Summary
Bladder cancer is the fifth most prevalent cancer in the U.S., yet is understudied and few laboratory models exist that reflect the biology of the human disease. Here we describe a biobank of patient-derived organoid lines that recapitulates the histopathological and molecular diversity of human bladder cancer. Organoid lines can be established efficiently from patient biopsies acquired before and after disease recurrence, and are interconvertible with orthotopic xenografts. Notably, organoid lines often retain parental tumor heterogeneity and exhibit a spectrum of genomic changes that are consistent with tumor evolution in culture. Analyses of drug response using bladder tumor organoids show partial correlations with mutational profiles as well as changes associated with treatment resistance, and specific responses can be validated using xenografts in vivo. Our studies indicate that patient-derived bladder tumor organoids represent a faithful model system for studying tumor evolution and treatment response in the context of precision cancer medicine.
ITI
A biobank of patient-derived bladder cancer organoids faithfully recapitulates features of human cancer and enables analysis of clonal evolution and drug responses.

Introduction
Most bladder cancers are urothelial carcinomas, with the majority of these being non-muscle invasive bladder cancers, which usually have a relatively favorable prognosis but are associated with considerable morbidity and high cost for managing treatment (Kamat et al., 2016; Knowles and Hurst, 2015; Lerner et al., 2016). These non-muscle invasive tumors can be classified as low-grade or high-grade, and encompass multiple growth patterns, including papillary tumors and carcinoma in situ (CIS), a flattened layer of dysplastic cells that is presumed to represent a common precursor of muscle invasive bladder cancer. In contrast, muscle invasive bladder cancers have a relatively poor prognosis (Kamat et al., 2016; Prasad et al., 2011).
Treatment regimens for bladder cancer and their efficacy vary depending on clinical stage and associated risk factors (Kamat et al., 2016; Lerner et al., 2016; Prasad et al., 2011; Resnick et al., 2013). The front-line treatment for non-muscle invasive bladder cancer is transurethral resection (TUR), with or without subsequent intravesical delivery (into the bladder lumen) of chemotherapy, such as mitomycin C, or bacillus Calmette-Guerin (BCG) (Redelman-Sidi et al., 2014), depending on disease stage, grade, and other clinical characteristics. In the case of muscle invasive bladder cancer, a current standard of care is cisplatin-based neoadjuvant chemotherapy followed by radical cystectomy (Grossman et al., 2003; Herr et al., 1998; Trialists et al., 2011). Although approximately 50% of patients who undergo cystectomy are alive after 5 years, the procedure affects quality of life and is consequently disfavored by many patients.
Several lines of evidence suggest that the distinct clinical outcomes of non-muscle invasive versus muscle invasive bladder tumors can be attributed to their different molecular profiles (Dyrskjot et al., 2003; Hurst et al., 2012; Lauss et al., 2010). For example, gain-of-function mutations of FGFR3 are more prevalent in low-grade non-muscle invasive bladder cancers, whereas loss or mutation of TP53 is more frequent in high-grade muscle invasive bladder cancers (Knowles and Hurst, 2015). However, molecular studies have shown that the relationship between these general categories of bladder cancer is complex (Lindgren et al., 2012). Moreover, an analysis of non-muscle invasive bladder cancer has identified multiple subtypes associated with disease outcome and clinical response, with the worst outcomes observed in patients with TP53 and ERBB2 (HER2) mutations (Hedegaard et al., 2016). Therefore, there is a need to develop models for both non-muscle invasive and muscle invasive bladder cancer to elucidate the biologic function of recurrent somatic mutations and their role in mediating transformation and promoting disease progression as a prelude to the development of rational treatment strategies.
Recently, several studies have shown the value of three-dimensional organoid culture systems for modeling varied aspects of cancer biology (Drost and Clevers, 2017; Weeber et al., 2017). In particular, patient-derived organoids derived from colorectal, pancreatic, liver, breast, and prostate cancers can model key features of parental tumors, and can also be used to investigate drug response (Boj et al., 2015; Broutier et al., 2017; Gao et al., 2014; Sachs et al., 2017; van de Wetering et al., 2015). Notably, a recent study on the establishment of patient-derived tumor organoid lines from a wide range of tumor types has also described the generation of eight bladder tumor organoid lines, which have not been extensively characterized (Pauli et al., 2017). Here, we report the generation and detailed analysis of 22 patient-derived bladder cancer organoid lines, and demonstrate their histopathological and molecular concordance with their corresponding parental tumors. We show that these organoid lines frequently retain tumor heterogeneity and display changes in their mutational profiles during culture and xenografting that are consistent with clonal evolution. Furthermore, we demonstrate the utility of these patient-derived organoid lines as a model system for investigating tumor evolution and its role in modulating drug response.
Results
Establishment of patient-derived bladder cancer organoid lines
To establish clinically-relevant models for human bladder cancer, we have generated a biobank of patient-derived organoid lines that can be readily propagated in three-dimensional culture. We established bladder cancer organoid lines using fresh patient TUR samples ranging from low-grade non-muscle invasive disease to high-grade muscle invasive cancer. Tumor tissues were transported directly from the cystoscopy suite or operating room to the laboratory for processing, where they were divided into pieces for organoid culture as well as for analysis of the parental tumor (Figure 1A). Our culture conditions are similar to those previously described by our group for mouse and human prostate organoids, which include Matrigel to support three-dimensional culture, hepatocyte medium, charcoal-stripped serum, and ROCK inhibitor to improve the survival of dissociated epithelial cells (Chua et al., 2014). Notably, these serum-containing conditions differ considerably from the defined media used by (Pauli et al., 2017), which are nearly identical to those previously utilized for prostate organoid culture (Gao et al., 2014).
Figure 1. Establishment of patient-derived bladder tumor organoids and xenografts.
(A) Overview of experimental plan. (B) Bright-field images of organoids together with H&E staining of parental tumors, patient-derived organoids, orthotopic xenografts generated from organoids, and organoids derived from the xenografts. Scale bars indicate 50 μm. (C) Ultrasound imaging (left) of orthotopic xenografts (dashed lines) and whole-mount images of corresponding bladders (right). Scale bars indicate 2 mm. See also Figure S1.
Using this approach, we have generated independent bladder tumor organoid lines (SCBO-1 through SCBO-16) corresponding to 16 different patients (Table 1; note that multiple lines have been established from three patients). These lines have been propagated by serial passaging, and have been successfully cryopreserved, allowing their long-term storage and retrieval. Our recent efficiency in establishing organoid lines has been approximately 70% (n=12/17 during the past nine months); we considered a line as successfully established when it had been serially passaged at least six times. Interestingly, an additional five primary organoid cultures initially appeared to be successful, but failed to propagate after 3–5 passages; based on their limited characterization, no obvious common features could be ascertained for these failed lines. In contrast, successfully established lines have been serially passaged for up to 26 passages and have been cryopreserved and recovered without any apparent diminution in their ability to propagate (Table 1).
Table 1.
Summary of patient-derived bladder tumor organoid lines and corresponding clinical data.
| Line | Overall tumor stage |
Pathologic classification of parental tumor sample | Sex | Ethnicity | Age | Smoking status |
Prior intravesical therapy |
Prior systemic therapy |
Passage number |
Orthotopic xenograft formation |
|---|---|---|---|---|---|---|---|---|---|---|
| SCBO-1 | Ta | High-grade papillary urothelial carcinoma, non-invasive | F | Caucasian | 62 | Never | Docetaxel | None | 12 | Yes |
| SCBO-2 | T2 | Low-grade papillary urothelial carcinoma, non-invasive | M | Caucasian | 79 | Former | None | None | 13 | Yes |
| SCBO-3 | T1 | Low-grade papillary urothelial carcinoma, non-invasive | M | African-American | 76 | Former | Docetaxel | None | 26 | Yes |
| SCBO-3.2 | Ta | Low-grade papillary urothelial carcinoma, non-invasive | M | African-American | 78 | Former | Docetaxel, MMC, BCG | None | 17 | Yes |
| SCBO-4 | T2 | Invasive high-grade urothelial carcinoma | F | Caucasian | 72 | Former | BCG, BCG/IFN | None | 13 | no growth |
| SCBO-5 | T1+CIS | High-grade urothelial carcinoma, non-invasive | M | Hispanic | 84 | Former | None | None | 26 | Yes |
| SCBO-6 | T1+CIS | Invasive high-grade urothelial carcinoma | M | Caucasian | 80 | Former | MMC, BCG | None | 16 | Yes |
| SCBO-7 | Ta | Low-grade papillary urothelial carcinoma, non-invasive | F | Caucasian | 77 | Former | None | None | 21 | Yes |
| SCBO-7.2 | Ta | High-grade papillary urothelial carcinoma, non-invasive | F | Caucasian | 78 | Former | None | None | 12 | Yes |
| SCBO-8 | Ta | High-grade urothelial carcinoma in situ (flat), non-invasive | M | Caucasian | 83 | Active | BCG | None | 20 | Yes |
| SCBO-9 | T1 | Low-grade papillary urothelial carcinoma, non-invasive | F | Caucasian | 68 | Never | None | None | 20 | Yes |
| SCBO-10 | T2 | High-grade urothelial carcinoma, non-invasive | M | Caucasian | 82 | Former | None | None | 21 | Yes |
| SCBO-11 | T1+CIS | Squamous cell carcinoma | M | Caucasian | 83 | Never | None | None | 21 | Yes |
| SCBO-11.2 | T1 | Squamous cell carcinoma, non-invasive | M | Caucasian | 84 | Never | None | None | 16 | Yes |
| SCBO-11.3 | Ta | Atypical squamous lesion (dysplasia) | M | Caucasian | 84 | Never | None | None | 10 | no growth |
| SCBO-12 | Ta | Low-grade papillary urothelial carcinoma, non-invasive | M | N/A | 58 | Former | None | None | 19 | no growth |
| SCBO-13 | T1+CIS | High-grade urothelial carcinoma, non-invasive | M | Asian | 48 | Former | None | None | 14 | N/D |
| SCBO-14 | Ta | Low-grade papillary urothelial carcinoma, non-invasive | M | Caucasian | 84 | Former | MMC | None | 22 | Yes |
| SCBO-15 | T3 | Invasive high-grade urothelial carcinoma | F | Caucasian | 81 | Active | None | None | 16 | Yes |
| SCBO-16 | Ta+CIS | High-grade papillary urothelial carcinoma, non-invasive | M | Caucasian | 62 | Never | None | None | 16 | N/D |
| SMBO-1 | T3 | High-grade urothelial carcinoma | M | Caucasian | 64 | Former | None | G/C, 4 cycles | 12 | Yes |
| SMBO-2 | T3 | Invasive high-grade urothelial carcinoma | M | Caucasian | 86 | Never | None | None | 13 | Yes |
Overall tumor stage is described for the patient tumor as a whole; pathologic classification is based on analysis of the specific parental tumor sample, which was adjacent to the samples used for organoid establishment and molecular analyses. In some cases, these evaluations are not completely concordant due to tumor heterogeneity and differences in sampling. Abbreviations: BCG, Bacillus Calmette–Guérin treatment; CIS, carcinoma in situ; G/C, gemcitabine/cisplatin; Hg, high-grade; IFN, interferon; Lg, low-grade; MMC, mitomycin C; N/A, not available; N/D, not done.
In culture, the organoid lines displayed morphologies ranging from spheroidal to asymmetric, and were sometimes comprised of relatively loose aggregates of cells (Figures 1B and S1). Next, we performed hematoxylin-eosin (H&E) staining of paraffin sections from the organoids as well as their corresponding parental tumors, which showed strong concordance in their histopathological features (Figures 1B and S1); importantly, the presence of benign tissue was never observed during serial passaging. Interestingly, we found that the SCBO-11 parental tumor and organoid line displayed the features of squamous cell carcinoma, an uncommon histologic subtype of bladder cancer (Rausch et al., 2014).
Since our current organoid lines were established from TUR specimens, they are enriched for non-muscle invasive bladder tumors, which represent the majority of urothelial cancers. However, since patients with muscle invasive bladder cancer undergo transurethral resection within the context of their standard diagnostic evaluation, we have also generated 4 organoid lines corresponding to muscle invasive disease (T2 and higher) (Table 1). In addition, 31% (n=5/16) of patients from whom organoid lines were established were female, consistent with the approximately three-fold lower incidence of bladder cancer in women relative to men (Lucca et al., 2015). We also established organoid lines from three patients from ethnic minorities (19%, n=3/16), consistent with the demographics of the patient population at NewYork-Presbyterian Hospital/Columbia University Medical Center. Taken together, these results suggest that our biobank of organoid lines is representative of the overall patient population from which it was established.
Since bladder cancer patients often undergo multiple biopsies during the course of their treatment, we have been able to derive organoid lines from chronologically distinct lesions from the same patient (designated in Table 1 by a decimal point followed by a number; e.g., SCBO-3.2). For example, we have established two independent lines (SCBO-3 and SCBO-3.2) using tumor samples collected 420 days apart from a patient with recurrent bladder cancer, before and after treatment with intravesical BCG and mitomycin C. In addition, we established three lines (SCBO-11, SCBO-11.2, and SCBO-11.3) from another patient at intervals of 91 and 98 days between tumor resections, in the absence of additional treatment.
To pursue analyses of patient-derived bladder tumor samples in culture and in an appropriate tissue microenvironment, we developed an optimized methodology to convert bladder tumor organoid lines into orthotopic xenografts in immunodeficient mice with high efficiency (83%; n=15/18) (Figure 1C), using ultrasound-guided implantation of cells between the bladder urothelium and lamina propria/muscle layer (Jager et al., 2013; Owczarek et al., 2017). Histological analyses of orthotopic xenografts as well as organoids derived from these xenografts demonstrated their similarity to the original parental tumor (Figure 1B). Furthermore, we successfully generated organoid lines (SMBO-1 and SMBO-2) from two patient-derived xenograft lines that were originally established from muscle invasive tumors. Thus, we can successfully interconvert organoids and xenografts with high efficiency.
Organoid lines recapitulate the mutational spectrum of human bladder cancer
To identify somatic mutations as well as DNA copy number alterations and to explore the genomic representativeness of our models, we performed targeted sequencing using the MSK-IMPACT platform, which examines all coding exons and selected intronic regions of 468 cancer-associated genes at a high depth of coverage (Cheng et al., 2015). These analyses identified numerous genomic alterations in the patient-derived organoid lines, including point mutations in putative oncogenic drivers as well as copy number alterations (Figure 2A). Furthermore, comparison of organoid lines and their corresponding parental tumors showed that their mutational profiles were highly concordant, with 11 of the lines analyzed showing greater than 80% concordance, and only 4 lines displaying less than 60% concordance (Figure 2B).
Figure 2. Molecular alterations in bladder organoid lines.
(A) Summary of mutations and DNA copy number alterations identified in organoid lines by deep targeted sequencing, together with their percentage representation in this dataset. Representative genes detected in the TCGA study and known to be mutated in bladder cancer are shown. Passage numbers of the organoid lines were: SCBO-1, P11; SCBO-2, P11; SCBO-3, P14; SCBO-3.2, P14; SCBO-4, P13; SCBO-5, P15; SCBO-6, P9; SCBO-7, P11; SCBO-7.2, P1; SCBO-8, P19; SCBO-9, P17; SCBO-10, P17; SCBO-11, P20; SCBO-11.2, P8; SCBO-11.3, P2; SCBO-12, P15; SCBO-13, P7; SCBO-14, P19; SCBO-15, P12; SCBO-16, P7; SMBO-1, P8; SMBO-2, P10. (B) Concordance of mutations detected in the parental tumor and corresponding organoid lines. Passage numbers are the same as in panel B. (C) Detection of a FGFR3-TACC3 fusion transcript in the SCBO-10 organoid line. Results from RT-PCR in SCBO-7, SCBO-8 and SCBO-10 organoids are shown (top), together with the junction sequences on the mRNA and the reading frame at the breakpoint (bottom).
Importantly, the mutational profiles of the patient-derived organoid lines recapitulated the majority of the common genomic alterations in human bladder cancer (Cancer Genome Atlas, 2014; Gui et al., 2011; Robertson et al., 2017). In particular, we observed that many organoid lines harbored mutations in epigenetic regulators such as ARID1A, KMT2C, KMT2D, and KDM6A, which are frequently observed in human bladder cancer (73%, n=11/15 non-recurrent lines examined). We found that 60% (n=9/15) of the lines harbored activating mutations in FGFR3, which arise in approximately 70% and 12% of non-muscle invasive and muscle invasive bladder cancers respectively (Al-Ahmadie et al., 2011; Cancer Genome Atlas, 2014). We also detected less common driver mutations characteristic of human bladder cancer such as alterations in STAG2, ERBB2, and EGFR (Figure 2A). Overall, the spectrum of mutations found in our organoid lines reflects their predominant origin from non-muscle invasive tumors. However, we observed that 33% (n=5/15) of the organoid lines carried mutations in TP53, which is deleted and/or mutated in more than 50% of muscle invasive bladder cancers (Cancer Genome Atlas, 2014). Only one line had an oncogenic mutation in RB1, in contrast with the higher frequency of these mutations in established bladder cancer cell lines.
Finally, our sequencing analyses indicated that the SCBO-10 line and its parental tumor harbored an FGFR3-TACC3 fusion, which occurs in approximately 5% of bladder tumors and results in FGFR3 activation (Cancer Genome Atlas, 2014; Williams et al., 2013). Given the potential biological significance of this rare event in bladder cancer, we validated the presence of this gene fusion with PCR amplification and sequencing, which identified a fusion between exon 17 of FGFR3 and exon 11 of TACC3 (Figure 2C). Interestingly, this exact FGFR3-TACC3 junction has been previously reported in a glioblastoma (GBM-021) (Di Stefano et al., 2015) as well as in the RT112 bladder cancer cell line (Williams et al., 2013).
Tumor evolution in organoid culture
To assess the genetic stability of the organoid lines in culture, as well as in orthotopic xenografts and xenograft-derived organoids, we performed similar deep sequencing to compare their mutational profiles. Notably, the high depth of coverage (approximately 500-fold) facilitated the detection of subclonal mutations (those arising in only a subset of tumor cells). For the most part, we found that mutational profiles were similar between samples within individual lines, indicating that the tumor genotype was largely maintained. However, we observed that a subset of mutations was either lost or gained during serial passaging in culture, and/or during grafting or reestablishment of organoids from grafts (Figure 3A). Upon analyzing the clonal composition of each line, we found that truncal mutations were retained, but subclonal mutations could be gained or lost (Figures 3B and S2). For example, the SCBO-3.2 line had an oncogenic CTNNB1(S45F) activating mutation, a common hotspot in human cancers (Morin et al., 1997), which was not detected in the parental tumor sample or in early-passage organoids (Figure 3B). The SCBO-5 line showed multiple examples of alleles that were lost during organoid culture, such as ERBB2(D227N) and JAK2(H538Y), or gained in culture, such as KMT2D(S831) (Figure 3B). Thus, many of the organoid lines displayed clonal evolution during serial passaging as well as subsequent xenografting and/or re-establishment of organoid cultures.
Figure 3. Tumor evolution during organoid culture and xenograft establishment.
(A) Summary of mutations detected by deep targeted sequencing of parental tumors (a), patient-derived organoids at early (b) and late passages (c), orthotopic xenografts generated from the organoids (d), and organoids derived from xenografts (e). See Figure S2 for details of variant allele fractions and passage numbers analyzed. (B) Variant allele fractions during clonal evolution of SCBO-3.2 (top) and SCBO-5 (bottom) as determined by targeted sequencing analysis. (C) Mutational signature decomposition analysis based on whole-exome sequencing. From the 29 signatures tested, the top signatures representing the majority of mutations with p < 0.05 are shown. (D) Phylogenetic trees based on whole-exome sequencing. Orthotopic xenografts were converted from SCBO-3 at P8, SCBO-3.2 at P4, SCBO-5 at P5, and SCBO-6 at P7 respectively. See also Figures S2 and S3 and Table S1.
To better elucidate the intratumoral heterogeneity leading to clonal evolution in culture, we performed whole-exome sequencing of 24 samples from five independent organoid lines. As expected, we found a strong correlation between the variant allele fractions derived from whole-exome sequencing data with those determined from targeted exome sequencing (Figure S3A). We also leveraged the broader exome sequencing to infer mutational signatures characteristic of underlying mutational processes (Figures 3C and S3B) (Alexandrov et al., 2013). These mutational signatures appeared to be stable, despite the ongoing clonal evolution during serial passaging and xenografting (Figure 3C). Most notably, we observed a signature characteristic of APOBEC gene editing in two of the organoid lines (SCBO-4 and SCBO-5), which is present in a large percentage of bladder tumors (Faltas et al., 2016; Kim et al., 2015; Robertson et al., 2017). We also identified a mutational signature characteristic of smoking in the SCBO-6 line, which was established from a patient who was a former smoker (Figure 3C and Table 1). Finally, we found that the overall tumor mutational burdens of the SCBO-3 and SCBO-3.2 lines were similar to those observed in low-grade non-muscle invasive bladder cancer, whereas the tumor mutational burdens in the SCBO-4, SCBO-5, and SCBO-6 lines were in the ranges reported for high-grade non-muscle invasive and muscle invasive bladder tumors (Table S1) (Pietzak et al., 2017), again supporting the similarity of the organoid lines to the overall spectrum of bladder cancers.
Phylogenetic analysis of exome sequencing data revealed patterns of linear and branched tumor evolution for the organoids, xenografts, and organoids established from xenografts (Figure 3D). However, the extent of tumor evolution differed between lines, with SCBO-5 displaying particularly prominent changes in the allelic fraction of numerous mutations. Taken together, these results show that tumor evolution readily occurs in bladder organoid culture, even in the absence of drug treatment, and that the patterns of clonal evolution resemble those described for recurrent primary human bladder cancer in vivo (Lamy et al., 2016).
Phenotypic stability and plasticity in organoid culture
Gene expression profiling analyses of muscle invasive bladder cancer have categorized a basal-like subtype that has a more aggressive phenotype, and a luminal-like subtype with a less aggressive phenotype (Cancer Genome Atlas, 2014; Choi et al., 2014; Damrauer et al., 2014). Therefore, to examine luminal and basal cell types within the bladder organoids, we performed immunofluorescence analyses of marker expression in each organoid line as well as their corresponding parental tumors, xenografts, and xenograft-derived organoids (Figures 4, S4, and S5). In these analyses, we examined expression of cytokeratin 7 (CK7), which is strongly expressed by all urothelial cells, as well as the basal epithelial marker CK5 and the luminal epithelial marker CK8. All but one (SCBO-11, which is a squamous carcinoma) of the parental tumors expressed CK7, as expected, and Ki67 immunoreactivity was readily detected in all samples. Nuclear immunostaining for p53 was concordant with the detection of TP53 mutations by targeted exome sequencing. In addition, nearly all of the organoid and parental tumor cells in each sample expressed either luminal and basal cytokeratins, while double-positive (“intermediate”) cells expressing both luminal and basal cytokeratins were infrequent.
Figure 4. Phenotypic stability and plasticity in organoid culture.
(A, B) H&E and immunostaining for the indicated markers in SCBO-10 (A) and SCBO-7 (B) parental tumors, organoids at the indicated passages, orthotopic xenografts generated from organoids, and organoids derived from xenografts. Scale bars indicate 50 μm. (C, D) Molecular subtypes of parental tumors and corresponding organoids at the indicated passages were analyzed using the BASE47 (C) and the MDACC classifiers (D). Heatmaps show normalized gene expression of organoid lines and parental tumors organized by the luminal and basal classifier genes. Unsupervised clustering analyses were performed by the z-score of normalized gene reads from RNA-seq data. (E) Summary of tumor subtypes as determined by the BASE47 and MDACC classifiers, as well as by immunofluorescence detection of markers. See also Figures S4, S5, S6, and Table S2.
Interestingly, these marker analyses revealed two general categories of organoid lines. In the first group, 36% (n=8/22) of organoid lines displayed strong phenotypic stability, showing similar marker expression profiles in the parental tumor, organoid line at different passages, xenograft, and xenograft-derived organoids (Figures S4 and S5). For example, the SCBO-10 line displayed consistent positive staining for p53, CK7 and the urothelial marker CK20 (Figure 4A). In addition, SCBO-10 was negative for the basal cytokeratins CK5 and CK14 and positive for the luminal markers CK8 and FOXA1, with a slight decrease in expression of the luminal marker GATA3 in organoids (Figure 4A). The second group, representing 64% (n=14/22) of the organoid lines, was characterized by substantial phenotypic differences between the parental tumors and organoids (Figures S4 and S5). In most of these cases (86%, n=12/14), for example SCBO-7, the parental tumor predominantly or exclusively expressed luminal markers, whereas the corresponding organoids lost luminal but gained basal marker expression in culture (Figure 4B). Notably, these basal organoid phenotypes often reverted back to the luminal phenotype of the parental tumor when grown as xenografts (88%; n=7/8), but then recurred in organoids derived from these xenografts (100%; n=5/5). Taken together, these results suggest that most phenotypic changes in organoid culture are associated with a transition towards a basal phenotype that is usually reversible in xenografts.
Classification of organoid lines into basal and luminal subtypes
To further characterize phenotypic instability in organoid culture, we performed RNA-sequencing analyses of parental tumors and organoid lines. Principal component analysis (PCA) of the resulting data showed that the parental tumors grouped together with bladder tumor samples from the TCGA dataset, whereas the organoid lines clustered together (Figure S6A). The overall similarity of the organoid lines to each other, rather than their corresponding parental tumors, is consistent with their rapid growth in culture as well as absence of stromal components. From supervised gene expression analysis, we derived a signature of genes whose expression distinguishes organoid lines from their parental tumors. Gene Set Enrichment Analyses (GSEA) using this signature demonstrated negative enrichment for gene sets corresponding to cell adhesion and extracellular molecules, consistent with the lack of a tumor microenvironment in organoid culture, as well as positive enrichment for gene sets corresponding to the cell cycle and ERBB signaling, most likely reflecting the proliferation of organoids in the presence of exogenous EGF (Figure S6B). These observations imply that the organoid lines may be difficult to categorize using subtype classifications that are dependent in part upon expression of cell cycle genes (e.g., (Hedegaard et al., 2016)), or expression of stromal and immune components (e.g., (Robertson et al., 2017)).
In contrast, we could successfully categorize the organoid lines into basal or luminal subtypes, using a molecular classifier (BASE47) based on studies of human bladder tumors (Damrauer et al., 2014). Our analysis showed that most of the organoid lines and corresponding parental tumors could be classified as either luminal or basal, although several did not display a clear luminal or basal subtype, which we considered as “mixed” (Figure 4C; Table S2). To validate these findings, we used a second independent classifier known as the MDACC classifier (Choi et al., 2014). In the majority of cases, the results using the MDACC classifier were concordant with those using the BASE47 classifier and with expression of luminal and basal markers as determined by immunofluorescence (Figure 4D; Table S2). Notably, we observed a general correlation between a mixed subtype in the parental tumor and the occurrence of a phenotypic shift towards a basal subtype in culture (Figures 4E, S4, and S5; Table S2), although two tumors had a strictly luminal phenotype yet also displayed this phenotypic transition.
Drug response of organoid lines
To explore the utility of the organoid lines as preclinical models for evaluation of drug response, we performed dose titration assays to examine the effects of 40 compounds using 9 organoid lines, and an additional 10 compounds using 11 lines (Figures 6A, S7, and S8; Table S3). Each organoid line was tested between passage 5 and 12 (in most cases at passage 8 or 9), so that lines displaying phenotypic instability had already completed their shift to a basal phenotype. Drugs were selected based on their clinical relevance for bladder cancer treatment, including standard-of-care therapies and investigational agents being tested in clinical trials, and/or their ability to target signaling pathways or molecules of interest.
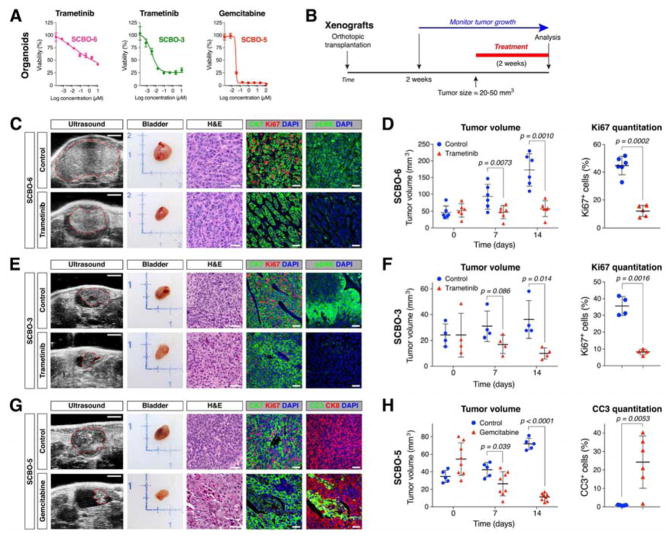
Figure 6. In vivo validation of drug response in orthotopic xenografts.
(A) Dose response curves for selected organoid lines treated with the indicated drugs in culture. (B) Overview of drug treatment of xenografts. Passage numbers of the organoid lines were: SCBO-6, P10; SCBO-3, P11; and SCBO-5, P9. (C, E, G) Ultrasound images of orthotopic tumors (dashed lines), whole-mount images of bladder, H&E-stained sections of xenografts, and immunofluorescence detection of the indicated markers in xenografts. Scale bars indicate 2 mm for ultrasound images, and 50 μm for H&E and immunofluorescence images. (D, F, H) Tumor volumes as determined by ultrasound imaging at the indicated time points, together with quantitation of Ki67 immunostaining (D, F) or of cleaved caspase-3 immunostaining (H).
The responses to these compounds revealed striking similarities and differences between organoid lines as well as partial correlations with their mutational profiles (Figure 5A). For example, SCBO-1 and SCBO-5 both displayed significant responses to treatment with the MEK inhibitor trametinib and the ERK inhibitor SCH772984, consistent with the presence of activating mutations in FGFR3 (Figure 2A). Conversely, SCBO-2 and SCBO-6 contain FGFR3 mutations, but did not display a significant response to trametinib or SCH772984, whereas the molecular basis of the response of SCBO-3 to these agents is unclear. Interestingly, none of the organoid lines tested displayed responses to three different FGFR inhibitors, perhaps suggesting their lack of potency under the conditions used or additional complexity in the molecular basis for response to these compounds. Overall, we noted a correlation between tumor progression and drug response, with the lines established from muscle invasive cancer (SCBO-4 and SCBO-6) and from recurrent disease after treatment failure (SCBO-3.2) displaying greatest drug resistance.
Figure 5. Drug response of organoid lines.
(A) Heatmap of logIC50 values calculated from drug response analyses of patient-derived organoids by applying nonlinear regression (curve fit). Putative drug targets of the tested compounds are listed at left. Specific values for IC50, Hill slope, and AUC are listed in Table S3. Passage numbers of the organoid lines were: SCBO-1, P8; SCBO-2, P9; SCBO-3, P8; SCBO-3.2, P9; SCBO-4, P10; SCBO-5, P8; SCBO-6, P9; SCBO-11, P5; SCBO-11.2, P6; SCBO-8, P12; SCBO-10, P8. (B) Dose response curves for SCBO-5 and SCBO-10 organoids treated with the MEK inhibitor trametinib, and SCBO-5 and SCBO-6 organoids with the mTOR inhibitor AZD8055. Each data point corresponds to three biological replicates; error bars correspond to one standard deviation. (C) Western blotting performed with the indicated antibodies of lysates from organoids treated for 8 hours with 10 nM or 500 nM of trametinib (left), or with 10 nM or 500 nM of AZD8055 (right). (D) Dose response curves for SCBO-3 and SCBO-3.2 organoids as well as SCBO-11 and SCBO-11.2 organoids treated with the indicated compounds. (E, F) Dose response curves for SCBO-6 organoids treated with the indicated drugs individually or in combination. See also Figures S7 and S8 and Table S3.
We used Western blotting to further examine the differential response of SCBO-5 and SCBO-10 to the MEK inhibitor trametinib, and of SCBO-5 and SCBO-6 to the mTOR inhibitor AZD8055 (Figures 5B and 5C). These analyses confirmed that trametinib potently inhibited MEK activation as shown by downregulation of phospho-ERK in both trametinib-sensitive SCBO-5 and trametinib-resistant SCBO-10 organoids (Figure 5C). Similarly, AZD8055 inhibited mTOR signaling as shown by downregulation of phospho-S6(S235/236) in both AZD8055-sensitive SCBO-6 as well as AZD8055-resistant SCBO-5 organoids (Figure 5C). In the case of AZD8055, drug sensitivity was consistent with the presence of a TSC1 mutation in SCBO-6, but not SCBO-5. However, trametinib sensitivity was only observed for SCBO-5, which contains an FGFR3 activating mutation, but not for SCBO-10, despite the presence of an FGFR3-TACC3 fusion.
Interestingly, we could compare the differential drug response of metachronous organoid lines established from patients with recurrent bladder cancer. We found that SCBO-3.2 was markedly more resistant to a wide range of drugs compared to SCBO-3, consistent with the emergence of a recurrent tumor after treatment with mitomycin C and BCG, whereas SCBO-11.2 displayed drug responses that were very similar to that of SCBO-11, consistent with the absence of intervening treatment (Figure 5D). These results suggest that drug responses in the SCBO-3.2 organoid line are likely to reflect changes in drug response of its parental tumor as a consequence of treatment failure.
In addition, we used the mutational profiles to predict potential additive drug responses. For example, we found that SCBO-6 displayed an additive response to treatment with the FGFR inhibitor JNJ-42756493 and the mTOR inhibitor AZD8055 and, consistent with the presence of both an activating FGFR3 mutation and a nonsense mutation in TSC1 (Figure 5E). We also found a similar additive interaction between JNJ-42756493 and the mTOR inhibitor sirolimus in SCBO-6 (Figure 5E). These results indicate that the molecular profiles of the organoid lines can be useful for identification of potential combinatorial therapies.
Finally, we sought to validate the drug responses identified in organoid culture by treatment of orthotopic xenografts in vivo. For this purpose, we examined three combinations of drugs and organoid lines, corresponding to the effects of trametinib treatment on SCBO-6 and SCBO-3, as well as for gemcitabine on SCBO-5, each of which displayed a response in organoid culture (Figure 6A). We observed a significant effect of each drug on tumor size in longitudinal assessments by ultrasound imaging (Figures 6B–H). We also found changes in tumor histology and a significant decrease in cellular proliferation following trametinib treatment as well as a significant increase in cleaved caspase-3 expression following gemcitabine treatment (Figures 6C–H). These findings suggest that the drug responses observed in organoid culture can be recapitulated when assayed in an in vivo context.
Discussion
Our studies demonstrate that patient-derived bladder tumor organoids can recapitulate the broad histopathological and molecular spectrum of both human non-muscle invasive and muscle invasive bladder cancers. The mutational profiles of the organoid lines are highly characteristic of not only the parental tumors from which they were derived, but of human bladder cancer overall. For example, we generated models harboring mutations in ERBB2 as well as FGFR3-TACC3 fusions. Notably, concordance of molecular profiles with those of parental tumors has been previously shown in analyses of colorectal organoids (van de Wetering et al., 2015; Weeber et al., 2015) as well as in a broad-based study of organoids from multiple tumor types (Pauli et al., 2017). Furthermore, we have shown that organoids and orthotopic xenografts can be interconverted with high efficiency, allowing each model to be used for its specific experimental advantages when appropriate, including for validating drug responses in vivo.
Patient-derived bladder tumor organoids often retain significant tumor heterogeneity, and can readily undergo clonal evolution in culture. In particular, our analysis of phylogenetic trees shows linear and branched patterns of clonal evolution (Figure 3D) that are similar to patterns described for urothelial cancers in vivo (Lamy et al., 2016). In principle, these patterns may correspond to a stochastic neutral drift, may represent a consequence of selection pressures in culture, and/or may reflect the activity of one or more oncogenic drivers within tumor subclones that mediate clonal competition. Future studies may address these mechanisms of clonal evolution using direct experimental manipulations in organoid culture.
Interestingly, we found that many of the organoid lines can change their marker phenotype and basal/luminal subtype in culture. Whereas all of the phenotypically stable lines have a strictly luminal phenotype, the lines that change phenotype in culture displayed a mixed or luminal subtype in the parental tumor, and then shifted to a basal subtype as organoids (Figure 5C; Table S2). Interestingly, there was no apparent correlation between the phenotypic stability or instability of the organoid lines with their pathology, mutational profile, or drug response. Since these marker expression patterns are usually reversible in xenografts derived from these organoid lines (Figures S4 and S5), and there are no clear correlations of phenotypic properties with changes in the frequencies of subclonal mutations as determined by analyses of variant allele fractions (Figure S2), we believe that these phenotypic changes in organoid culture are likely to be due to cellular plasticity. Such plasticity may arise through changes in patterns of epigenetic modifications under conditions of organoid culture versus xenografts. It is conceivable that this phenotypic plasticity may reflect a natural disease process in which progression of some bladder tumors is associated with luminal to basal subtype switching (Choi et al., 2017). Another possibility is that the observed gain of basal marker expression may recapitulate the early pre-microscopic stages of tumor differentiation towards a squamous phenotype (which expresses basal markers) that is often observed in bladder tumors in vivo (Adam and DeGraff, 2015; Gellert et al., 2015). Furthermore, the observed plasticity of luminal cells in bladder tumor organoids may suggest that maintenance of luminal phenotypes often requires a stromal microenvironment, which is lacking in organoid culture but restored in xenografts.
A notable feature of primary bladder cancer that differs from most solid tumors is the ability to obtain tumor samples from the same patient at multiple distinct disease states. In many cases, patients will undergo resections before and after treatment, and each resection provides an opportunity to generate a corresponding organoid line, which is feasible due to our relatively high efficiency of organoid establishment. In future studies, these organoid lines from patients with recurrent bladder cancer provide an excellent model system to investigate tumor evolution and drug response in primary solid tumors.
The promise of patient-derived organoid models in precision cancer medicine relies upon the notion that characterization of their mutational profiles in combination with high-throughput screening with a library of therapeutic compounds can elucidate druggable targets. In the case of bladder cancer, patients are often diagnosed early in disease progression, and patients with non-muscle invasive cancer frequently undergo multiple resections and treatments to avoid cystectomy and its deleterious effects on quality of life. Thus, positive drug responses identified by screening in organoid culture could be validated in organoid-derived xenografts, and in principle could then be used to guide intravesical therapies. Notably, previous studies using genetically-engineered mouse models have already established the conceptual basis for translating pre-clinical laboratory findings into clinical trials of intravesical therapies (Delto et al., 2013; Seager et al., 2009). Consequently, as a next step, it will be essential to pursue co-clinical studies to determine whether the response of patient-derived bladder tumor organoids to drug treatment in culture recapitulates patient response to the same treatments in vivo.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Michael M. Shen (mshen@columbia.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human specimens
Bladder tissues and blood were obtained from patients undergoing transurethral resection of bladder tumors (TURBT) at Columbia University Medical Center. Two patient-derived xenograft lines were established from nephroureterectomies performed at Memorial Sloan Kettering Cancer Center. All patients gave informed consent under the auspices of Institutional Review Board-approved protocols.
Patient clinical characteristics
Samples collected from 18 patients at Columbia University Medical Center were included in the study, 17 of whom were diagnosed with urothelial carcinoma and 1 of whom was diagnosed with squamous cell carcinoma. These patients consisted of 13 males and 5 females and ranged from ages 48 to 86 years of age, with a median of 72.7 years. Samples from Memorial Sloan Kettering Cancer Center were males at 64 and 86 years of age.
Organoid xenograft studies
All experiments using animals were performed according to protocols approved by the Institutional Animal Care and Use Committee (IACUC) at Columbia University Medical Center. 6–8 week old male NOG (NOD.Cg-PrkdcscidIl2rgtm1Sug/JicTac; Taconic) mice were used for ultrasound-guided delivery of organoids diluted in 50% Matrigel/media into the mouse bladder wall. Mice were observed for clinical signs of disease daily, and mice were euthanized according to IACUC-approved criteria.
METHOD DETAILS
Sample Collection
Fresh tumor samples were collected endoscopically by cold cup biopsy or using the resectoscope loop without electrical current (“cold loop resection”), or by cystectomy. Tissue samples were placed on ice in hepatocyte culture medium supplemented with 10 ng/ml epidermal growth factor (EGF; Corning), 5% charcoal-stripped fetal bovine serum (CS-FBS, Gemini), and 100 μg/ml Primocin (InvivoGen) and transported directly to the laboratory. Tissue samples were used for the establishment of primary organoid cultures and for analysis of parental tumors.
Tissue dissociation and organoid culture
Organoid culture was performed as previously described (Chua et al., 2014), with minor modifications to the tissue dissociation protocol to improve cell viability. For tissue dissociation, tumor tissues from patients or xenografts were washed in organoid culture media (hepatocyte media with 10 ng/ml EGF, 5% CS-FBS, 10 μM Y-27632 (STEMCELL Technologies), and 1X Glutamax (Gibco)), supplemented with 100 μg/ml Primocin, and minced with scissors. Tumor tissues were then incubated in 10 ml of the organoid culture media supplemented with 100 μg/ml Primocin and 1:10 dilution of collagenase/hyaluronidase (STEMCELL Technologies) at 37 °C for 15 min. Dissociated tissues were spun down at 350 g for 5 min, resuspended in 10 ml of PBS, and spun down again. The tissues were resuspended in 5 ml of TrypLE Express (Invitrogen), followed by incubation at room temperature for 3 min. Trypsinization was stopped by addition of 10 ml modified Hank’s balanced salt solution (HBSS; STEMCELL Technologies) supplemented with 5% CS-FBS, 10 μM of the ROCK inhibitor Y-27632 and 100 μg/ml Primocin, followed by centrifugation at 350 g. Dissociated tissues were resuspended with 10 ml of HBSS supplemented with 5% CS-FBS, 10 μM Y-27632 and 100 μg/ml Primocin, and passed through a 100 μm cell strainer (Corning). Dissociated cell clusters (approximately 2–10 cells per cluster, and 1 x 106 cells in total) were spun down and resuspended in 60% Matrigel (Corning)/organoid culture media, and plated in a 250 μl drop in the middle of one well of a pre-coated 6-well plate (Corning) with 60% Matrigel. The drop was solidified by a 30-minute incubation at 37°C and 5% CO2. After solid drops formed, 1.5 ml of the organoid culture media was added to the well, and the medium was changed every 3–4 days. Typically, approximately 50–80% of the cell clusters would form organoids, although there was considerable variation between lines and not all organoids could propagate after passaging.
For passaging, 1 mg/ml dispase (STEMCELL Technologies) was added to the medium followed by incubation for 60 min at 37°C to digest the Matrigel. Subsequently, organoids were centrifuged at 350 g for 5 min, washed in PBS and spun down. 5 ml TrypLE Express (Invitrogen) was added, and organoids were incubated at room temperature for 3 min, followed by mechanical dissociation to small cell clusters by pipetting. Organoids were passaged at a 1:2-3 dilution every 2–3 weeks. To generate stocks, organoids were snap-frozen in 90% CS-FBS and 10% DMSO and stored in liquid nitrogen. Cryopreserved stocks have been successfully recovered for up to at least 18 months after freezing.
Histology and immunostaining
Tissues and organoids were processed for paraffin sectioning using standard protocols. For processing organoids, organoids were harvested using 1 mg/ml dispase to digest the Matrigel as for passaging, washed in PBS, fixed in 10% formalin (Fisher Chemical) for 1 h, and placed in rat tail collagen I (Corning) before tissue processing and embedding. Haematoxylin–eosin staining was performed using standard protocols on 5 μm paraffin sections.
Histologic evaluation of hematoxylin-eosin stained sections of parental tumor samples, organoids, and orthotopic xenografts was performed by a urological pathologist (H.A.-A.) to classify grade and stage according to the most recent World Health Organization (WHO) classification system (Humphrey et al., 2016). The organoids and xenografts were evaluated according to how closely they resembled the parental tumors since there are no classification systems for such specimens. Table 1 shows the staging of the overall tumor, which describes the histology of the tumor sample as a whole, as well as a summary of the pathology of the specific parental tumor sample, which was adjacent to the samples used for organoid establishment and molecular analyses. As a result, the histology of the overall tumor may not always correspond to that of the parental tumor sample. In our analyses, we report the concordance of histology and molecular profiles between the organoid lines to their corresponding parental tumor sample.
For immunostaining, 5 μm paraffin sections were deparaffinized in xylene, followed by boiling in antigen unmasking solution (Vector Labs). Slides were blocked in 5% normal goat serum (NGS; Vector Labs), and incubated with primary antibodies (see Key Resources Table) diluted in 1% NGS overnight at 4°C. Samples were then incubated with secondary antibodies labelled with Alexa Fluor 488, 555 or 647 (Invitrogen) diluted in 1% NGS at RT for 1 h. Slides were mounted with VECTASHIELD mounting medium with DAPI (Vector Labs). Immunofluorescence staining was imaged using a Leica TCS SP5 spectral confocal microscope.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Chicken polyclonal anti-CK5 | BioLegend | Cat#905901 |
| Rabbit monoclonal anti-CK7 | Abcam | Cat#ab181598 |
| Rat monoclonal anti-CK8 | Developmental Studies Hybridoma Bank (DSHB) | Cat#TROMA-I |
| Rabbit polyclonal anti-CK14 | BioLegend | Cat#905301 |
| Mouse monoclonal anti-CK20 | Dako | Cat#M7019 |
| Rabbit polyclonal anti-p53 | Santa Cruz | Cat#SC-6243 |
| Rat monoclonal anti-Ki67 | eBioscience | Cat#14-5698-82 |
| Rabbit polyclonal anti-cleaved caspase 3 | BD | Cat#559565 |
| Goat anti-mouse IgG, Alexa Fluor 488 | Invitrogen | Cat#A11029 |
| Goat anti-rabbit IgG, Alexa Fluor 488 | Invitrogen | Cat#A11008 |
| Goat anti-rat IgG, Alexa Fluor 555 | Invitrogen | Cat#A21434 |
| Goat anti-rat IgG, Alexa Fluor 647 | Invitrogen | Cat#A21247 |
| Goat anti-chicken IgG, Alexa Fluor 555 | Invitrogen | Cat#A21437 |
| Goat anti-chicken IgG, Alexa Fluor 647 | Invitrogen | Cat#A21449 |
| Mouse monoclonal anti-FOXA1 | Abcam | Cat#55178 |
| Rabbit monoclonal anti-GATA3 | Cell Signaling Technology | Cat#5852 |
| Rabbit monoclonal anti-phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) | Cell Signaling Technology | Cat#4370 |
| Rabbit monoclonal anti-p44/42 MAPK (Erk1/2) | Cell Signaling Technology | Cat#4695 |
| Rabbit monoclonal anti-phospho-Akt (Ser473) | Cell Signaling Technology | Cat#4060 |
| Rabbit polyclonal anti-Akt | Cell Signaling Technology | Cat#9272 |
| Rabbit monoclonal anti-phospho-S6 (Ser235/236) | Cell Signaling Technology | Cat#4858 |
| Rabbit monoclonal anti-S6 | Cell Signaling Technology | Cat#2217 |
| Rabbit monoclonal beta-actin | Cell Signaling Technology | Cat#8457 |
| Goat anti-rabbit IgG, HRP-linked | Cell Signaling Technology | Cat#7074 |
| Bacterial and Virus Strains | ||
| Biological Samples | ||
| Human bladder tissues and blood | Columbia University Medical Center | http://www.cumc.columbia.edu |
| Patient-derived xenografts (PDXs) | Memorial Sloan Kettering Cancer Center | http://www.mskcc.org |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Hepatocyte media with epidermal growth factor (EGF) | Corning | Cat#355056 |
| Matrigel | Corning | Cat#354234 |
| Rat tail collagen I | Corning | Cat#354249 |
| Y-27632 | STEMCELL Tech | Cat#07171 |
| Collagenase/hyaluronidase | STEMCELL Tech | Cat#07912 |
| Modified Hank’s balanced salt solution (HBSS) | STEMCELL Tech | Cat#37150 |
| Dispase | STEMCELL Tech | Cat#07913 |
| Glutamax | Gibco | Cat#35050 |
| Primocin | InvivoGen | Cat#ant-pm-1 |
| TrypLE Express | Invitrogen | Cat#12605036 |
| 10% formalin | Fisher Chemical | Cat#SF100-4 |
| Halt protease and phosphatase inhibitor cocktail | Thermo Fisher Scientific | Cat#78441 |
| Tween 80 | Sigma-Aldrich | Cat#P8074 |
| Hydroxypropylmethyl cellulose (HEC) | Sigma-Aldrich | Cat#H7509 |
| Dimethyl Sulfoxide (DMSO) | Sigma-Aldrich | Cat#D2650 |
| Antigen unmasking solution | Vector Labs | Cat#H3300 |
| SuperSignal West Pico PLUS Chemiluminescent Substrate | Thermo Fisher Scientific | Cat#34577 |
| Neratinib (HKI-272) | Selleck Chemicals | Cat#S2150 |
| Erlotinib | Cayman Chemical | Cat#10483 |
| Sapitinib (AZD8931) | Selleck Chemicals | Cat#S2192 |
| PD-173074 | Cayman Chemical | Cat#13032 |
| Dovitinib | Cayman Chemical | Cat#15220 |
| JNJ-42756493 | ActiveBiochem | Cat#A-1278 |
| Trametanib (GSK1120212) | Selleck Chemicals | Cat#S2673 |
| Selumetinib (AZD6244) | Cayman Chemical | Cat#11599 |
| SCH772984 | Selleck Chemicals | Cat#S7101 |
| Gemcitabine | Selleck Chemicals | Cat#S1714 |
| Cisplatin | Selleck Chemicals | Cat#S1166 |
| Doxorubicin (Adriamycin) | Cayman Chemical | Cat#15007 |
| 5-Fluorouracil | Cayman Chemical | Cat#14416 |
| Ifosfamide | Cayman Chemical | Cat#17562 |
| Methotrexate | Cayman Chemical | Cat#13960 |
| Mitomycin C | Sigma-Aldrich | Cat#M4287 |
| Vinblastine | Selleck Chemicals | Cat#S1248 |
| Paclitaxel | Cayman Chemical | Cat#10461 |
| Docetaxel | Cayman Chemical | Cat#11637 |
| Cabazitaxel | Selleck Chemicals | Cat#S3022 |
| Sirolimus (Rapamycin) | Selleck Chemicals | Cat#S1039 |
| AZD8055 | Cayman Chemical | Cat#16978 |
| Gedatolisib (PF-05212384) | Cayman Chemical | Cat#14567 |
| Pictilisib (GDC0941) | Selleck Chemicals | Cat#S1065 |
| MK-2206 | Cayman Chemical | Cat#11593 |
| (+)-JQ1 | Cayman Chemical | Cat#11187 |
| GSK126 | Cayman Chemical | Cat#15415 |
| Mocetinostat | Selleck Chemicals | Cat#S1122 |
| Veliparib (ABT-888) | Selleck Chemicals | Cat#S1004 |
| Talazoparib (BMN-673) | Selleck Chemicals | Cat#S7048 |
| Alisertib (MLN8237) | Selleck Chemicals | Cat#S1133 |
| Doramapimod (BIRB 0796) | Cayman Chemical | Cat#10460 |
| Enzastaurin (LY317615) | Cayman Chemical | Cat#11601 |
| BI-2536 | Cayman Chemical | Cat#17385 |
| Palbociclib (PD-0332991) | Cayman Chemical | Cat#16273 |
| PF477736 | Cayman Chemical | Cat#17859 |
| Lestaurtinib (CEP-701) | Cayman Chemical | Cat#12094 |
| Ruxolitinib (INCB-18424) | Cayman Chemical | Cat#11609 |
| Linsitinib (OSI-906) | Selleck Chemicals | Cat#S1091 |
| PLX4720 | Cayman Chemical | Cat#15142 |
| Motesanib (AMG-706) | Selleck Chemicals | Cat#S1032 |
| Navitoclax (ABT-263) | Selleck Chemicals | Cat#S1001 |
| PAC-1 | Selleck Chemicals | Cat#S2738 |
| XAV 939 | Cayman Chemical | Cat#13596 |
| FK866 (APO866, Daporinad) | Selleck Chemicals | Cat#S2799 |
| Nutlin-3a | Cayman Chemical | Cat#18585 |
| Avagacestat (BMS-708163) | Cayman Chemical | Cat#16711 |
| Tanespimycin (17-AAG) | Cayman Chemical | Cat#11039 |
| Ganetespib | Selleck Chemicals | Cat#S1159 |
| Ixazomib (MLN2238) | Selleck Chemicals | Cat#S2180 |
| Critical Commercial Assays | ||
| MagMAX-96 Total RNA Isolation Kit | Invitrogen | Cat#AM1839 |
| TruSeq RNA Library Prep Kit v2 | Illumina | Cat#RS-122-2001 |
| TruSeq Stranded Total RNA Library Prep Kit with Ribo-Zero Human Kit | Illumina | Cat#RS-122-2201 |
| CellTiter-Glo 3D Cell Viability Assay | Promega | Cat#G9683 |
| DNeasy Blood and Tissue kit | Qiagen | Cat#69504 |
| SureSelect Human All Exon V4 | Agilent | Cat#5190-4668 |
| TruSeq SBS kit v5 | Illumina | Cat#FC-104-5001 |
| Deposited Data | ||
| Raw Data Files from RNA-seq | This paper | GEO: GSE103990 |
| Experimental Models: Cell Lines | ||
| Human: SCBO-1 organoids | This paper | N/A |
| Human: SCBO-2 organoids | This paper | N/A |
| Human: SCBO-3 organoids | This paper | N/A |
| Human: SCBO-3.2 organoids | This paper | N/A |
| Human: SCBO-4 organoids | This paper | N/A |
| Human: SCBO-5 organoids | This paper | N/A |
| Human: SCBO-6 organoids | This paper | N/A |
| Human: SCBO-7 organoids | This paper | N/A |
| Human: SCBO-7.2 organoids | This paper | N/A |
| Human: SCBO-8 organoids | This paper | N/A |
| Human: SCBO-9 organoids | This paper | N/A |
| Human: SCBO-10 organoids | This paper | N/A |
| Human: SCBO-11 organoids | This paper | N/A |
| Human: SCBO-11.2 organoids | This paper | N/A |
| Human: SCBO-11.3 organoids | This paper | N/A |
| Human: SCBO-12 organoids | This paper | N/A |
| Human: SCBO-13 organoids | This paper | N/A |
| Human: SCBO-14 organoids | This paper | N/A |
| Human: SCBO-15 organoids | This paper | N/A |
| Human: SCBO-16 organoids | This paper | N/A |
| Human: SMBO-1 organoids | This paper | N/A |
| Human: SMBO-2 organoids | This paper | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: NOD.Cg- Prkdcscid Il2rgtm1Sug/JicTac (NOG) | Taconic | Cat#NOG-M |
| Oligonucleotides | ||
| Primer: FGFR3_ex12 Forward: CGTGAAGATGCTGAAAGACGATG | (Di Stefano et al., 2015) | N/A |
| Primer: TACC3_ex14 Reverse: AAACGCTTGAAGAGGTCGGAG | (Di Stefano et al., 2015) | N/A |
| Primer: GAPDH Forward: GGACCTGACCTGCCGTCTAGAA | This paper | N/A |
| Primer: GAPDH Reverse: GGTGTCGCTGTTGAAGTCAGAG | This paper | N/A |
| Recombinant DNA | ||
| Software and Algorithms | ||
| Prism version 7 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Burrows-Wheeler Aligner: bwa-0.7.12 | (Li and Durbin, 2009) | https://sourceforge.net/projects/bio-bwa/files/ |
| Picard suite: picard-tools-1.124 | (McKenna et al., 2010) | https://broadinstitute.github.io/picard/ |
| Genome Analysis Toolkit: GenomeAnalysisTK-3.4-0-g7e26428 | (DePristo et al., 2011) | https://software.broadinstitute.org/gatk/download/ |
| MuTect: muTect-1.1.7 | (Cibulskis et al., 2013) | https://github.com/broadinstitute/mutect |
| Pindel | (Yae et al., 2009) | https://github.com/genome/pindel |
| Variant Effect Predictor version 86 | N/A | https://github.com/Ensembl/ensembl-vep |
| Vcfmaf version 1.6 | N/A | https://github.com/mskcc/vcf2maf |
| ExAC r0.3 | N/A | http://exac.broadinstitute.org/downloads |
| RSEM | (Li and Dewey, 2011) | https://github.com/deweylab/RSEM |
| STAR_2.5.0a | N/A | https://github.com/alexdobin/STAR |
| DESeq2 | (Love et al., 2014) | https://github.com/mikelove/DESeq2 |
| Gene Set Enrichment Analysis, v2-2.2.0 | (Subramanian et al., 2005) | http://software.broadinstitute.org/gsea/ |
| R version 3.3.3 | (Team, 2016) | https://www.r-project.org/ |
| Other | ||
| VEVO 2100 Ultrasound Imaging System | Visualsonics | N/A |
Targeted sequencing
A capture-based assay (MSK-IMPACT) for a panel of 468 genes of recurrent mutations, copy number changes and structural rearrangements in cancer was used to examine critical mutations in tumors and organoid samples (Zehir et al., 2017). Synthetic DNA probes were designed to capture protein-coding exons of these genes, promoter regions of TERT, and also the introns of 17 recurrently rearranged genes. Tumor and matched normal blood samples were paired for simultaneous sequencing. Once accessioned, an H&E-stained slide was reviewed and annotated for relevant specimen information, including tumor type, tumor purity and whether macrodissection of the indicated tumor region was necessary before nucleic acid extraction. Sequence data were deposited in the cBioPortal Browser for Cancer Genomics (Cerami et al., 2012).
Sequence data from orthotopic xenografts received special handling to eliminate potential contaminating mouse sequences. When appropriate, we aligned reads to an indexed hybrid genome of both human and mouse sequences in order to find the optimal position in either genome, and when reads could not be unambiguously placed, they were flagged as degenerate. Our standard pipeline was then used for reads aligned only to the human genome.
Whole-exome sequencing
Whole-genome DNA samples were prepared with DNeasy Blood and Tissue kit (Qiagen) following the manufacturer’s instructions. Normal (n =5), primary tumors (n = 5), and tumor-derived organoid or xenograft samples (n =19) were analyzed using SureSelect Human All Exon V4 (Agilent Technologies). In brief, we used 1.5 to 2 μg genomic DNA, and pre- and post-capture PCR amplification carried with six and ten cycles respectively. Samples were barcoded and run on a HiSeq 2500 instrument in 100-bp paired-end runs using TruSeq SBS kit v5 (Illumina). The average aligned reads per samples were 96 million, with 240-fold coverage on targeted regions, and the average duplication rate was 16.0%.
Sequence alignment and mutation identification
Sequence alignment and mutation identification were performed as previously described (Johnson et al., 2014). In brief, paired-end sequence data were aligned to the human genome (hg19) using Burrows-Wheeler Aligner (Li and Durbin, 2009). Read duplication, base quality recalibration, and multiple-sequence realignment were performed by the Picard suite and the Genome Analysis Toolkit before mutation detection (DePristo et al., 2011; McKenna et al., 2010). Bam files were coordinate sorted and processed for detection of point mutations by MuTect (Cibulskis et al., 2013) and of small indels less than 50 bp in length by Pindel (Ye et al., 2009). Mutations were annotated to gene transcripts in Ensembl release 75 (Genecode release 19) by Variant Effect Predictor (VEP) version 86 and vcf2maf version 1.6.11. To exclude putative germline variants in the results, we excluded any variant identified in ExAC r0.3 with a minor population allele frequency greater than 0.04%. Low confidence calls were excluded if the number of mutated reads less than 3. All analyses were conducted in R software version 3.3.3 (R Core Development Team).
Mutational signatures and phylogenetic analysis
We analyzed the presence of 29 mutational signatures based on previous work (Alexandrov et al., 2013), excluding the aflatoxin signature due to its similarity to the smoking signature. Mutation process compositions were calculated based on SNPs detected by whole-exome sequencing. Confidence intervals of the signatures were determined by resampling of the mutations in each specimen. Final results shown in Figure 3C were simplified by combining signatures with p values less than 0.05 that represent a small number of mutations as “other”.
For phylogenetic analysis, sample distances between pairs of samples from each line were calculated based on non-shared mutations, followed by hierarchical clustering using the average agglomeration method. We used the stringlist package in R software to count the non-shared mutations, and used the ape package to plot the tree. The resulting phylogenetic trees were revised and denoted with recurrent mutations.
Detection of FGFR3-TACC3 fusion transcript
Total RNA was extracted from organoids using the MagMAX-96 Total RNA Isolation Kit (Invitrogen) according to manufacturer instructions. 500 ng of total RNA was reverse-transcribed using the Maxima H Minus Reverse Transcriptase (Thermo Fisher Scientific). RT-PCR was performed as described previously (Di Stefano et al., 2015) using Phusion DNA Polymerase (NEB), and products subjected to Sanger sequencing.
Gene expression profiling
Gene expression profiling analyses were performed using patient tissue samples and corresponding organoids. Total RNA from fresh tissues or organoids was isolated using the MagMAX-96 Total RNA Isolation Kit (Invitrogen), and total RNA from formalin-fixed paraffin-embedded (FFPE) tissue sections was purified using Qiagen QIAsymphony SP automated RNA extraction (Qiagen). The quantity and quality of each sample were measured using an Agilent 2100 Bioanalyzer. RNA sequencing was performed at the JP Sulzberger Columbia Genome Center at Columbia University Medical Center (New York, NY). A TruSeq RNA Library Prep Kit v2 (Illumina) for RNA from fresh samples and a TruSeq Stranded Total RNA Library Prep Kit with Ribo-Zero Human Kit (Illumina) for RNA from FFPE samples were used for library preparation, followed by sequencing (30 million single-end reads, or 60 million paired-end reads) on an Illumina HiSeq 2500.
Read numbers for genes were extracted by RSEM (with STAR alignment program) (Li and Dewey, 2011). Mean effect reads per sample was 25 million. Normalized counts were used to analyze the expression patterns of luminal and basal classifier genes. The raw and normalized data files are deposited in Gene Expression Omnibus (GEO) with series entry number GSE103990.
Principal component analysis was performed using the top 1000 differentially expressed genes. Differentially expressed genes in organoid samples were identified compared to their primary tumors using the DESeq2 package (Love et al., 2014). Gene Set Enrichment Analysis was applied to identify significantly changed pathways (Subramanian et al., 2005).
Organoid drug response assay
To plate organoids for analyses of drug response, organoids were collected 4–5 days after passaging and passed through a 100 μm cell strainer (Corning no. 352360) to eliminate large organoids. Subsequently, organoids were resuspended in 2% Matrigel/organoid culture medium (15–20,000 organoids/ml) and dispensed into ultralow-attachment 96-well plates (Corning) in triplicate. At 24 h after plating, a 7-point 5-fold dilution series of each compound was dispensed. Drug concentrations ranged from 10 μmol/l to 128 pmol/l or from 100 μmol/l to 1.28 nmol/l, depending on individual drug properties (maximal DMSO concentration used was 1%). Cell viability was assayed using CellTiter-Glo 3D (Promega) according to the manufacturer’s instructions following 6 days of drug incubation, and results were normalized to vehicle controls.
Data analyses were performed using GraphPad Prism 7.0b software, and the values of IC50, Hill slope, and AUC were calculated by applying nonlinear regression (curve fit) and the equation log(inhibitor) vs. normalized response (variable slope).
Western blotting
To obtain total cell extracts, approximately 3,000 organoids were lysed in RIPA buffer (10 mM Tris-HCl pH 8, 140 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1% Triton, 0.1% SDS) supplemented with protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific) for 15 min on ice, followed by centrifugation at 15,000 g for 10 min at 4 °C. Proteins were resolved by SDS–PAGE, transferred to PVDF membrane and analyzed by immunoblot using primary antibodies shown in the Key Resources table. Anti-rabbit IgG conjugated to horseradish peroxidase was used as secondary antibody (Cell Signaling). Detection was performed with chemiluminescent substrate (Thermo Fisher Scientific) followed by exposure to ECL Hyperfilm (GE Healthcare Amersham).
Organoid xenograft studies
For xenograft establishment, organoids were implanted orthotopically into the submucosal layer of the bladder wall of male NOG mice (6–8 weeks old; Taconic) using ultrasound guidance (VEVO 2100 Ultrasound Imaging System) (Jager et al., 2013; Owczarek et al., 2017). Briefly, under anesthesia, the lamina propria of the bladder was delaminated from the detrusor muscle by ultrasound-guided delivery of PBS using a 1 ml syringe with a 30 G 0.5 inch needle, followed by delivery of a 50 μl of organoid suspension (1 × 106 cells) in 50% Matrigel/50% organoid culture media into the resulting submucosal pocket using a 25 G 5/8 inch needle. Tumors were monitored by ultrasound imaging every two weeks. The tumors were harvested when their volume reached >200 mm3. Fresh tumor samples were used for the establishment of xenograft-derived organoids or fixed for 24 h in 10% formalin.
For in vivo drug studies, 6 mice per treatment arm were used for SCBO-6, 4 mice per treatment arm for SCBO-3, and 8 mice per treatment arm for SCBO-5. Trametinib (Selleck Chemicals) was administered by oral gavage (0.5 mg/kg; five days on, two days off) after tumors reached a volume of approximately 20–50 mm3. Trametinib was resuspended in 0.5% hydroxypropyl methylcellulose (Sigma) and 0.2% Tween-80 (Sigma) in distilled water. Gemcitabine (Selleck Chemicals) was resuspended in 0.9% sodium chloride and administered by intraperitoneal injection (100 mg/kg; twice weekly) after tumors reached a volume of approximately 20–50 mm3. Tumors were monitored by ultrasound imaging once per week. Mouse weight was monitored at the beginning of the treatment, then weekly, and at the end of the treatment. Fresh tumors were harvested after two weeks of drug treatment.
QUANTIFICATION AND STATISTICAL ANALYSIS
Student’s paired t test was carried out using GraphPad Prism 7.0b to calculate significance. P-values are indicated on plots. Results are expressed as mean ± standard error (S.D.).
DATA AND SOFTWARE AVAILABILITY
Targeted sequencing data are available through cBioPortal (Cerami et al., 2012). RNA-sequencing data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through accession number GSE 103990 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE103990).
Organoid lines described in this study will be distributed by the Precision Xenograft and Organoid Shared Resource of the Herbert Irving Comprehensive Cancer Center. This core facility will provide organoid lines for a modest fee to cover distribution costs to interested investigators under the terms of a Materials Transfer Agreement with Columbia University Medical Center.
Supplementary Material
H&E-stained sections of parental tumors, patient-derived organoids, orthotopic xenografts generated from the organoids, and organoids derived from the xenografts, together with bright-field images of the patient-derived organoids. Each line is shown except for those in Figure 1B. Scale bars indicate 50 μm.
Summary of whole-exome sequencing data. Related to Figure 3.
Stability of luminal and basal phenotypes. Related to Figure 4.
Plots show variant allele fractions for selected mutations in parental tumors, patient-derived organoids at the indicated passages, orthotopic xenografts generated from the organoids, and organoids derived from the xenografts. Each line is shown except for those in Figure 3B.
(A) Variant allele fractions (VAF) detected by both whole-exome sequencing and targeted IMPACT-Seq are well-correlated. (B) SNP mutation patterns are shown for the combined results from each line. A total of 25,447 SNPs were detected, with 31% C>G transversions and 48% C>T transitions.
Shown is marker expression in parental tumors, patient-derived organoids at early and late passages, orthotopic xenografts generated from the organoids, and organoids derived from the xenografts. Data for SCBO-7 and SCBO-10 are shown in Figure 4. (left) Immunofluorescence staining for CK7 (green), Ki67 (red) and DAPI (blue). (right) Immunofluorescence staining for p53 (green), CK8 (red), CK5 (white) and DAPI (blue). Scale bars indicate 50 μm.
Shown is marker expression in parental tumors, patient-derived organoids at early and late passages, orthotopic xenografts generated from the organoids, and organoids derived from the xenografts. (left) Immunofluorescence staining for CK7 (green), Ki67 (red) and DAPI (blue). (right) Immunofluorescence staining for p53 (green), CK8 (red), CK5 (white) and DAPI (blue). Scale bars indicate 50 μm.
(A) Principal components analysis (PCA) for patient-derived organoids (orange), parental tumors (green), and bladder tumors analyzed by the TCGA (blue). (B) Gene Set Enrichment Analysis (GSEA) plots using a signature comparing organoids to their parental tumors shows negative enrichment for gene sets corresponding to cell adhesion and extracellular molecules, and positive enrichment for gene sets corresponding to cell cycle and ERBB signaling.
Dose response curves are shown for 11 independent patient-derived organoid lines treated with 10 compounds, and 9 lines treated with 16 compounds. Cell viability was measured by CellTiter Glo assay after 6 days of drug treatment. Each data point corresponds to three biological replicates; error bars correspond to one standard deviation.
Dose response curves are shown for 9 independent patient-derived organoid lines treated with 24 compounds. Cell viability was measured by CellTiter Glo assay after 6 days of drug treatment. Each data point corresponds to three biological replicates; error bars correspond to one standard deviation.
IC50, Hill slope, and AUC values for drug response of organoid lines. Related to Figure 5.
Highlights.
Efficient generation of a biobank of patient-derived bladder cancer organoids
Organoids recapitulate the histological and molecular spectrum of human bladder cancer
Bladder tumor organoids display clonal evolution in culture and as xenografts
Drug response of organoids can be validated in xenografts
Acknowledgments
We thank all of the patients who consented to donate their tumor tissues for this study, as well as the surgical teams who facilitated this work. This work was supported by grants from the NIH (C.A.-S., D.B.S., M.M.S.), Urology Care Foundation Research Scholar Award (S.H.L., T.B.O.), Urology Care Foundation Residency Research Award (L.J.B.), Cycle for Survival (D.B.S.), Bladder Cancer Advocacy Network (C.A.-S.), TJ Martell Foundation (M.C.B., C.A.-S., M.M.S.), and by philantrophic support provided by Linda and Ilan Kaufthal and the Russell Berrie Foundation (J.M.M., C.A.-S., H.A.-A., D.B.S., M.M.S.).
Footnotes
Author contributions
S.H.L., J.T.M., M.V.S., and A.B.W. generated and cultured organoid lines, S.H.L. performed H&E staining, immunostaining, and drug response assays, C.W.C., L.J.B., and M.V.S. developed the culture protocol, J.M.M., C.B.A., and M.C.B. provided patient tumor specimens, K.K. and J.A.C. provided patient-derived xenografts, K.K. and E.J.P. assisted with targeted exome sequencing, W.H. and C.K. performed bioinformatic analyses, T.B.O., S.H.L., and S.K.B. generated and analyzed orthotopic xenografts, H.A.-A. analyzed histology, J.T.M. and J.M.M. maintained clinical records, S.H.L., W.H., B.S.T., C.A.-S., H.A.-A., D.B.S., and M.M.S. interpreted data, and S.H.L., W.H., and M.M.S. wrote the manuscript.
Declaration of interests
D.B.S. is a consultant for Pfizer and Loxo Oncology. M.M.S., L.J.B., and C.W.C. are inventors of U.S. patent application 15/288,871, which is related to this work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adam RM, DeGraff DJ. Molecular mechanisms of squamous differentiation in urothelial cell carcinoma: a paradigm for molecular subtyping of urothelial cell carcinoma of the bladder. Urol Oncol. 2015;33:444–450. doi: 10.1016/j.urolonc.2015.06.006. [DOI] [PubMed] [Google Scholar]
- Al-Ahmadie HA, Iyer G, Janakiraman M, Lin O, Heguy A, Tickoo SK, Fine SW, Gopalan A, Chen YB, Balar A, et al. Somatic mutation of fibroblast growth factor receptor-3 (FGFR3) defines a distinct morphological subtype of high-grade urothelial carcinoma. J Pathol. 2011;224:270–279. doi: 10.1002/path.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 2013;3:246–259. doi: 10.1016/j.celrep.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boj SF, Hwang CI, Baker LA, Chio II, Engle DD, Corbo V, Jager M, Ponz-Sarvise M, Tiriac H, Spector MS, et al. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160:324–338. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broutier L, Mastrogiovanni G, Verstegen MM, Francies HE, Gavarro LM, Bradshaw CR, Allen GE, Arnes-Benito R, Sidorova O, Gaspersz MP, et al. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat Med. 2017;23:1424–1435. doi: 10.1038/nm.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas RN. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–322. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, Chandramohan R, Liu ZY, Won HH, Scott SN, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi W, Ochoa A, McConkey DJ, Aine M, Hoglund M, Kim WY, Real FX, Kiltie AE, Milsom I, Dyrskjot L, et al. Genetic alterations in the molecular subtypes of bladder cancer: Illustration in the Cancer Genome Atlas dataset. Eur Urol. 2017;72:354–365. doi: 10.1016/j.eururo.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi W, Porten S, Kim S, Willis D, Plimack ER, Hoffman-Censits J, Roth B, Cheng T, Tran M, Lee IL, et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell. 2014;25:152–165. doi: 10.1016/j.ccr.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua CW, Shibata M, Lei M, Toivanen R, Barlow LJ, Bergren SK, Badani KK, McKiernan JM, Benson MC, Hibshoosh H, et al. Single luminal epithelial progenitors can generate prostate organoids in culture. Nat Cell Biol. 2014;16:951–961. doi: 10.1038/ncb3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damrauer JS, Hoadley KA, Chism DD, Fan C, Tiganelli CJ, Wobker SE, Yeh JJ, Milowsky MI, Iyer G, Parker JS, et al. Intrinsic subtypes of high-grade bladder cancer reflect the hallmarks of breast cancer biology. Proc Natl Acad Sci U S A. 2014;111:3110–3115. doi: 10.1073/pnas.1318376111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delto JC, Kobayashi T, Benson M, McKiernan J, Abate-Shen C. Preclinical analyses of intravesical chemotherapy for prevention of bladder cancer progression. Oncotarget. 2013;4:269–276. doi: 10.18632/oncotarget.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Stefano AL, Fucci A, Frattini V, Labussiere M, Mokhtari K, Zoppoli P, Marie Y, Bruno A, Boisselier B, Giry M, et al. Detection, characterization, and inhibition of FGFR-TACC fusions in IDH wild-type glioma. Clin Cancer Res. 2015;21:3307–3317. doi: 10.1158/1078-0432.CCR-14-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost J, Clevers H. Translational applications of adult stem cell-derived organoids. Development. 2017;144:968–975. doi: 10.1242/dev.140566. [DOI] [PubMed] [Google Scholar]
- Dyrskjot L, Thykjaer T, Kruhoffer M, Jensen JL, Marcussen N, Hamilton-Dutoit S, Wolf H, Orntoft TF. Identifying distinct classes of bladder carcinoma using microarrays. Nat Genet. 2003;33:90–96. doi: 10.1038/ng1061. [DOI] [PubMed] [Google Scholar]
- Faltas BM, Prandi D, Tagawa ST, Molina AM, Nanus DM, Sternberg C, Rosenberg J, Mosquera JM, Robinson B, Elemento O, et al. Clonal evolution of chemotherapy-resistant urothelial carcinoma. Nat Genet. 2016;48:1490–1499. doi: 10.1038/ng.3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, Vela I, Sboner A, Iaquinta PJ, Karthaus WR, Gopalan A, Dowling C, Wanjala JN, Undvall EA, Arora VK, et al. Organoid cultures derived from patients with advanced prostate cancer. Cell. 2014;159:176–187. doi: 10.1016/j.cell.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert LL, Warrick J, Al-Ahmadie HA. Urothelial carcinoma with squamous differentiation--the pathologists perspective. Urol Oncol. 2015;33:437–443. doi: 10.1016/j.urolonc.2015.07.018. [DOI] [PubMed] [Google Scholar]
- Grossman HB, Natale RB, Tangen CM, Speights VO, Vogelzang NJ, Trump DL, deVere White RW, Sarosdy MF, Wood DP, Jr, Raghavan D, et al. Neoadjuvant chemotherapy plus cystectomy compared with cystectomy alone for locally advanced bladder cancer. N Engl J Med. 2003;349:859–866. doi: 10.1056/NEJMoa022148. [DOI] [PubMed] [Google Scholar]
- Gui Y, Guo G, Huang Y, Hu X, Tang A, Gao S, Wu R, Chen C, Li X, Zhou L, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet. 2011;43:875–878. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedegaard J, Lamy P, Nordentoft I, Algaba F, Hoyer S, Ulhoi BP, Vang S, Reinert T, Hermann GG, Mogensen K, et al. Comprehensive transcriptional analysis of early-stage urothelial carcinoma. Cancer Cell. 2016;30:27–42. doi: 10.1016/j.ccell.2016.05.004. [DOI] [PubMed] [Google Scholar]
- Herr HW, Bajorin DF, Scher HI. Neoadjuvant chemotherapy and bladder-sparing surgery for invasive bladder cancer: ten-year outcome. J Clin Oncol. 1998;16:1298–1301. doi: 10.1200/JCO.1998.16.4.1298. [DOI] [PubMed] [Google Scholar]
- Humphrey PA, Moch H, Cubilla AL, Ulbright TM, Reuter VE. The 2016 WHO Classification of Tumours of the Urinary System and Male Genital Organs-Part B: Prostate and Bladder Tumours. Eur Urol. 2016;70:106–119. doi: 10.1016/j.eururo.2016.02.028. [DOI] [PubMed] [Google Scholar]
- Hurst CD, Platt FM, Taylor CF, Knowles MA. Novel tumor subgroups of urothelial carcinoma of the bladder defined by integrated genomic analysis. Clin Cancer Res. 2012;18:5865–5877. doi: 10.1158/1078-0432.CCR-12-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager W, Moskalev I, Janssen C, Hayashi T, Awrey S, Gust KM, So AI, Zhang K, Fazli L, Li E, et al. Ultrasound-guided intramural inoculation of orthotopic bladder cancer xenografts: a novel high-precision approach. PLoS One. 2013;8:e59536. doi: 10.1371/journal.pone.0059536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, McLean CY, Fouse SD, Yamamoto S, Ueda H, Tatsuno K, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343:189–193. doi: 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamat AM, Hahn NM, Efstathiou JA, Lerner SP, Malmstrom PU, Choi W, Guo CC, Lotan Y, Kassouf W. Bladder cancer. Lancet. 2016;388:2796–2810. doi: 10.1016/S0140-6736(16)30512-8. [DOI] [PubMed] [Google Scholar]
- Kim J, Akbani R, Creighton CJ, Lerner SP, Weinstein JN, Getz G, Kwiatkowski DJ. Invasive bladder cancer: genomic insights and therapeutic promise. Clin Cancer Res. 2015;21:4514–4524. doi: 10.1158/1078-0432.CCR-14-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles MA, Hurst CD. Molecular biology of bladder cancer: new insights into pathogenesis and clinical diversity. Nat Rev Cancer. 2015;15:25–41. doi: 10.1038/nrc3817. [DOI] [PubMed] [Google Scholar]
- Lamy P, Nordentoft I, Birkenkamp-Demtroder K, Thomsen MB, Villesen P, Vang S, Hedegaard J, Borre M, Jensen JB, Hoyer S, et al. Paired exome analysis reveals clonal evolution and potential therapeutic targets in urothelial carcinoma. Cancer Res. 2016;76:5894–5906. doi: 10.1158/0008-5472.CAN-16-0436. [DOI] [PubMed] [Google Scholar]
- Lauss M, Ringner M, Hoglund M. Prediction of stage, grade, and survival in bladder cancer using genome-wide expression data: a validation study. Clin Cancer Res. 2010;16:4421–4433. doi: 10.1158/1078-0432.CCR-10-0606. [DOI] [PubMed] [Google Scholar]
- Lerner SP, Bajorin DF, Dinney CP, Efstathiou JA, Groshen S, Hahn NM, Hansel D, Kwiatkowski D, O'Donnell M, Rosenberg J, et al. Summary and recommendations from the National Cancer Institute's Clinical Trials Planning Meeting on novel therapeutics for non-muscle invasive bladder cancer. Bladder Cancer. 2016;2:165–202. doi: 10.3233/BLC-160053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren D, Sjodahl G, Lauss M, Staaf J, Chebil G, Lovgren K, Gudjonsson S, Liedberg F, Patschan O, Mansson W, et al. Integrated genomic and gene expression profiling identifies two major genomic circuits in urothelial carcinoma. PLoS One. 2012;7:e38863. doi: 10.1371/journal.pone.0038863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucca I, Klatte T, Fajkovic H, de Martino M, Shariat SF. Gender differences in incidence and outcomes of urothelial and kidney cancer. Nat Rev Urol. 2015;12:653. doi: 10.1038/nrurol.2015.257. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- Owczarek TB, Kobayashi T, Ramirez R, Rong L, Puzio-Kuter AM, Iyer G, Teo MY, Sanchez-Vega F, Wang J, Schultz N, et al. ARF confers a context-dependent response to chemotherapy in muscle-invasive bladder cancer. Cancer Res. 2017;77:1035–1046. doi: 10.1158/0008-5472.CAN-16-2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli C, Hopkins BD, Prandi D, Shaw R, Fedrizzi T, Sboner A, Sailer V, Augello M, Puca L, Rosati R, et al. Personalized in vitro and in vivo cancer models to guide precision medicine. Cancer Discov. 2017;7:462–477. doi: 10.1158/2159-8290.CD-16-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietzak EJ, Bagrodia A, Cha EK, Drill EN, Iyer G, Isharwal S, Ostrovnaya I, Baez P, Li Q, Berger MF, et al. Next-generation sequencing of nonmuscle invasive bladder cancer reveals potential biomarkers and rational therapeutic targets. Eur Urol. 2017;72:952–959. doi: 10.1016/j.eururo.2017.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad SM, Decastro GJ, Steinberg GD Medscape. Urothelial carcinoma of the bladder: definition, treatment and future efforts. Nat Rev Urol. 2011;8:631–642. doi: 10.1038/nrurol.2011.144. [DOI] [PubMed] [Google Scholar]
- Rausch S, Lotan Y, Youssef RF. Squamous cell carcinogenesis and squamous cell carcinoma of the urinary bladder: a contemporary review with focus on nonbilharzial squamous cell carcinoma. Urol Oncol. 2014;32:32e11–36. doi: 10.1016/j.urolonc.2012.11.020. [DOI] [PubMed] [Google Scholar]
- Redelman-Sidi G, Glickman MS, Bochner BH. The mechanism of action of BCG therapy for bladder cancer--a current perspective. Nat Rev Urol. 2014;11:153–162. doi: 10.1038/nrurol.2014.15. [DOI] [PubMed] [Google Scholar]
- Resnick MJ, Bassett JC, Clark PE. Management of superficial and muscle-invasive urothelial cancers of the bladder. Curr Opin Oncol. 2013;25:281–288. doi: 10.1097/CCO.0b013e32835eb583. [DOI] [PubMed] [Google Scholar]
- Robertson AG, Kim J, Al-Ahmadie H, Bellmunt J, Guo G, Cherniack AD, Hinoue T, Laird PW, Hoadley KA, Akbani R, et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell. 2017;171:540–556. e525. doi: 10.1016/j.cell.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs N, de Ligt J, Kopper O, Gogola E, Bounova G, Weeber F, Balgobind AV, Wind K, Gracanin A, Begthel H, et al. A living biobank of breast cancer organoids captures disease heterogeneity. Cell. 2017 doi: 10.1016/j.cell.2017.11.010. [DOI] [PubMed] [Google Scholar]
- Seager CM, Puzio-Kuter AM, Patel T, Jain S, Cordon-Cardo C, Mc Kiernan J, Abate-Shen C. Intravesical delivery of rapamycin suppresses tumorigenesis in a mouse model of progressive bladder cancer. Cancer Prev Res (Phila Pa) 2009;2:1008–1014. doi: 10.1158/1940-6207.CAPR-09-0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trialists, I.C.o., Party, M.R.C.A.B.C.W., Research, E.O.f., Group, T.o.C.G.-U.T.C., Group, A.B.C.S., Group, N.C.I.o.C.C.T., Finnbladder, Group, N.B.C.S., Group, C.U.E.d.T.O. Griffiths G, et al. International phase III trial assessing neoadjuvant cisplatin, methotrexate, and vinblastine chemotherapy for muscle-invasive bladder cancer: long-term results of the BA06 30894 trial. J Clin Oncol. 2011;29:2171–2177. doi: 10.1200/JCO.2010.32.3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, van Houdt W, van Gorp J, Taylor-Weiner A, Kester L, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161:933–945. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeber F, Ooft SN, Dijkstra KK, Voest EE. Tumor organoids as a pre-clinical cancer model for drug discovery. Cell Chem Biol. 2017;24:1092–1100. doi: 10.1016/j.chembiol.2017.06.012. [DOI] [PubMed] [Google Scholar]
- Weeber F, van de Wetering M, Hoogstraat M, Dijkstra KK, Krijgsman O, Kuilman T, Gadellaa-van Hooijdonk CG, van der Velden DL, Peeper DS, Cuppen EP, et al. Preserved genetic diversity in organoids cultured from biopsies of human colorectal cancer metastases. Proc Natl Acad Sci U S A. 2015;112:13308–13311. doi: 10.1073/pnas.1516689112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SV, Hurst CD, Knowles MA. Oncogenic FGFR3 gene fusions in bladder cancer. Hum Mol Genet. 2013;22:795–803. doi: 10.1093/hmg/dds486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, Srinivasan P, Gao J, Chakravarty D, Devlin SM, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
H&E-stained sections of parental tumors, patient-derived organoids, orthotopic xenografts generated from the organoids, and organoids derived from the xenografts, together with bright-field images of the patient-derived organoids. Each line is shown except for those in Figure 1B. Scale bars indicate 50 μm.
Summary of whole-exome sequencing data. Related to Figure 3.
Stability of luminal and basal phenotypes. Related to Figure 4.
Plots show variant allele fractions for selected mutations in parental tumors, patient-derived organoids at the indicated passages, orthotopic xenografts generated from the organoids, and organoids derived from the xenografts. Each line is shown except for those in Figure 3B.
(A) Variant allele fractions (VAF) detected by both whole-exome sequencing and targeted IMPACT-Seq are well-correlated. (B) SNP mutation patterns are shown for the combined results from each line. A total of 25,447 SNPs were detected, with 31% C>G transversions and 48% C>T transitions.
Shown is marker expression in parental tumors, patient-derived organoids at early and late passages, orthotopic xenografts generated from the organoids, and organoids derived from the xenografts. Data for SCBO-7 and SCBO-10 are shown in Figure 4. (left) Immunofluorescence staining for CK7 (green), Ki67 (red) and DAPI (blue). (right) Immunofluorescence staining for p53 (green), CK8 (red), CK5 (white) and DAPI (blue). Scale bars indicate 50 μm.
Shown is marker expression in parental tumors, patient-derived organoids at early and late passages, orthotopic xenografts generated from the organoids, and organoids derived from the xenografts. (left) Immunofluorescence staining for CK7 (green), Ki67 (red) and DAPI (blue). (right) Immunofluorescence staining for p53 (green), CK8 (red), CK5 (white) and DAPI (blue). Scale bars indicate 50 μm.
(A) Principal components analysis (PCA) for patient-derived organoids (orange), parental tumors (green), and bladder tumors analyzed by the TCGA (blue). (B) Gene Set Enrichment Analysis (GSEA) plots using a signature comparing organoids to their parental tumors shows negative enrichment for gene sets corresponding to cell adhesion and extracellular molecules, and positive enrichment for gene sets corresponding to cell cycle and ERBB signaling.
Dose response curves are shown for 11 independent patient-derived organoid lines treated with 10 compounds, and 9 lines treated with 16 compounds. Cell viability was measured by CellTiter Glo assay after 6 days of drug treatment. Each data point corresponds to three biological replicates; error bars correspond to one standard deviation.
Dose response curves are shown for 9 independent patient-derived organoid lines treated with 24 compounds. Cell viability was measured by CellTiter Glo assay after 6 days of drug treatment. Each data point corresponds to three biological replicates; error bars correspond to one standard deviation.
IC50, Hill slope, and AUC values for drug response of organoid lines. Related to Figure 5.