Atgl) J Biol Chem. 2009;284:34538–34544. doi: 10.1074/jbc.M109.068478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pagnon J, Matzaris M, Stark R, Meex RC, Macaulay SL, Brown W, O'Brien PE, Tiganis T, Watt MJ. Identification and functional characterization of protein kinase A phosphorylation sites in the major lipolytic protein, adipose triglyceride lipase. Endocrinology. 2012;153:4278–4289. doi: 10.1210/en.2012-1127. [DOI] [PubMed] [Google Scholar]
- 51.Schweiger M, Lass A, Zimmermann R, Eichmann TO, Zechner R. Neutral lipid storage disease: genetic disorders caused by mutations in adipose triglyceride lipase/PNPLA2 or CGI-58/ABHD5. Am J Physiol Endocrinol Metab. 2009;297:E289–296. doi: 10.1152/ajpendo.00099.2009. [DOI] [PubMed] [Google Scholar]
- 52.Yang X, Heckmann BL, Zhang X, Smas CM, Liu J. Distinct mechanisms regulate ATGL-mediated adipocyte lipolysis by lipid droplet coat proteins. Molecular endocrinology. 2013;27:116–126. doi: 10.1210/me.2012-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schweiger M, Paar M, Eder C, Brandis J, Moser E, Gorkiewicz G, Grond S, Radner FP, Cerk I, Cornaciu I, Oberer M, Kersten S, Zechner R, Zimmermann R, Lass A. G0/G1 switch gene-2 regulates human adipocyte lipolysis by affecting activity and localization of adipose triglyceride lipase. J Lipid Res. 2012;53:2307–2317. doi: 10.1194/jlr.M027409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cawthorn WP, Sethi JK. TNF-alpha and adipocyte biology. FEBS Lett. 2008;582:117–131. doi: 10.1016/j.febslet.2007.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen X, Xun K, Chen L, Wang Y. TNF-alpha, a potent lipid metabolism regulator. Cell Biochem Funct. 2009;27:407–416. doi: 10.1002/cbf.1596. [DOI] [PubMed] [Google Scholar]
- 56.Uysal KT, Wiesbrock SM, Marino MW, Hotamisligil GS. Protection from obesity-induced insulin resistance in mice lacking TNF-alpha function. Nature. 1997;389:610–614. doi: 10.1038/39335. [DOI] [PubMed] [Google Scholar]
- 57.Yang X, Zhang X, Heckmann BL, Lu X, Liu J. Relative contribution of adipose triglyceride lipase and hormone-sensitive lipase to tumor necrosis factor-alpha (TNF-alpha)-induced lipolysis in adipocytes. J Biol Chem. 2011;286:40477–40485. doi: 10.1074/jbc.M111.257923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang X, Xie X, Heckmann BL, Saarinen AM, Czyzyk TA, Liu J. Targeted disruption of g0/g1 switch gene 2 enhances adipose lipolysis, alters hepatic energy balance, and alleviates high-fat diet-induced liver steatosis. Diabetes. 2014;63:934–946. doi: 10.2337/db13-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ma T, Lopez-Aguiar AG, Li A, Lu Y, Sekula D, Nattie EE, Freemantle S, Dmitrovsky E. Mice lacking G0S2 are lean and cold-tolerant. Cancer biology & therapy. 2014;15 doi: 10.4161/cbt.28251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.El-Assaad W, El-Kouhen K, Mohammad AH, Yang J, Morita M, Gamache I, Mamer O, Avizonis D, Hermance N, Kersten S, Tremblay ML, Kelliher MA, Teodoro JG. Deletion of the gene encoding G0/G 1 switch protein 2 (G0s2) alleviates high-fat-diet-induced weight gain and insulin resistance, and promotes browning of white adipose tissue in mice. Diabetologia. 2015;58:149–157. doi: 10.1007/s00125-014-3429-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Choi H, Lee H, Kim TH, Kim HJ, Lee YJ, Lee SJ, Yu JH, Kim D, Kim KS, Park SW, Kim JW. G0/G1 switch gene 2 has a critical role in adipocyte differentiation. Cell death and differentiation. 2014;21:1071–1080. doi: 10.1038/cdd.2014.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heckmann BL, Zhang X, Xie X, Saarinen A, Lu X, Yang X, Liu J. Defective adipose lipolysis and altered global energy metabolism in mice with adipose overexpression of the lipolytic inhibitor G0/G1 switch gene 2 (G0S2) The Journal of biological chemistry. 2014;289:1905–1916. doi: 10.1074/jbc.M113.522011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shin S, Choi YM, Han JY, Lee K. Inhibition of lipolysis in the novel transgenic quail model overexpressing G0/G1 switch gene 2 in the adipose tissue during feed restriction. PloS one. 2014;9:e100905. doi: 10.1371/journal.pone.0100905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang Y, Zhang Y, Qian H, Lu J, Zhang Z, Min X, Lang M, Yang H, Wang N, Zhang P. The g0/g1 switch gene 2 is an important regulator of hepatic triglyceride metabolism. PloS one. 2013;8:e72315. doi: 10.1371/journal.pone.0072315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sugaya Y, Satoh H. Liver-specific G0 /G1 switch gene 2 (G0s2) expression promotes hepatic insulin resistance by exacerbating hepatic steatosis in male Wistar rats. Journal of diabetes. 2016 doi: 10.1111/1753-0407.12482. [DOI] [PubMed] [Google Scholar]
- 66.Heier C, Radner FP, Moustafa T, Schreiber R, Grond S, Eichmann TO, Schweiger M, Schmidt A, Cerk IK, Oberer M, Theussl HC, Wojciechowski J, Penninger JM, Zimmermann R, Zechner R. G0/G1 Switch Gene 2 Regulates Cardiac Lipolysis. The Journal of biological chemistry. 2015;290:26141–26150. doi: 10.1074/jbc.M115.671842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen PR, Shin S, Choi YM, Kim E, Han JY, Lee K. Overexpression of G0/G1 Switch Gene 2 in Adipose Tissue of Transgenic Quail Inhibits Lipolysis Associated with Egg Laying. International journal of molecular sciences. 2016;17:384. doi: 10.3390/ijms17030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jessen N, Nielsen TS, Vendelbo MH, Viggers R, Stoen OG, Evans A, Frobert O. Pronounced expression of the lipolytic inhibitor G0/G1 Switch Gene 2 (G0S2) in adipose tissue from brown bears (Ursus arctos) prior to hibernation. Physiological reports. 2016;4 doi: 10.14814/phy2.12781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Laurens C, Badin PM, Louche K, Mairal A, Tavernier G, Marette A, Tremblay A, Weisnagel SJ, Joanisse DR, Langin D, Bourlier V, Moro C. G0/G1 Switch Gene 2 controls adipose triglyceride lipase activity and lipid metabolism in skeletal muscle. Molecular metabolism. 2016;5:527–537. doi: 10.1016/j.molmet.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Turnbull PC, Longo AB, Ramos SV, Roy BD, Ward WE, Peters SJ. Increases in skeletal muscle ATGL and its inhibitor G0S2 following 8 weeks of endurance training in metabolically different rat skeletal muscles. American journal of physiology. Regulatory, integrative and comparative physiology. 2016;310:R125–133. doi: 10.1152/ajpregu.00062.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bachner D, Ahrens M, Schroder D, Hoffmann A, Lauber J, Betat N, Steinert P, Flohe L, Gross G. Bmp-2 downstream targets in mesenchymal development identified by subtractive cloning from recombinant mesenchymal progenitors (C3H10T1/2) Developmental dynamics : an official publication of the American Association of Anatomists. 1998;213:398–411. doi: 10.1002/(SICI)1097-0177(199812)213:4<398::AID-AJA5>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 72.Zandbergen F, Mandard S, Escher P, Tan NS, Patsouris D, Jatkoe T, Rojas-Caro S, Madore S, Wahli W, Tafuri S, Muller M, Kersten S. The G0/G1 switch gene 2 is a novel PPAR target gene. Biochem J. 2005;392:313–324. doi: 10.1042/BJ20050636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oh SA, Suh Y, Pang MG, Lee K. Cloning of avian G(0)/G(1) switch gene 2 genes and developmental and nutritional regulation of G(0)/G(1) switch gene 2 in chicken adipose tissue. Journal of animal science. 2011;89:367–375. doi: 10.2527/jas.2010-3339. [DOI] [PubMed] [Google Scholar]
- 74.Zeng F, Xie L, Pang X, Liu W, Nie Q, Zhang X. Complementary deoxyribonucleic acid cloning of avian G0/G1 switch gene 2, and its expression and association with production traits in chicken. Poultry science. 2011;90:1548–1554. doi: 10.3382/ps.2010-01204. [DOI] [PubMed] [Google Scholar]
- 75.Schweiger M, Paar M, Eder C, Brandis J, Moser E, Gorkiewicz G, Grond S, Radner FP, Cerk I, Cornaciu I, Oberer M, Kersten S, Zechner R, Zimmermann R, Lass A. G0/G1 switch gene-2 regulates human adipocyte lipolysis by affecting activity and localization of adipose triglyceride lipase. J Lipid Res. 2012:35. doi: 10.1194/jlr.M027409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ahn J, Li X, Choi YM, Shin S, Oh SA, Suh Y, Nguyen TH, Baik M, Hwang S, Lee K. Differential expressions of G0/G1 switch gene 2 and comparative gene identification-58 are associated with fat content in bovine muscle. Lipids. 2014;49:1–14. doi: 10.1007/s11745-013-3866-3. [DOI] [PubMed] [Google Scholar]
- 77.Skopp A, May M, Janke J, Kielstein H, Wunder R, Flade-Kuthe R, Kuthe A, Jordan J, Engeli S. Regulation of G0/G1 switch gene 2 (G0S2) expression in human adipose tissue. Archives of physiology and biochemistry. 2016;122:47–53. doi: 10.3109/13813455.2015.1122066. [DOI] [PubMed] [Google Scholar]
- 78.Nielsen TS, Vendelbo MH, Jessen N, Pedersen SB, Jorgensen JO, Lund S, Moller N. Fasting, but not exercise, increases adipose triglyceride lipase (ATGL) protein and reduces G(0)/G(1) switch gene 2 (G0S2) protein and mRNA content in human adipose tissue. The Journal of clinical endocrinology and metabolism. 2011;96:E1293–1297. doi: 10.1210/jc.2011-0149. [DOI] [PubMed] [Google Scholar]
- 79.Heckmann BL, Zhang X, Saarinen AM, Liu J. Regulation of G0/G1 Switch Gene 2 (G0S2) Protein Ubiquitination and Stability by Triglyceride Accumulation and ATGL Interaction. PloS one. 2016;11:e0156742. doi: 10.1371/journal.pone.0156742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jaeger D, Schoiswohl G, Hofer P, Schreiber R, Schweiger M, Eichmann TO, Pollak NM, Poecher N, Grabner GF, Zierler KA, Eder S, Kolb D, Radner FP, Preiss-Landl K, Lass A, Zechner R, Kershaw EE, Haemmerle G. Fasting-induced G0/G1 switch gene 2 and FGF21 expression in the liver are under regulation of adipose tissue derived fatty acids. Journal of hepatology. 2015 doi: 10.1016/j.jhep.2015.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Heckmann BL, Zhang X, Saarinen AM, Schoiswohl G, Kershaw EE, Zechner R, Liu J. Liver X receptor alpha mediates hepatic triglyceride accumulation through upregulation of G0/G1 Switch Gene 2 expression. JCI insight. 2017;2:e88735. doi: 10.1172/jci.insight.88735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ma L, Robinson LN, Towle HC. ChREBP*Mlx is the principal mediator of glucose-induced gene expression in the liver. J Biol Chem. 2006;281:28721–28730. doi: 10.1074/jbc.M601576200. [DOI] [PubMed] [Google Scholar]
- 83.Hong C, Tontonoz P. Liver X receptors in lipid metabolism: opportunities for drug discovery. Nature reviews. Drug discovery. 2014;13:433–444. doi: 10.1038/nrd4280. [DOI] [PubMed] [Google Scholar]
- 84.Ducheix S, Montagner A, Theodorou V, Ferrier L, Guillou H. The liver X receptor: a master regulator of the gut-liver axis and a target for non alcoholic fatty liver disease. Biochemical pharmacology. 2013;86:96–105. doi: 10.1016/j.bcp.2013.03.016. [DOI] [PubMed] [Google Scholar]
- 85.Oosterveer MH, van Dijk TH, Grefhorst A, Bloks VW, Havinga R, Kuipers F, Reijngoud DJ. Lxralpha deficiency hampers the hepatic adaptive response to fasting in mice. The Journal of biological chemistry. 2008;283:25437–25445. doi: 10.1074/jbc.M801922200. [DOI] [PubMed] [Google Scholar]
- 86.Repa JJ, Liang G, Ou J, Bashmakov Y, Lobaccaro JM, Shimomura I, Shan B, Brown MS, Goldstein JL, Mangelsdorf DJ. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes & development. 2000;14:2819–2830. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yoshikawa T, Shimano H, Amemiya-Kudo M, Yahagi N, Hasty AH, Matsuzaka T, Okazaki H, Tamura Y, Iizuka Y, Ohashi K, Osuga J, Harada K, Gotoda T, Kimura S, Ishibashi S, Yamada N. Identification of liver X receptor-retinoid X receptor as an activator of the sterol regulatory element-binding protein 1c gene promoter. Molecular and cellular biology. 2001;21:2991–3000. doi: 10.1128/MCB.21.9.2991-3000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Joseph SB, Laffitte BA, Patel PH, Watson MA, Matsukuma KE, Walczak R, Collins JL, Osborne TF, Tontonoz P. Direct and indirect mechanisms for regulation of fatty acid synthase gene expression by liver X receptors. The Journal of biological chemistry. 2002;277:11019–11025. doi: 10.1074/jbc.M111041200. [DOI] [PubMed] [Google Scholar]
- 89.Shimano H, Horton JD, Shimomura I, Hammer RE, Brown MS, Goldstein JL. Isoform 1c of sterol regulatory element binding protein is less active than isoform 1a in livers of transgenic mice and in cultured cells. The Journal of clinical investigation. 1997;99:846–854. doi: 10.1172/JCI119248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sun X, Haas ME, Miao J, Mehta A, Graham MJ, Crooke RM, de Barros JP, Wang JG, Aikawa M, Masson D, Biddinger SB. Insulin Dissociates the Effects of Liver X Receptor on Lipogenesis, Endoplasmic Reticulum Stress, and Inflammation. The Journal of biological chemistry. 2016;291:1115–1122. doi: 10.1074/jbc.M115.668269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hong C, Tontonoz P. Liver X receptors in lipid metabolism: opportunities for drug discovery. Nature reviews. Drug discovery. 2014;13:433–444. doi: 10.1038/nrd4280. [DOI] [PubMed] [Google Scholar]
- 92.Lengqvist J, Mata De Urquiza A, Bergman AC, Willson TM, Sjovall J, Perlmann T, Griffiths WJ. Polyunsaturated fatty acids including docosahexaenoic and arachidonic acid bind to the retinoid X receptor alpha ligand-binding domain. Mol Cell Proteomics. 2004;3:692–703. doi: 10.1074/mcp.M400003-MCP200. [DOI] [PubMed] [Google Scholar]
- 93.Nakamura MT, Yudell BE, Loor JJ. Regulation of energy metabolism by long-chain fatty acids. Prog Lipid Res. 2014;53:124–144. doi: 10.1016/j.plipres.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 94.Steineger HH, Arntsen BM, Spydevold O, Sorensen HN. Gene transcription of the retinoid X receptor alpha (RXRalpha) is regulated by fatty acids and hormones in rat hepatic cells. J Lipid Res. 1998;39:744–754. [PubMed] [Google Scholar]
- 95.Fan YY, Spencer TE, Wang N, Moyer MP, Chapkin RS. Chemopreventive n-3 fatty acids activate RXRalpha in colonocytes. Carcinogenesis. 2003;24:1541–1548. doi: 10.1093/carcin/bgg110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Boergesen M, Pedersen TA, Gross B, van Heeringen SJ, Hagenbeek D, Bindesboll C, Caron S, Lalloyer F, Steffensen KR, Nebb HI, Gustafsson JA, Stunnenberg HG, Staels B, Mandrup S. Genome-wide profiling of liver X receptor, retinoid X receptor, and peroxisome 38 proliferator-activated receptor alpha in mouse liver reveals extensive sharing of binding sites. Molecular and cellular biology. 2012;32:852–867. doi: 10.1128/MCB.06175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ducheix S, Montagner A, Polizzi A, Lasserre F, Marmugi A, Bertrand-Michel J, Podechard N, Al Saati T, Chetiveaux M, Baron S, Boue J, Dietrich G, Mselli-Lakhal L, Costet P, Lobaccaro JM, Pineau T, Theodorou V, Postic C, Martin PG, Guillou H. Essential fatty acids deficiency promotes lipogenic gene expression and hepatic steatosis through the liver X receptor. Journal of hepatology. 2013;58:984–992. doi: 10.1016/j.jhep.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 98.Colin S, Bourguignon E, Boullay AB, Tousaint JJ, Huet S, Caira F, Staels B, Lestavel S, Lobaccaro JM, Delerive P. Intestine-specific regulation of PPARalpha gene transcription by liver X receptors. Endocrinology. 2008;149:5128–5135. doi: 10.1210/en.2008-0637. [DOI] [PubMed] [Google Scholar]
- 99.Inagaki T, Dutchak P, Zhao G, Ding X, Gautron L, Parameswara V, Li Y, Goetz R, Mohammadi M, Esser V, Elmquist JK, Gerard RD, Burgess SC, Hammer RE, Mangelsdorf DJ, Kliewer SA. Endocrine regulation of the fasting response by PPARalpha-mediated induction of fibroblast growth factor 21. Cell Metab. 2007;5:415–425. doi: 10.1016/j.cmet.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 100.Ide T, Shimano H, Yoshikawa T, Yahagi N, Amemiya-Kudo M, Matsuzaka T, Nakakuki M, Yatoh S, Iizuka Y, Tomita S, Ohashi K, Takahashi A, Sone H, Gotoda T, Osuga J, Ishibashi S, Yamada N. Cross-talk between peroxisome proliferator-activated receptor (PPAR) alpha and liver X receptor (LXR) in nutritional regulation of fatty acid metabolism. II. LXRs suppress lipid degradation gene promoters through inhibition of PPAR signaling. Mol Endocrinol. 2003;17:1255–1267. doi: 10.1210/me.2002-0191. [DOI] [PubMed] [Google Scholar]
- 101.Yoshikawa T, Ide T, Shimano H, Yahagi N, Amemiya-Kudo M, Matsuzaka T, Yatoh S, Kitamine T, Okazaki H, Tamura Y, Sekiya M, Takahashi A, Hasty AH, Sato R, Sone H, Osuga J, Ishibashi S, Yamada N. Cross-talk between peroxisome proliferator-activated 39 receptor (PPAR) alpha and liver X receptor (LXR) in nutritional regulation of fatty acid metabolism. I. PPARs suppress sterol regulatory element binding protein-1c promoter through inhibition of LXR signaling. Mol Endocrinol. 2003;17:1240–1254. doi: 10.1210/me.2002-0190. [DOI] [PubMed] [Google Scholar]
- 102.Gao M, Bu L, Ma Y, Liu D. Concurrent activation of liver X receptor and peroxisome proliferator-activated receptor alpha exacerbates hepatic steatosis in high fat diet-induced obese mice. PLoS One. 2013;8:e65641. doi: 10.1371/journal.pone.0065641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang W, Bu SY, Mashek MT, I OS, Sibai Z, Khan SA, Ilkayeva O, Newgard CB, Mashek DG, Unterman TG. Integrated Regulation of Hepatic Lipid and Glucose Metabolism by Adipose Triacylglycerol Lipase and FoxO Proteins. Cell reports. 2016;15:349–359. doi: 10.1016/j.celrep.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Deng X, Zhang W, I OS, Williams JB, Dong Q, Park EA, Raghow R, Unterman TG, Elam MB. FoxO1 inhibits sterol regulatory element-binding protein-1c (SREBP-1c) gene expression via transcription factors Sp1 and SREBP-1c. The Journal of biological chemistry. 2012;287:20132–20143. doi: 10.1074/jbc.M112.347211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu X, Qiao A, Ke Y, Kong X, Liang J, Wang R, Ouyang X, Zuo J, Chang Y, Fang F. FoxO1 represses LXRalpha-mediated transcriptional activity of SREBP-1c promoter in HepG2 cells. FEBS letters. 2010;584:4330–4334. doi: 10.1016/j.febslet.2010.09.027. [DOI] [PubMed] [Google Scholar]
- 106.Nielsen TS, Kampmann U, Nielsen RR, Jessen N, Orskov L, Pedersen SB, Jorgensen JO, Lund S, Moller N. Reduced mRNA and protein expression of perilipin A and G0/G1 switch gene 2 (G0S2) in human adipose tissue in poorly controlled type 2 diabetes. The Journal of clinical endocrinology and metabolism. 2012;97:E1348–1352. doi: 10.1210/jc.2012-1159. [DOI] [PubMed] [Google Scholar]
- 107.Svart M, Kampmann U, Voss T, Pedersen SB, Johannsen M, Rittig N, Poulsen PL, Nielsen TS, Jessen N, Moller N. Combined Insulin Deficiency and Endotoxin Exposure Stimulate Lipid Mobilization and Alter Adipose Tissue Signaling in an Experimental Model of 40 Ketoacidosis in Subjects With Type 1 Diabetes: A Randomized Controlled Crossover Trial. Diabetes. 2016;65:1380–1386. doi: 10.2337/db15-1645. [DOI] [PubMed] [Google Scholar]
- 108.Gibson G. Human evolution: thrifty genes and the dairy queen. Current biology : CB. 2007;17:R295–296. doi: 10.1016/j.cub.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 109.Hara K, Kubota N, Tobe K, Terauchi Y, Miki H, Komeda K, Tamemoto H, Yamauchi T, Hagura R, Ito C, Akanuma Y, Kadowaki T. The role of PPARgamma as a thrifty gene both in mice and humans. The British journal of nutrition. 2000;84(Suppl 2):S235–239. doi: 10.1079/096582197388608. [DOI] [PubMed] [Google Scholar]
- 110.Genne-Bacon EA. Thinking evolutionarily about obesity. The Yale journal of biology and medicine. 2014;87:99–112. [PMC free article] [PubMed] [Google Scholar]
- 111.Campbell LV. The thrifty gene hypothesis: maybe everyone is right? International journal of obesity. 2008;32:723–724. doi: 10.1038/sj.ijo.0803772. author reply 725–726. [DOI] [PubMed] [Google Scholar]
- 112.Joffe B, Zimmet P. The thrifty genotype in type 2 diabetes: an unfinished symphony moving to its finale? Endocrine. 1998;9:139–141. doi: 10.1385/ENDO:9:2:139. [DOI] [PubMed] [Google Scholar]
- 113.Sharma AM. The thrifty-genotype hypothesis and its implications for the study of complex genetic disorders in man. Journal of molecular medicine. 1998;76:568–571. doi: 10.1007/s001090050251. [DOI] [PubMed] [Google Scholar]
- 114.Kurat CF, Natter K, Petschnigg J, Wolinski H, Scheuringer K, Scholz H, Zimmermann R, Leber R, Zechner R, Kohlwein SD. Obese yeast: triglyceride lipolysis is functionally conserved from mammals to yeast. The Journal of biological chemistry. 2006;281:491–500. doi: 10.1074/jbc.M508414200. [DOI] [PubMed] [Google Scholar]
- 115.Gronke S, Mildner A, Fellert S, Tennagels N, Petry S, Muller G, Jackle H, Kuhnlein RP. Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell metabolism. 2005;1:323–330. doi: 10.1016/j.cmet.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 116.Briand F, Treguier M, Andre A, Grillot D, Issandou M, Ouguerram K, Sulpice T. Liver X receptor activation promotes macrophage-to-feces reverse cholesterol transport in a dyslipidemic hamster model. J Lipid Res. 2010;51:763–770. doi: 10.1194/jlr.M001552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kannisto K, Gafvels M, Jiang ZY, Slatis K, Hu X, Jorns C, Steffensen KR, Eggertsen G. LXR driven induction of HDL-cholesterol is independent of intestinal cholesterol absorption and ABCA1 protein expression. Lipids. 2014;49:71–83. doi: 10.1007/s11745-013-3853-8. [DOI] [PubMed] [Google Scholar]
- 118.Ou X, Dai X, Long Z, Tang Y, Cao D, Hao X, Hu Y, Li X, Tang C. Liver X receptor agonist T0901317 reduces atherosclerotic lesions in apoE−/− mice by up-regulating NPC1 expression, Science in China. Series C. Life sciences / Chinese Academy of Sciences. 2008;51:418–429. doi: 10.1007/s11427-008-0054-4. [DOI] [PubMed] [Google Scholar]
- 119.Plosch T, Kok T, Bloks VW, Smit MJ, Havinga R, Chimini G, Groen AK, Kuipers F. Increased hepatobiliary and fecal cholesterol excretion upon activation of the liver X receptor is independent of ABCA1. J Biol Chem. 2002;277:33870–33877. doi: 10.1074/jbc.M206522200. [DOI] [PubMed] [Google Scholar]
- 120.Yasuda T, Grillot D, Billheimer JT, Briand F, Delerive P, Huet S, Rader DJ. Tissue-specific liver X receptor activation promotes macrophage reverse cholesterol transport in vivo. Arterioscler Thromb Vasc Biol. 2010;30:781–786. doi: 10.1161/ATVBAHA.109.195693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zanotti I, Poti F, Pedrelli M, Favari E, Moleri E, Franceschini G, Calabresi L, Bernini F. The LXR agonist T0901317 promotes the reverse cholesterol transport from macrophages by increasing plasma efflux potential. J Lipid Res. 2008;49:954–960. doi: 10.1194/jlr.M700254-JLR200. [DOI] [PubMed] [Google Scholar]
- 122.Cha JY, Repa JJ. The liver X receptor (LXR) and hepatic lipogenesis. The carbohydrate-response element-binding protein is a target gene of LXR. J Biol Chem. 2007;282:743–751. doi: 10.1074/jbc.M605023200. [DOI] [PubMed] [Google Scholar]
- 123.Zhang Y, Breevoort SR, Angdisen J, Fu M, Schmidt DR, Holmstrom SR, Kliewer SA, Mangelsdorf DJ, Schulman IG. Liver LXRalpha expression is crucial for whole body cholesterol homeostasis and reverse cholesterol transport in mice. The Journal of clinical investigation. 2012;122:1688–1699. doi: 10.1172/JCI59817. [DOI] [PMC free article] [PubMed] [Google Scholar]