Abstract
Over 1.4 million people are diagnosed with colorectal cancer (CRC) each year, making it the third most common cancer in the world. Increased screening and therapeutic modalities including improved combination treatments have reduced CRC mortality, although incidence and mortality rates are still increasing in some areas. Serum-based biomarkers are mainly used for follow-up of cancer, and are ideal due to the ease and minimally invasive nature of sample collection. Unfortunately, CEA and other serum markers have too low sensitivity for screening and preoperative diagnostic purposes. Increasing interest is focused on the possible use of biomarkers for predicting treatment response and prognosis in cancer. In this study, we have performed mass spectrometry analysis (UPLC-UDMSE) of serum samples from 19 CRC patients. Increased levels of C-reactive protein (CRP), which occur during local inflammation and the presence of a systemic inflammatory response, have been linked to poor prognosis in CRC patients. We chose to analyze samples according to CRP values by dividing them into the categories CRP <30 and >30, and, separately, according to short and long 5-year survival. The aim was to discover differentially expressed proteins associated with poor prognosis and shorter survival. We quantified 256 proteins and performed detailed statistical analyses and pathway analysis. We discovered multiple proteins that are up- or downregulated in patients with CRP >30 as compared to CRP <30 and in patients with short as compared to long 5-year survival. Pathways that were enriched include LXR/RXR activation, FXR/RXR activation, complement and coagulation cascades and acute phase signaling response, with some of the proteins we identified having roles in these pathways. In this study, we have identified multiple proteins, of which a few have been previously identified as potential biomarkers, and others that have been identified as potential biomarkers for CRC for the first time, to the best of our knowledge. While these proteins still need to be validated in larger patient series, this pilot study will pave the way for future studies aiming to provide better biomarkers for patients with CRC.
Introduction
Colorectal cancer (CRC) is the third most common cancer worldwide, accounting for almost 10% of the global cancer burden with over 1.4 million new cases and 700 000 deaths each year. While the 5-year survival rate for CRC is around 60%, only 40% of CRC patients are diagnosed when the disease occurs locally [1, 2]. This is partly due to limited resources in many countries as well as a lack of widespread screening programs [3]. Even for patients who undergo curative resection the prognosis is quite poor, with only 50% of patients surviving 5 years [4]. Progress has been made to reduce CRC incidence and mortality over the past decade through increased screening and improved treatments such as the development of new combination therapies [2, 5]. For example, the addition of either bevacizumab or panitumumab to chemotherapy consisting of oxaliplatin, fluorouracil and leucovorin (FOLFOX4) has contributed to better survival in CRC patients [6, 7]. However, CRC still remains a major cause of cancer death.
Biomarkers are molecules that change upon transition to a pathological state and can be used to indicate the presence of disease. They can be used for screening and detection of cancer, as well as for monitoring of treatment and during follow-up. While the TNM staging system provides a standard basis for staging and a prediction of survival, biomarkers can provide information concerning the subdivision of tumor classes into subgroups that exhibit different behavior [8, 9]. Proteins detectable in serum are routinely used as biomarkers, such as carcinoembryonic antigen (CEA), which is the most widely used biomarker for CRC. However, CEA is not useful for the detection of early CRC due to lack of sensitivity and specificity, which limits its usefulness [10, 11].
Proteomics is the study of the complete proteome of a biological sample, and proteomic techniques such as mass spectrometry are widely used to search for biomarkers. Due to the need for new biomarkers with better sensitivity and specificity, searches to identify new potential proteins that could serve as biomarkers with clinical use are ongoing. Through proteomic analysis several serum proteins with diagnostic potential for CRC have already been identified, and there are likely more awaiting discovery [10, 12]. In our study, we decided to analyze serum samples from CRC patients according to C-reactive protein (CRP) values and, separately, according to 5-year survival. Increased levels of CRP, an acute-phase plasma protein whose concentration increases during both local inflammation and during a systemic inflammatory response (SIR), have been linked to poorer survival in CRC patients [13, 14]. Therefore, it is of interest to identify proteins that are differentially expressed in patients with high CRP values and a concomitant poorer prognosis, as well as in patients with short 5-year survival. Identifying patients with a poor prognosis would help select those who would benefit from more aggressive treatment and assist patients with a good prognosis in avoiding unnecessarily harsh treatment.
In this pilot study, we have used Ultra Performance Liquid Chromatography-Ultra Definition Mass Spectrometry (UPLC-UDMSE)-based proteomics to compare serum samples from 19 CRC patients. The samples were first analyzed after dividing them into two categories based on CRP, CRP <30 and CRP >30, and the same samples were analyzed again after being divided into the categories short and long 5-year survival. In this study, we have quantified 256 proteins with two or more unique peptides. Data were further analyzed by ANOVA, principal component analysis (PCA), Orthogonal Projections to Latent Structures Discriminant Analysis (OPLS-DA)-modeling and OPLS-DA-associated S-plot. Pathway analysis was performed using Integrated Molecular Pathway Level Analysis (IMPaLa) and Ingenuity Pathway Analysis (IPA). In our pilot study, we propose several potential biomarkers, which have good statistical significance.
Material and methods
Patient samples
The study included preoperative serum samples from 19 patients with colon cancer who underwent hemicolectomy resection with curative intent at Department of Surgery, Helsinki University Hospital between September 1999 and June 2006. Eight patients had an elevated CRP value, without any signs or symptoms of infection, but were considered to have a systemic inflammatory response due to their cancer. Eleven patients had an extremely low CRP value of 0. The patient details are given in S1 Table. The CRP values were measured according to the routine method of the clinical laboratory, Helsinki University Hospital. The clinical data came from patient records, the survival data from Population Register Centre of Finland and the cause of death for all those deceased from Statistics Finland. The study was approved by the Surgical Ethics Committee of Helsinki University Hospital (Dnro HUS 226/E6/06, extension TMK02 §66 17.4.2013). A written informed consent was obtained from all the participants in this study.
Serum sample processing and trypsin digestion
Serum samples were processed essentially as previously described [15] and the protocol was repeated here. Briefly, serum samples were thawed and TOP 12 proteins were depleted using the TOP12 protein depletion kit (Pierce, ThermoFisher) according to the manufacturer’s instructions. Total protein concentration was estimated with Pierce BCA assay kit (Pierce, ThermoFisher). Total proteins (100 μg) from TOP12 depleted serum were aliquoted and dried by speedvac (Savant, ThermoFisher). Dried proteins were dissolved in 35 μL of 50 mmol/L Tris buffer, pH 7.8 containing 6M urea. Further, 1.8 μL of 200 mmol/L DTT was added to the samples and mixture was incubated at RT for 1 h with shaking. Iodoacetamide (7 μL of 200 mmol/L stock solution) was added to the total protein mixture with shaking at RT for 1 h. To quench excess iodoacetamide, DTT (7 μL of 200 mmol/L) was added to protein samples with shaking for 1 h at RT. After diluting the samples with 270 μL of MQ water, trypsin was added at 1:50 trypsin:protein ratio and protein mixture was digested at 37°C overnight. 30 μg of tryptic peptides were cleaned with C18 spin columns (Pierce, ThermoFisher). Cleaned peptides were dissolved to reach final concentration of 1.4μg/4μL in 0.1% formic acid. 12.5 fmol/μL of Hi3 spike-in standard peptides (Waters, MA, USA) were added to facilitate quantification.
Liquid chromatography-mass spectrometry (LC-MS) and quantification
UPLC-MS
UPLC-MS was performed as described previously [15]. Briefly, four μL samples (equivalent to ~1.4 μg total protein) were injected to nano Acquity UPLC (Ultra Performance Liquid Chromatography)–system (Waters Corporation, MA, USA). TRIZAIC nanoTile 85 μm × 100 mm HSS-T3u wTRAP was used as separation device. Samples were loaded, trapped and washed for two minutes with 8.0 μL/min with 1% B. The analytical gradient used is as follows: 0–1 min 1% B, at 2 min 5% B, at 65 min 30% B, at 78 min 50% B, at 80 min 85% B, at 83 min 85% B, at 84 min 1% B and at 90 min 1% B with 450 nL/min. Buffer A was 0.1% formic acid in water and buffer B was 0.1% formic acid in acetonitrile. Data were acquired using HDMSE mode with Synapt G2-S HDMS (Waters Corporation, MA, USA). Data was collected in the range of 100–2000 m/z, scan time one-second, IMS wave velocity 650 m/s. Collision energy was ramped from 20 to 60 V. Calibration was performed with Glu1-Fibrinopeptide B MS2 fragments. Glu1-Fibrinopeptide B precursor ion was used as a lock mass during the runs. The samples were run in triplicates.
Data analysis
Data analysis [15, 16] and label-free quantification [17] were performed as previously described. The information will be repeated here. Briefly, the raw files were imported to Progenesis QI for proteomics software (Nonlinear Dynamics, Newcastle, UK). Lock mass of 785.8426 m/z, (doubly charged Glu1-Fibrinopeptide B) was used for mass correction. Peak picking and alignment were performed with default parameters of the algorithm. The peptide identification was done against Uniprot human FASTA sequences (UniprotKB Release 2015_09, 20205 sequence entries) which included ClpB protein sequence (CLPB_ECOLI (P63285)), which was inserted for label-free quantification. Fixed modification at cysteine (carbamidomethyl) and variable at methionine (oxidation) were used. Trypsin was specified as digesting enzyme with one missed cleavage allowed. False discovery rate (FDR) was set to less than 4% and auto error tolerances for fragment and precursor were used. Minimum one ion fragments per peptide, minimum three fragments per protein and minimum one peptide per protein were marked as “required” for ion matching.
Parsimony principle was used to group the proteins however, peptides unique to the proteins are also given as output. According to the parsimony principle, protein hits are reported as the minimum set comprising of all observed peptides. However, Progenesis QI for proteomics does not follow a strict parsimonious approach because of over-stringency [18]. However, to resolve conflicts, if two proteins are found with common peptides, protein with fewer peptides is immersed into the protein with more number of peptides. All relevant proteins are given in the output as a group under the lead protein having highest coverage or the highest score if the coverages of proteins are equal. Lead identity peptide data is used for quantitation and further details of this approach are given on the company website (www.nonlinear.com). The ANOVA relies on assumption that the conditions are independent and the means of the conditions are equal. Every peptide injection also contained 50 fmol of six CLPB_ECOLI (P63285, ClpB protein) peptides (Hi3 E. coli Standard, Waters). Peptide abundances were normalized with Hi3 spiked standard and relative quantitation was done with the non-conflicting peptides found. Signal of the protein is the average of the abundances of the comprised peptides. Details of the Progenesis software can be found on the software website (www.nonlinear.com) and in published literature [19]. Protein-wise differences between controls and cases were tested with ANOVA. Progenesis QI for proteomics was used for performing principle component analysis. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [20] partner repository with the dataset identifier PXD008583.
Results
Metadata
Serum samples from 19 CRC patients were analyzed in this study. The patients’ age ranged from 41 to 95 years old. CRP values ranged from 0–196 mg/L and were subsequently divided into the categories CRP <30 or >30 mg/L, based on a previous study [14]. Categories for survival were determined as long (alive five years post-surgery) or short (deceased within five years post-surgery) 5-year survival.
Proteomic analysis
In this study we analyzed CRC samples by UPLC-UDMSE and quantified 387 proteins from serum with a minimum of one unique peptide. When filtered to proteins with two or more unique peptides, this number was reduced to 256 proteins, which were subsequently used for analysis (S2 Table). Confidence score of identification ranged from 3733,5 to 9,1 and fold changes ranged from 1063,3 to 1,4.
CRP dataset
Altogether, 45 proteins passed the cut-off of ANOVA p-value 0.05 when the highest mean was set to CRP <30 and 26 proteins when the highest mean was set to CRP >30. The first 20 proteins to pass the cut-off are presented in Table 1, and all 71 proteins are listed in S3 Table. Our main criterion for identifying proteins different between the classes of samples was ANOVA p-values and therefore, proteins with p-values greater than 0.05 were not considered to be different.
Table 1. The first 20 proteins with 2 or more unique peptides analyzed according to CRP values and passing the cut-off of 0.05 for ANOVA.
Accession, peptide count, unique peptides, confidence score, ANOVA p-value, maximum fold change and highest and lowest mean condition as well as the full protein name are given in the table.
| Accession | Peptide count | Unique peptides | Confidence score | Anova (p) | Max fold change | Highest mean condition | Lowest mean condition | Description |
|---|---|---|---|---|---|---|---|---|
| P18065 | 5 | 5 | 33.0 | 0.000063 | 2.41 | CRP >30 | CRP < 30 | Insulin-like growth factor-binding protein 2 OS = Homo sapiens GN = IGFBP2 PE = 1 SV = 2 |
| P35372 | 2 | 2 | 11.3 | 8.94E-05 | 6.74 | CRP >30 | CRP < 30 | Mu-type opioid receptor OS = Homo sapiens GN = OPRM1 PE = 1 SV = 2 |
| P02741 | 3 | 2 | 29.3 | 0.0001 | 2.85 | CRP >30 | CRP < 30 | C-reactive protein OS = Homo sapiens GN = CRP PE = 1 SV = 1 |
| P0DJI8 | 30 | 19 | 189.2 | 0.0003 | 5.22 | CRP >30 | CRP < 30 | Serum amyloid A-1 protein OS = Homo sapiens GN = SAA1 PE = 1 SV = 1 |
| Q96PD5 | 51 | 46 | 510.5 | 0.0006 | 1.54 | CRP < 30 | CRP >30 | N-acetylmuramoyl-L-alanine amidase OS = Homo sapiens GN = PGLYRP2 PE = 1 SV = 1 |
| P02655 | 9 | 8 | 116.2 | 0.0006 | 3.43 | CRP < 30 | CRP >30 | Apolipoprotein C-II OS = Homo sapiens GN = APOC2 PE = 1 SV = 1 |
| Q8WVJ2 | 3 | 3 | 30.1 | 0.0011 | 2.99 | CRP < 30 | CRP >30 | NudC domain-containing protein 2 OS = Homo sapiens GN = NUDCD2 PE = 1 SV = 1 |
| P08887 | 2 | 2 | 9.6 | 0.0013 | 4.48 | CRP >30 | CRP < 30 | Interleukin-6 receptor subunit alpha OS = Homo sapiens GN = IL6R PE = 1 SV = 1 |
| Q9Y5I1 | 2 | 2 | 16.0 | 0.0014 | 1.56 | CRP >30 | CRP < 30 | Protocadherin alpha-11 OS = Homo sapiens GN = PCDHA11 PE = 2 SV = 1 |
| P78380 | 2 | 2 | 9.3 | 0.0015 | 4.03 | CRP >30 | CRP < 30 | Oxidized low-density lipoprotein receptor 1 OS = Homo sapiens GN = OLR1 PE = 1 SV = 1 |
| Q9Y6K1 | 5 | 3 | 40.8 | 0.0017 | 1.35 | CRP < 30 | CRP >30 | DNA (cytosine-5)-methyltransferase 3A OS = Homo sapiens GN = DNMT3A PE = 1 SV = 4 |
| P42081 | 3 | 2 | 15.3 | 0.0023 | 1.66 | CRP < 30 | CRP >30 | T-lymphocyte activation antigen CD86 OS = Homo sapiens GN = CD86 PE = 1 SV = 2 |
| Q6ZSA8 | 2 | 2 | 16.4 | 0.0029 | 3.53 | CRP >30 | CRP < 30 | Putative uncharacterized protein FLJ45684 OS = Homo sapiens PE = 5 SV = 2 |
| P07225 | 61 | 49 | 549.2 | 0.0030 | 1.28 | CRP >30 | CRP < 30 | Vitamin K-dependent protein S OS = Homo sapiens GN = PROS1 PE = 1 SV = 1 |
| P37088 | 5 | 5 | 39.7 | 0.0033 | 3.29 | CRP >30 | CRP < 30 | Amiloride-sensitive sodium channel subunit alpha OS = Homo sapiens GN = SCNN1A PE = 1 SV = 1 |
| P04180 | 9 | 9 | 52.5 | 0.0038 | 1.45 | CRP < 30 | CRP >30 | Phosphatidylcholine-sterol acyltransferase OS = Homo sapiens GN = LCAT PE = 1 SV = 1 |
| P00734 | 103 | 78 | 785.3 | 0.0039 | 1.24 | CRP < 30 | CRP >30 | Prothrombin OS = Homo sapiens GN = F2 PE = 1 SV = 2 |
| P19823 | 115 | 97 | 858.1 | 0.0041 | 1.59 | CRP < 30 | CRP >30 | Inter-alpha-trypsin inhibitor heavy chain H2 OS = Homo sapiens GN = ITIH2 PE = 1 SV = 2 |
| P08294 | 3 | 3 | 21.8 | 0.0041 | 2.68 | CRP < 30 | CRP >30 | Extracellular superoxide dismutase [Cu-Zn] OS = Homo sapiens GN = SOD3 PE = 1 SV = 2 |
| P15169 | 26 | 21 | 248.7 | 0.0044 | 1.62 | CRP >30 | CRP < 30 | Carboxypeptidase N catalytic chain OS = Homo sapiens GN = CPN1 PE = 1 SV = 1 |
Principal component analysis (PCA)
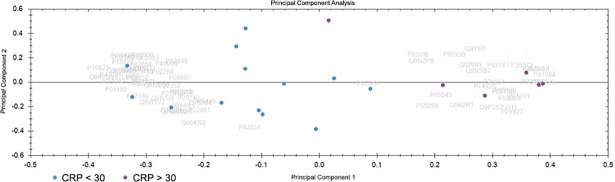
PCA was performed using the software Progenesis QI for Proteomics. It determines the main axes of variations in a dataset and points out outliers. The PCA biplot helps to establish the differences between two or more classes of samples and visualize them. The PCA biplot of CRP <30 and >30 samples is shown in Fig 1. Only proteins with 2 or more unique peptides and an ANOVA p-value of less than 0.05 were considered for this PCA. A separation between the two groups can be seen here.
Fig 1. Principal component analysis (PCA).
Blue dots are CRC samples with CRP <30 and purple dots are samples with CRP >30. The PCA was performed on proteins with 2 or more unique peptides that passed the cut-off of 0.05 for ANOVA.
Orthogonal Projections to Latent Structures Discriminant Analysis (OPLS-DA)
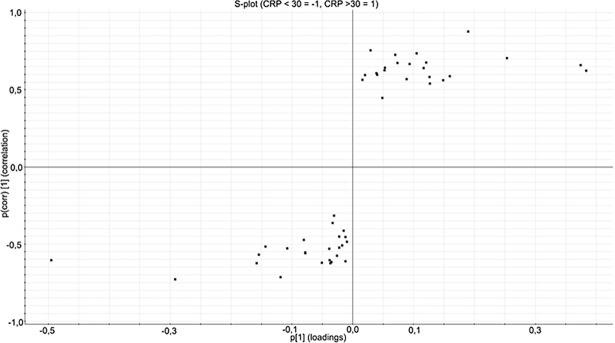
OPLS-DA is a modeling technique used to model the data from two classes in order to present the differences between these classes. Only proteins with 2 or more unique peptides and an ANOVA p-value of less than 0.05 were used for OPLS-DA modeling. From this model, an S-plot can be generated, where the x-axis is the measure of change in a particular analyte and the y-axis is the significance of the analyte in the comparison between two groups. The S-plot can be used to find the proteins that are most significantly different in the two groups. An S-plot was generated for the proteins found in the analysis of the CRP <30 and >30 categories. Proteins passing the cut-off value of +0.7 or -0.7 for p(corr) values were considered to be significantly different and are presented in Table 2. Insulin-like growth factor-binding protein 2 (IGFBP2), Protocadherin alpha-11 (PCDHA11), Mu-type opioid receptor (OPRM1) and Serum amyloid A-1 protein (SAA1) were found to be upregulated in the CRP >30 category. Apolipoprotein C-II (APOC2) and N-acetylmuramoyl-L-alanine amidase (PGLYRP2) were found to be downregulated in the CRP >30 category.
Table 2. Proteins significantly different in the S-plot generated by comparing CRP <30 and >30 categories.
Peptide count is the total number of peptides found for the given protein and unique peptides are the number of peptides unique to that protein out of the total peptides. Confidence score, ANOVA p-value, highest and lowest mean condition, the full name of the protein, covariance (p[1]) and correlation (p(corr)[1]) are shown in the table.
| Primary accession | Peptide count | Unique peptides | Confidence score | Anova (p) | Max fold change | Highest mean condition | Lowest mean condition | Description | p[1] | p(corr)[1] |
|---|---|---|---|---|---|---|---|---|---|---|
| P18065 | 5 | 5 | 33.0439 | 6.31E-05 | 2.4114 | CRP >30 | CRP < 30 | Insulin-like growth factor-binding protein 2 OS = Homo sapiens GN = IGFBP2 PE = 1 SV = 2 | 0.189875 | 0.875512 |
| Q9Y5I1 | 2 | 2 | 15.9705 | 0.001471215 | 1.5643 | CRP >30 | CRP < 30 | Protocadherin alpha-11 OS = Homo sapiens GN = PCDHA11 PE = 2 SV = 1 | 0.0294086 | 0.75569 |
| Q6ZSA8 | 2 | 2 | 16.4398 | 0.002917281 | 3.5307 | CRP >30 | CRP < 30 | Putative uncharacterized protein FLJ45684 OS = Homo sapiens PE = 5 SV = 2 | 0.105306 | 0.735447 |
| P35372 | 2 | 2 | 11.3036 | 8.94E-05 | 6.7394 | CRP >30 | CRP < 30 | Mu-type opioid receptor OS = Homo sapiens GN = OPRM1 PE = 1 SV = 2 | 0.070283 | 0.724809 |
| P0DJI8 | 30 | 19 | 189.2047 | 0.000380854 | 5.2211 | CRP >30 | CRP < 30 | Serum amyloid A-1 protein OS = Homo sapiens GN = SAA1 PE = 1 SV = 1 | 0.253761 | 0.705353 |
| P02655 | 9 | 8 | 116.1691 | 0.000684848 | 3.4281 | CRP < 30 | CRP >30 | Apolipoprotein C-II OS = Homo sapiens GN = APOC2 PE = 1 SV = 1 | -0.118406 | -0.715632 |
| Q96PD5 | 51 | 46 | 510.5228 | 0.000633028 | 1.5371 | CRP < 30 | CRP >30 | N-acetylmuramoyl-L-alanine amidase OS = Homo sapiens GN = PGLYRP2 PE = 1 SV = 1 | -0.291687 | -0.729485 |
The proteins passing the cut-off from the samples divided into CRP <30 or >30 are shown in Fig 2, visualized on an S-plot.
Fig 2. S-plot of the CRC samples with CRP <30 and >30 obtained from the OPLS-DA analysis.
Only proteins with an ANOVA p-value of less than 0.05 are shown. The S-plot shows the relationship between the correlation (p(corr)) and the covariance (p), where variables having a p(corr) value higher than 0.7 or lower than -0.7 are considered significantly different. Data are log10-transformed and mean-centered. The proteins in the upper right section of the plot are downregulated and the proteins in the lower left section upregulated in the CRP >30 category as compared to the CRP <30 category.
Pathway analysis
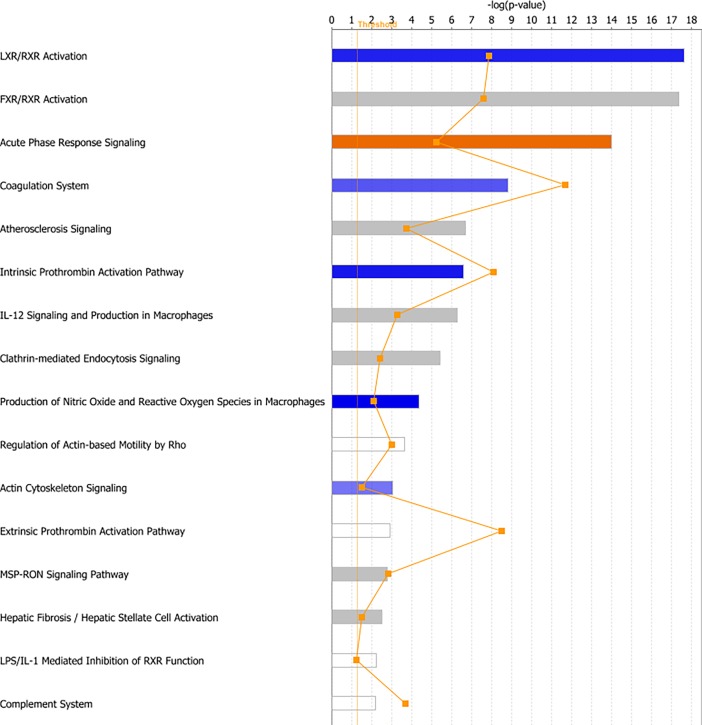
In this study, we used two tools for pathway analysis: Integrated Molecular Pathway Level Analysis (IMPaLa) and Ingenuity Pathway Analysis (IPA). IMPaLa was used for pathway over-representation analysis and the results when data were analyzed between categories CRP <30 and >30 are given in S4 Table. Complement and coagulation cascades, as well as vitamin B12 metabolism and the selenium micronutrient network were found to be enriched. We also performed pathway analysis using IPA and found multiple canonical pathways, molecular and cellular functions, as well as networks that were enriched in the CRP >30 dataset. Among the canonical pathways that were enriched were LXR/RXR activation, FXR/RXR activation, acute phase response signaling and, similar to the results from the IMPaLa analysis, the complement system. Some of the top canonical pathways are shown in Fig 3.
Fig 3. Canonical pathways found by core analysis in IPA of the CRP categories.
The top canonical pathways enriched by Ingenuity Pathway Analysis (IPA) are shown above. The straight orange vertical line running through the bars is the threshold for the p-value for the particular pathway’s enrichment. The horizontal axis is the–log(p-value) and the vertical axis shows the given pathways.
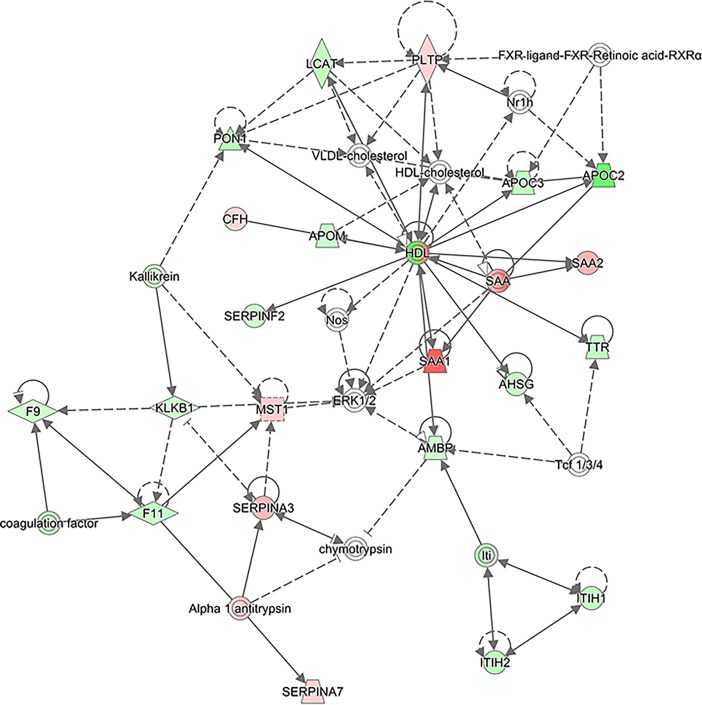
IPA also revealed protein networks enriched in the CRP >30 category when compared to the CRP <30 category, and the top network is presented in Fig 4. As can be seen, several serum amyloid A (SAA) proteins such as SAA1 and SAA2 are upregulated here. Several members of the serpin family, SERPINA3, alpha 1 antitrypsin (encoded by the SERPINA1 gene) and SERPINA7, are also upregulated in this network. The top six networks found by IPA analysis and the full list of proteins involved in these networks are given in S5 Table.
Fig 4. The top network of proteins found to be up- or downregulated by IPA when analyzed according to the CRP categories.
Only proteins passing the cut-off of 0.05 for ANOVA were used. Upregulated proteins are shown in red and downregulated proteins in green. White proteins are proteins involved in these pathways but not detected in our dataset.
Survival dataset
The serum samples used in this study were also analyzed by comparing long and short 5-year survival categories. When data were analyzed according to 5-year survival, 19 proteins passed the cut-off of ANOVA p-value 0.05 when the highest mean was set to long and 12 proteins when the highest mean was set to short. The first 20 proteins to pass the cut-off are presented in Table 3, and all 31 proteins are listed in S6 Table. Again, our main criterion for identifying proteins different between the classes of samples was ANOVA p-values and therefore, proteins with p-values greater than 0.05 were not considered to be different.
Table 3. The first 20 proteins with 2 or more unique peptides analyzed according to 5-year survival and passing the cut-off of 0.05 for ANOVA.
Accession, peptide count, unique peptides, confidence score, ANOVA p-value, maximum fold change and highest and lowest mean condition as well as the full protein name are given in the table.
| Accession | Peptide count | Unique peptides | Confidence score | Anova (p) | Max fold change | Highest mean condition | Lowest mean condition | Description |
|---|---|---|---|---|---|---|---|---|
| P02750 | 42 | 34 | 358.9 | 0.0019 | 2.23 | Survival < 5 a | Survival > 5 a | Leucine-rich alpha-2-glycoprotein OS = Homo sapiens GN = LRG1 PE = 1 SV = 2 |
| P00747 | 148 | 118 | 1396.1 | 0.004178 | 1.28 | Survival > 5 a | Survival < 5 a | Plasminogen OS = Homo sapiens GN = PLG PE = 1 SV = 2 |
| P02656 | 24 | 21 | 150.2 | 0.0049 | 1.48 | Survival > 5 a | Survival < 5 a | Apolipoprotein C-III OS = Homo sapiens GN = APOC3 PE = 1 SV = 1 |
| P05452 | 23 | 17 | 245.4 | 0.0050 | 1.51 | Survival > 5 a | Survival < 5 a | Tetranectin OS = Homo sapiens GN = CLEC3B PE = 1 SV = 3 |
| P42081 | 3 | 2 | 15.3 | 0.0089 | 1.52 | Survival > 5 a | Survival < 5 a | T-lymphocyte activation antigen CD86 OS = Homo sapiens GN = CD86 PE = 1 SV = 2 |
| Q6ZS52 | 2 | 2 | 10.5 | 0.0109 | 2.05 | Survival < 5 a | Survival > 5 a | Putative uncharacterized protein FLJ45825 OS = Homo sapiens PE = 5 SV = 1 |
| P78380 | 2 | 2 | 9.3 | 0.0115 | 3.31 | Survival < 5 a | Survival > 5 a | Oxidized low-density lipoprotein receptor 1 OS = Homo sapiens GN = OLR1 PE = 1 SV = 1 |
| Q96BN8 | 3 | 2 | 13.4 | 0.0129 | 1.70 | Survival > 5 a | Survival < 5 a | Ubiquitin thioesterase otulin OS = Homo sapiens GN = OTULIN PE = 1 SV = 3 |
| O14986 | 7 | 7 | 48.0 | 0.0155 | 1.48 | Survival > 5 a | Survival < 5 a | Phosphatidylinositol 4-phosphate 5-kinase type-1 beta OS = Homo sapiens GN = PIP5K1B PE = 1 SV = 2 |
| P49908 | 13 | 8 | 95.9 | 0.0186 | 1.32 | Survival > 5 a | Survival < 5 a | Selenoprotein P OS = Homo sapiens GN = SEPP1 PE = 1 SV = 3 |
| P00734 | 103 | 78 | 785.3 | 0.0209 | 1.19 | Survival > 5 a | Survival < 5 a | Prothrombin OS = Homo sapiens GN = F2 PE = 1 SV = 2 |
| P02655 | 9 | 8 | 116.2 | 0.0214 | 2.25 | Survival > 5 a | Survival < 5 a | Apolipoprotein C-II OS = Homo sapiens GN = APOC2 PE = 1 SV = 1 |
| P35527 | 15 | 13 | 108.1 | 0.0225 | 1.67 | Survival < 5 a | Survival > 5 a | Keratin. type I cytoskeletal 9 OS = Homo sapiens GN = KRT9 PE = 1 SV = 3 |
| P02741 | 3 | 2 | 29.3 | 0.0233 | 2.08 | Survival < 5 a | Survival > 5 a | C-reactive protein OS = Homo sapiens GN = CRP PE = 1 SV = 1 |
| P15169 | 26 | 21 | 248.7 | 0.0235 | 1.51 | Survival < 5 a | Survival > 5 a | Carboxypeptidase N catalytic chain OS = Homo sapiens GN = CPN1 PE = 1 SV = 1 |
| P01042 | 68 | 58 | 697.7 | 0.0275 | 1.11 | Survival > 5 a | Survival < 5 a | Kininogen-1 OS = Homo sapiens GN = KNG1 PE = 1 SV = 2 |
| P24385 | 2 | 2 | 18.9 | 0.0302 | 1.47 | Survival > 5 a | Survival < 5 a | G1/S-specific cyclin-D1 OS = Homo sapiens GN = CCND1 PE = 1 SV = 1 |
| P29622 | 2 | 2 | 13.3 | 0.0324 | 1.77 | Survival > 5 a | Survival < 5 a | Kallistatin OS = Homo sapiens GN = SERPINA4 PE = 1 SV = 3 |
| Q02410 | 4 | 2 | 18.5 | 0.0330 | 1.68 | Survival > 5 a | Survival < 5 a | Amyloid beta A4 precursor protein-binding family A member 1 OS = Homo sapiens GN = APBA1 PE = 1 SV = 3 |
| P10909 | 61 | 45 | 556.9 | 0.0345 | 1.19 | Survival > 5 a | Survival < 5 a | Clusterin OS = Homo sapiens GN = CLU PE = 1 SV = 1 |
PCA
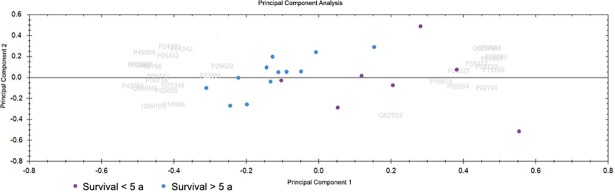
The PCA biplot showing long and short 5-year survival samples is shown in Fig 5. Only proteins with 2 or more unique peptides and an ANOVA p-value of less than 0.05 were considered for this PCA. A separation between the two groups can be seen here, with very few samples overlapping slightly.
Fig 5. PCA where blue dots are CRC samples with a long 5-year survival and purple dots are CRC samples with a short 5-year survival.
The PCA was performed on proteins with 2 or more unique peptides that passed the cut-off of 0.05 for ANOVA.
OPLS-DA
An S-plot was also generated for the proteins found in the analysis of long and short 5-year survival. Again, proteins passing the cut-off value of +0.7 or -0.7 for p(corr) values were considered to be significantly different and are presented in Table 4. Apolipoprotein C-I (APOC1), Oxidized low-density lipoprotein receptor 1 (OLR1) and CRP were found to be upregulated in the short 5-year survival category, while tetranectin and V-type proton ATPase subunit G 1 (ATP6V1G1) were found to be downregulated.
Table 4. Proteins significantly different in the S-plot generated by comparing long and short 5-year survival.
Peptide count is the total number of peptides found for the given protein and unique peptides are the number of peptides unique to that protein out of the total peptides. Confidence score, ANOVA p-value, highest and lowest mean condition, the full name of the protein, covariance (p[1]) and correlation (p(corr)[1]) are shown in the table.
| Primary accession | Peptide count | Unique peptides | Confidence score | Anova (p) | Max fold change | Highest mean condition | Lowest mean condition | Description | p[1] | p(corr)[1] |
|---|---|---|---|---|---|---|---|---|---|---|
| P02654 | 9 | 5 | 60.50 | 0.0345 | 2.0448 | SHORT | LONG | Apolipoprotein C-I OS = Homo sapiens GN = APOC1 PE = 1 SV = 1 | 0.0528039 | 0.759156 |
| Q6ZSA8 | 2 | 2 | 16.43 | 0.0484 | 2.5360 | SHORT | LONG | Putative uncharacterized protein FLJ45684 OS = Homo sapiens PE = 5 SV = 2 | 0.12572 | 0.729375 |
| P78380 | 2 | 2 | 9.25 | 0.0115 | 3.3056 | SHORT | LONG | Oxidized low-density lipoprotein receptor 1 OS = Homo sapiens GN = OLR1 PE = 1 SV = 1 | 0.122362 | 0.720305 |
| P02741 | 3 | 2 | 29.30 | 0.0233 | 2.0843 | SHORT | LONG | C-reactive protein OS = Homo sapiens GN = CRP PE = 1 SV = 1 | 0.0936024 | 0.712781 |
| P05452 | 23 | 17 | 245.35 | 0.0050 | 1.5138 | LONG | SHORT | Tetranectin OS = Homo sapiens GN = CLEC3B PE = 1 SV = 3 | -0.100494 | -0.734962 |
| O75348 | 3 | 2 | 21.63 | 0.0486 | 1.4632 | LONG | SHORT | V-type proton ATPase subunit G 1 OS = Homo sapiens GN = ATP6V1G1 PE = 1 SV = 3 | -0.0176084 | -0.737745 |
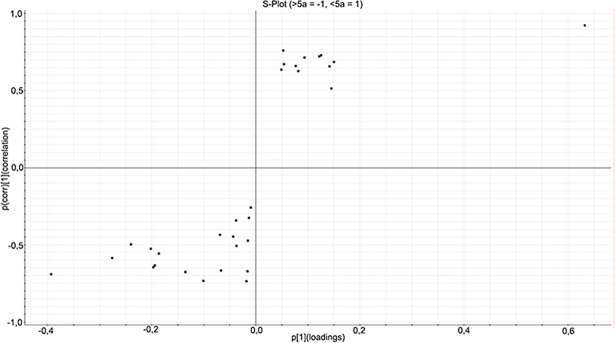
The proteins passing the cut-off from the samples divided into long and short 5-year survival are shown in Fig 6, visualized on an S-plot.
Fig 6. S-plot of the CRC samples with long or short 5-year survival obtained from the OPLS-DA analysis.
Only proteins with an ANOVA p-value of less than 0.05 are shown. Variables having a p(corr) value higher than 0.7 or lower than -0.7 are considered significantly different. Data are log10-transformed and mean-centered. The proteins in the upper right section of the plot are downregulated and the proteins in the lower left section upregulated in the short 5-year survival category as compared to the long 5-year survival category.
Pathway analysis
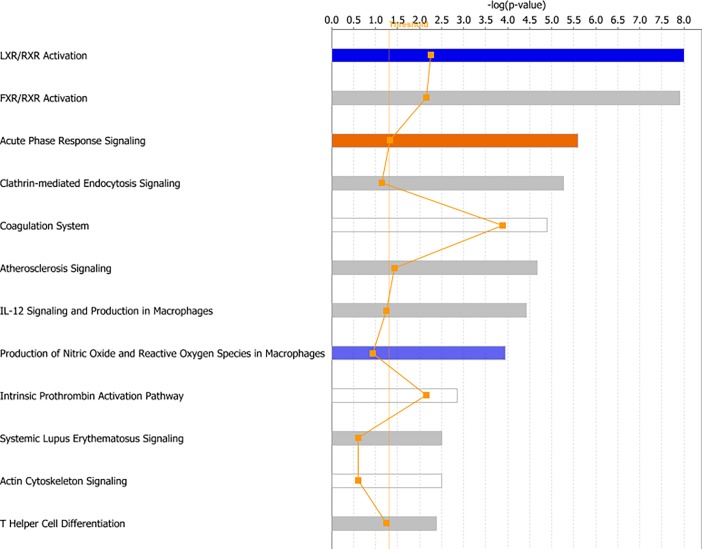
IMPaLa was again used for pathway over-representation analysis and the results when data were analyzed according to long and short 5-year survival are given in S7 Table. Signaling cascades involving platelet degranulation, activation, signaling and aggregation were found to be enriched. Complement and coagulation cascades and the statin pathway were also enriched. We also performed pathway analysis using IPA on this dataset and found multiple canonical pathways, molecular and cellular functions, as well as networks that were enriched in the short 5-year survival dataset. The canonical pathways enriched were similar to those found in the CRP >30 dataset, such as LXR/RXR activation, FXR/RXR activation and acute phase response signaling. Some of the top canonical pathways are shown in Fig 7.
Fig 7. Canonical pathways found by core analysis in IPA of the 5-year survival categories.
The top canonical pathways enriched by Ingenuity Pathway Analysis (IPA) are shown above. The straight orange vertical line running through the bars is the threshold for the p-value for the particular pathway’s enrichment. The horizontal axis is the–log(p-value) and the vertical axis shows the given pathways.
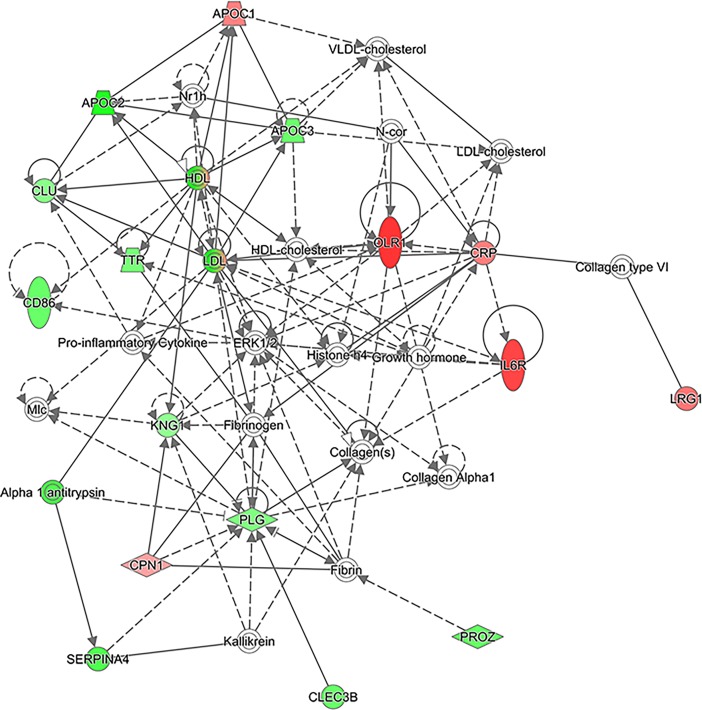
IPA also revealed protein networks enriched in the short 5-year survival category when compared to the long 5-year survival category, and the top network is presented in Fig 8. The top four networks found by IPA analysis and the full list of proteins involved in these networks are given in S8 Table. As can be seen, high-density lipoprotein (HDL) and low-density lipoprotein (LDL) are downregulated here, while OLR1 and CRP are upregulated. Several apolipoproteins were also involved in this network: APOC1 is upregulated while APOC2 and APOC3 are downregulated in the short 5-year survival category as compared to the long 5-year survival category.
Fig 8. The top network of proteins found to be up- or downregulated by IPA when analyzed according to the survival categories.
Only proteins passing the cut-off of 0.05 for ANOVA were used. Upregulated proteins are shown in red and downregulated proteins in green. White proteins are proteins involved in these pathways but not detected in our dataset.
Discussion
In recent years, the incidence of CRC has only significantly decreased in the United States, while both incidence and death rates are rapidly increasing in other, less-developed countries [1, 3]. CEA is the most widely used biomarker in CRC but is of little help in detecting early CRC, which limits its usefulness. Due to a lack of effective, non-invasive biomarkers, new biomarkers are needed to efficiently diagnose and screen for CRC [11].
In this study, we retrospectively recruited 19 CRC patients and studied preoperative serum samples by UPLC-UDMSE. We quantified 256 proteins with 2 or more unique peptides, which were used for analysis. An elevated concentration of CRP is linked to poor prognosis in CRC patients [14], and we therefore decided to analyze data according to CRP values to find differentially expressed proteins. We also analyzed the same samples according to long and short 5-year survival in order to discover proteins that could potentially be of use in predicting outcome.
By using proteomic methods, serum proteins can be studied to discover new potential biomarkers. The discovery of a biomarker that could be measured from blood samples is ideal due to the minimal invasiveness and ease of obtaining such samples. Abundant proteins such as albumin represent more than 99% of the proteins found in serum. This complicates the discovery of novel protein biomarkers, and the abundant proteins were removed to reduce the complexity of serum samples. This enables the discovery of low-abundance proteins, some of which may be of clinical importance [21, 22].
PCA analysis of the samples divided into categories according to CRP (CRP <30 and >30) and 5-year survival (short and long), showed a separation on the PCA biplot. We continued with OPLS-DA modeling and generation of an S-plot. Proteins passing the cut-off value of +0.7 or -0.7 for p(corr) values were considered to be significantly different (Table 2 and Table 4). Among the proteins upregulated in the CRP >30 category are IGFBP2 and SAA1 (also abbreviated SAA).
IGFBP2 is one of the six binding proteins that modulate the interactions of Insulin-like growth factors (IGFs) with their receptor. IGFs have important roles in growth and cell proliferation [23]. IGFBP2 has been identified as a potential biomarker for CRC in previous studies and elevated levels of IGFBP2 have been shown to be associated with poor survival in CRC patients [24, 25]. Elevated serum levels of IGFBP2 have also been discovered in patients with breast [26], ovarian [27] and prostate cancer [28]. Our findings further support the role of IGFBP2 as a biomarker for CRC, although further validation is required.
SAA is a lipoprotein whose concentration increases during the acute phase of the inflammatory response (APR) [29]. SAA has been proposed as a biomarker for CRC, albeit in a small study [30]. Elevated serum levels of SAA have also been identified in patients with gastric [31], lung [32], nasopharyngeal [33], renal cell [34] and endometrial cancer [35], where they correlated with poor prognosis. In our study, we also found that PCDHA11 and OPRM1 were upregulated in the CRP >30 category when compared to the CRP <30 category. These proteins are, to the best of our knowledge, identified here as potential biomarkers for the first time.
Pathway analysis by IPA and IMPaLa mostly found pathways involved in lipid metabolism, as well as complement and coagulation cascades and acute phase response signaling to be enriched, implying that these are the main perturbed pathways in CRC patients with high CRP. IPA pathway analysis showed LXR/RXR activation and FXR/RXR activation as the most enriched pathways (Fig 3). LXR/RXR activation is suggested to have a role in the absorption of cholesterol, and LXRs also regulate the biliary excretion of cholesterol [36, 37]. Farnesoid X receptors (FXRs) also form heterodimers with RXRs, and FXR/RXR activation plays a role in the regulation of both cholesterol and bile acid metabolism [38, 39]. Deregulation of lipid metabolism has increasingly been recognized as a feature of cancer cells [40, 41]. SAA has a role in lipid metabolism, linking the results of the IPA pathway analysis to one of the proteins found by OPLS-DA and S-plot analysis.
Enhanced levels of coagulation markers have been shown to occur in CRC patients and advanced cancer is also associated with a hypercoagulable state, which is in line with our findings of enriched coagulation cascades in pathway analysis [42, 43]. The enrichment of complement cascades and acute phase response signaling in the CRP >30 category is logical, due to the presence of an inflammatory response in these patients.
The same analyses were performed for the short and long 5-year survival categories. Among the proteins upregulated in the short 5-year survival category are APOC1, OLR1 and CRP. APOC1 is a lipoprotein that, among other functions, helps to maintain HDL structure [44]. High preoperative serum levels of APOC1 have been shown to correlate with poor prognosis in pancreatic cancer patients [45]. Serum APOC1 needs to be further investigated to evaluate its use as a biomarker, especially in CRC, for which there are no studies. OLR1, a receptor for low-density lipoproteins, has been proposed to function as an oncogene [46], although the role of OLR1 in CRC is unknown. CRP was also upregulated, which was to be expected, as we deliberately included patients with elevated CRP levels in this study.
Pathway analysis by IPA found similar pathways to be enriched in the category with short 5-year survival as in the category with high CRP. Pathway analysis by IMPaLa showed pathways involving platelets as well as complement and coagulation cascades to be enriched. Platelets have long been implicated in the spread of cancer, and cancer patients often have a high platelet count and turnover. An elevated platelet count has also been shown to be indicative of poor prognosis in CRC patients [47, 48].
In this study, we analyzed samples according to CRP (<30 vs. >30) and 5-year survival (short vs. long) to identify differences in serum proteins within these categories and to find proteins that are linked to patient outcome and prognosis. These proteins could be used to select patients with a poor prognosis that would benefit from a more aggressive treatment regimen and help those with a good prognosis to avoid it. Here, we have identified multiple potential biomarkers, although they require further validation.
This study was limited due to its small size, as only 19 patients were included in this study. CRC is a heterogeneous disease that can be divided into several subtypes characterized by distinct molecular pathologies and clinical features. For example, microsatellite instability (MSI) is associated with CRC prognosis, with MSI-high CRC showing better survival than microsatellite stable (MSS) CRC [49, 50]. Lack of tumor molecular data for our dataset therefore leads to some additional limitations of this study. Due to the small size of this study, it was not feasible to subdivide our material into subcategories: for example, only around 15% of CRCs are MSI-high [49], giving us very few patients with MSI-high CRC that could be used for comparisons.
Another limitation of the study could be that multiple hypothesis testing correction was not employed. We realize that it might lead to a few results being falsely significant, but it has to be considered that multiple testing correction is often over-stringent, and significant inferences are usually missed in trying to control a Type I error. This is called committing a Type II error and it’s always the trade-off between Type I and II errors that a researcher has to decide between. Moreover, to establish a useful prognostic biomarker, one usually starts with a number of lead candidates, and at the validation stage, in a very large cohort, multiple hypothesis correction can be employed. Also, popular methods of multiple testing corrections, such as Bonferroni and Sidak methods, are under-powered when variable measurements are correlated [51], which is often the case with proteomic measurements. Some of the proteins we identified here have been recognized as potential biomarkers previously, whereas some have been identified as potential biomarkers for the first time, to the best of our knowledge. This pilot study will therefore pave the way for further studies, with the aim to provide better biomarkers for CRC patients.
Supporting information
The table shows patients’ gender, age, stage and location of cancer, preoperative CRP value, survival, and cause of death or alive.
(XLSX)
Accession, peptide count, unique peptides, confidence score, ANOVA p-value, maximum fold change and highest and lowest mean condition as well as the full protein name are given in the table.
(XLSX)
Accession, peptide count, unique peptides, confidence score, ANOVA p-value, maximum fold change and highest and lowest mean condition as well as the full protein name are given in the table.
(XLSX)
Accession, peptide count, unique peptides, confidence score, ANOVA p-value, maximum fold change and highest and lowest mean condition as well as the full protein name are given in the table.
(XLSX)
Pathway name, source, number of overlapping genes, number of all pathway genes and P and Q values of enrichment are listed.
(XLSX)
Only proteins passing the cut-off of 0.05 for ANOVA were used.
(XLS)
Pathway name, source, number of overlapping genes, number of all pathway genes and P and Q values of enrichment are listed.
(XLSX)
Only proteins passing the cut-off of 0.05 for ANOVA were used.
(XLS)
Data Availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD008583 (http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD008583).
Funding Statement
CH and AR received funding in the form of grants from the Sigrid Jusélius Foundation and the Finnish Cancer Foundation. CH and RR also received funding from Helsinki University Hospital and RR also received funding from the University of Helsinki. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Stewart BW, Wild CP, editors (2014). World Cancer Report 2014. Lyon, France: International Agency for Research on Cancer. [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67(3):177–93. doi: 10.3322/caac.21395 [DOI] [PubMed] [Google Scholar]
- 3.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 4.McArdle CS, Hole DJ. Outcome following surgery for colorectal cancer: analysis by hospital after adjustment for case-mix and deprivation. Br J Cancer. 2002. February 1;86(3):331–5. doi: 10.1038/sj.bjc.6600120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edwards BK, Ward E, Kohler BA, Eheman C, Zauber AG, Anderson RN, et al. Annual report to the nation on the status of cancer, 1975–2006, featuring colorectal cancer trends and impact of interventions (risk factors, screening, and treatment) to reduce future rates. Cancer. 2010;116(3):544–73. doi: 10.1002/cncr.24760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giantonio BJ, Catalano PJ, Meropol NJ, O'Dwyer PJ, Mitchell EP, Alberts SR, et al. Bevacizumab in combination with oxaliplatin, fluorouracil, and leucovorin (FOLFOX4) for previously treated metastatic colorectal cancer: results from the Eastern Cooperative Oncology Group Study E3200. J Clin Oncol. 2007;25(12):1539–44. doi: 10.1200/JCO.2006.09.6305 [DOI] [PubMed] [Google Scholar]
- 7.Douillard JY, Siena S, Cassidy J, Tabernero J, Burkes R, Barugel M, et al. Randomized, phase III trial of panitumumab with infusional fluorouracil, leucovorin, and oxaliplatin (FOLFOX4) versus FOLFOX4 alone as first-line treatment in patients with previously untreated metastatic colorectal cancer: the PRIME study. J Clin Oncol. 2010;28(31):4697–705. doi: 10.1200/JCO.2009.27.4860 [DOI] [PubMed] [Google Scholar]
- 8.Srinivas PR, Kramer BS, Srivastava S. Trends in biomarker research for cancer detection. The Lancet Oncology. 2001;2(11):698–704. doi: 10.1016/S1470-2045(01)00560-5 [DOI] [PubMed] [Google Scholar]
- 9.Ludwig JA, Weinstein JN. Biomarkers in cancer staging, prognosis and treatment selection. Nat Rev Cancer. 2005;5(11):845–56. doi: 10.1038/nrc1739 [DOI] [PubMed] [Google Scholar]
- 10.Hanash SM, Pitteri SJ, Faca VM. Mining the plasma proteome for cancer biomarkers. Nature. 2008;452(7187):571–9. doi: 10.1038/nature06916 [DOI] [PubMed] [Google Scholar]
- 11.Duffy MJ. Carcinoembryonic antigen as a marker for colorectal cancer: is it clinically useful? Clin Chem. 2001. April;47(4):624–30. [PubMed] [Google Scholar]
- 12.Ward DG, Suggett N, Cheng Y, Wei W, Johnson H, Billingham LJ, et al. Identification of serum biomarkers for colon cancer by proteomic analysis. Br J Cancer. 2006;94(12):1898–905. doi: 10.1038/sj.bjc.6603188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Black S, Kushner I, Samols D. C-reactive Protein. J Biol Chem. 2004;279(47):48487–90. doi: 10.1074/jbc.R400025200 [DOI] [PubMed] [Google Scholar]
- 14.Kersten C, Louhimo J, Algars A, Lahdesmaki A, Cvancerova M, Stenstedt K, et al. Increased C-reactive protein implies a poorer stage-specific prognosis in colon cancer. Acta Oncol. 2013;52(8):1691–8. doi: 10.3109/0284186X.2013.835494 [DOI] [PubMed] [Google Scholar]
- 15.Saraswat M, Joenvaara S, Seppanen H, Mustonen H, Haglund C, Renkonen R. Comparative proteomic profiling of the serum differentiates pancreatic cancer from chronic pancreatitis. Cancer Med. 2017;6(7):1738–51. doi: 10.1002/cam4.1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saraswat M, Joenvaara S, Jain T, Tomar AK, Sinha A, Singh S, et al. Human Spermatozoa Quantitative Proteomic Signature Classifies Normo- and Asthenozoospermia. Mol Cell Proteomics. 2017;16(1):57–72. doi: 10.1074/mcp.M116.061028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silva JC, Gorenstein MV, Li GZ, Vissers JP, Geromanos SJ. Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol Cell Proteomics. 2006. January;5(1):144–56. Epub 2005 Oct 11. doi: 10.1074/mcp.M500230-MCP200 [DOI] [PubMed] [Google Scholar]
- 18.Serang O, Moruz L, Hoopmann MR, Kall L. Recognizing uncertainty increases robustness and reproducibility of mass spectrometry-based protein inferences. J Proteome Res. 2012;11(12):5586–91. doi: 10.1021/pr300426s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Luca A, Henry M, Meleady P, O'Connor R. Label-free LC-MS analysis of HER2+ breast cancer cell line response to HER2 inhibitor treatment. Daru. 2015;23:40 doi: 10.1186/s40199-015-0120-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deutsch EW, Csordas A, Sun Z, Jarnuczak A, Perez-Riverol Y, Ternent T, et al. The ProteomeXchange Consortium in 2017: supporting the cultural change in proteomics public data deposition. Nucleic Acids Res 54(D1):D1100–D1106 doi: 10.1093/nar/gkw936 (PubMed PMID: 27924013) 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim HJ, Yu MH, Kim H, Byun J, Lee C. Noninvasive molecular biomarkers for the detection of colorectal cancer. BMB Rep. 2008. October 31;41(10):685–92. [DOI] [PubMed] [Google Scholar]
- 22.Dayon L, Kussmann M. Proteomics of human plasma: A critical comparison of analytical workflows in terms of effort, throughput and outcome. EuPA Open Proteomics. 2013;1:8–16. [Google Scholar]
- 23.Sandhu MS, Dunger DB, Giovannucci EL. Insulin, insulin-like growth factor-I (IGF-I), IGF binding proteins, their biologic interactions, and colorectal cancer. J Natl Cancer Inst. 2002. July 3;94(13):972–80. [DOI] [PubMed] [Google Scholar]
- 24.Renehan AG, Jones J, Potten CS, Shalet SM, O'Dwyer ST. Elevated serum insulin-like growth factor (IGF)-II and IGF binding protein-2 in patients with colorectal cancer. Br J Cancer. 2000. November;83(10):1344–50. doi: 10.1054/bjoc.2000.1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liou JM, Shun CT, Liang JT, Chiu HM, Chen MJ, Chen CC, et al. Plasma insulin-like growth factor-binding protein-2 levels as diagnostic and prognostic biomarker of colorectal cancer. J Clin Endocrinol Metab. 2010;95(4):1717–25. doi: 10.1210/jc.2009-2668 [DOI] [PubMed] [Google Scholar]
- 26.Busund LT, Richardsen E, Busund R, Ukkonen T, Bjornsen T, Busch C, et al. Significant expression of IGFBP2 in breast cancer compared with benign lesions. J Clin Pathol. 2005;58(4):361–6. doi: 10.1136/jcp.2004.020834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flyvbjerg A, Mogensen O, Mogensen B, Nielsen OS. Elevated serum insulin-like growth factor-binding protein 2 (IGFBP-2) and decreased IGFBP-3 in epithelial ovarian cancer: correlation with cancer antigen 125 and tumor-associated trypsin inhibitor. J Clin Endocrinol Metab. 1997. July;82(7):2308–13. doi: 10.1210/jcem.82.7.4085 [DOI] [PubMed] [Google Scholar]
- 28.Kanety H, Madjar Y, Dagan Y, Levi J, Papa MZ, Pariente C, et al. Serum insulin-like growth factor-binding protein-2 (IGFBP-2) is increased and IGFBP-3 is decreased in patients with prostate cancer: correlation with serum prostate-specific antigen. J Clin Endocrinol Metab. 1993. July;77(1):229–33. doi: 10.1210/jcem.77.1.7686915 [DOI] [PubMed] [Google Scholar]
- 29.Steel DM, Whitehead AS. The major acute phase reactants: C-reactive protein, serum amyloid P component and serum amyloid A protein. Immunol Today. 1994. February;15(2):81–8. doi: 10.1016/0167-5699(94)90138-4 [DOI] [PubMed] [Google Scholar]
- 30.Glojnarić I1, Casl MT, Simić D, Lukac J. Serum amyloid A protein (SAA) in colorectal carcinoma. Clin Chem Lab Med. 2001. February;39(2):129–33. doi: 10.1515/CCLM.2001.022 [DOI] [PubMed] [Google Scholar]
- 31.Chan DC, Chen CJ, Chu HC, Chang WK, Yu JC, Chen YJ, et al. Evaluation of serum amyloid A as a biomarker for gastric cancer. Ann Surg Oncol. 2007;14(1):84–93. doi: 10.1245/s10434-006-9091-z [DOI] [PubMed] [Google Scholar]
- 32.Cho WC, Yip TT, Cheng WW, Au JS. Serum amyloid A is elevated in the serum of lung cancer patients with poor prognosis. Br J Cancer. 2010;102(12):1731–5. doi: 10.1038/sj.bjc.6605700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cho WC, Yip TT, Yip C, Yip V, Thulasiraman V, Ngan RK, et al. Identification of serum amyloid a protein as a potentially useful biomarker to monitor relapse of nasopharyngeal cancer by serum proteomic profiling. Clin Cancer Res. 2004. January 1;10(1 Pt 1):43–52. [DOI] [PubMed] [Google Scholar]
- 34.Kimura M, Tomita Y, Imai T, Saito T, Katagiri A, Ohara-Mikami Y, et al. Significance of serum amyloid A on the prognosis in patients with renal cell carcinoma. Cancer. 2001. October 15;92(8):2072–5. [DOI] [PubMed] [Google Scholar]
- 35.Cocco E, Bellone S, El-Sahwi K, Cargnelutti M, Buza N, Tavassoli FA, et al. Serum amyloid A: a novel biomarker for endometrial cancer. Cancer. 2010;116(4):843–51. doi: 10.1002/cncr.24838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murthy S, Born E, Mathur SN, Field FJ. LXR/RXR activation enhances basolateral efflux of cholesterol in CaCo-2 cells. Journal of Lipid Research. 2002;43(7):1054–64. [DOI] [PubMed] [Google Scholar]
- 37.Chuu CP. Modulation of liver X receptor signaling as a prevention and therapy for colon cancer. Med Hypotheses. 2011;76(5):697–9. doi: 10.1016/j.mehy.2011.01.037 [DOI] [PubMed] [Google Scholar]
- 38.Kalaany NY, Mangelsdorf DJ. LXRS and FXR: the yin and yang of cholesterol and fat metabolism. Annu Rev Physiol. 2006;68:159–91. doi: 10.1146/annurev.physiol.68.033104.152158 [DOI] [PubMed] [Google Scholar]
- 39.Kassam A, Miao B, Young PR, Mukherjee R. Retinoid X receptor (RXR) agonist-induced antagonism of farnesoid X receptor (FXR) activity due to absence of coactivator recruitment and decreased DNA binding. J Biol Chem. 2003;278(12):10028–32. doi: 10.1074/jbc.M208312200 [DOI] [PubMed] [Google Scholar]
- 40.Santos CR, Schulze A. Lipid metabolism in cancer. FEBS J. 2012;279(15):2610–23. doi: 10.1111/j.1742-4658.2012.08644.x [DOI] [PubMed] [Google Scholar]
- 41.Zhang F, Du G. Dysregulated lipid metabolism in cancer. World J Biol Chem. 2012;3(8):167–74. doi: 10.4331/wjbc.v3.i8.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Duijnhoven EM, Lustermans FA, van Wersch JW. Evaluation of the coagulation/fibrinolysis balance in patients with colorectal cancer. Haemostasis. 1993. May-Jun;23(3):168–72. doi: 10.1159/000216870 [DOI] [PubMed] [Google Scholar]
- 43.Belting M, Ahamed J, Ruf W. Signaling of the tissue factor coagulation pathway in angiogenesis and cancer. Arterioscler Thromb Vasc Biol. 2005;25(8):1545–50. doi: 10.1161/01.ATV.0000171155.05809.bf [DOI] [PubMed] [Google Scholar]
- 44.Wroblewski MS, Wilson-Grady JT, Martinez MB, Kasthuri RS, McMillan KR, Flood-Urdangarin C, et al. A functional polymorphism of apolipoprotein C1 detected by mass spectrometry. FEBS J. 2006;273(20):4707–15. doi: 10.1111/j.1742-4658.2006.05473.x [DOI] [PubMed] [Google Scholar]
- 45.Takano S, Yoshitomi H, Togawa A, Sogawa K, Shida T, Kimura F, et al. Apolipoprotein C-1 maintains cell survival by preventing from apoptosis in pancreatic cancer cells. Oncogene. 2008. May 1;27(20):2810–22. Epub 2007 Nov 26. doi: 10.1038/sj.onc.1210951 [DOI] [PubMed] [Google Scholar]
- 46.Khaidakov M, Mitra S, Kang BY, Wang X, Kadlubar S, Novelli G, et al. Oxidized LDL receptor 1 (OLR1) as a possible link between obesity, dyslipidemia and cancer. PLoS One. 2011;6(5):e20277 doi: 10.1371/journal.pone.0020277 Epub 2011 May 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nash GF, Turner LF, Scully MF, Kakkar AK. Platelets and cancer. The Lancet Oncology. 2002;3(7):425–30. [DOI] [PubMed] [Google Scholar]
- 48.Lin MS, Huang JX, Zhu J, Shen HZ. Elevation of platelet count in patients with colorectal cancer predicts tendency to metastases and poor prognosis. Hepatogastroenterology. 2012. September;59(118):1687–90. doi: 10.5754/hge12277 [DOI] [PubMed] [Google Scholar]
- 49.Kocarnik JM, Shiovitz S, Phipps AI. Molecular phenotypes of colorectal cancer and potential clinical applications. Gastroenterol Rep (Oxf). 2015;3(4):269–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Inamura K. Colorectal Cancers: An Update on Their Molecular Pathology. Cancers (Basel). 2018;10(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blakesley RE, Mazumdar S, Dew MA, Houck PR, Tang G, Reynolds CF 3rd, et al. Comparisons of methods for multiple hypothesis testing in neuropsychological research. Neuropsychology. 2009;23(2):255–64. doi: 10.1037/a0012850 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The table shows patients’ gender, age, stage and location of cancer, preoperative CRP value, survival, and cause of death or alive.
(XLSX)
Accession, peptide count, unique peptides, confidence score, ANOVA p-value, maximum fold change and highest and lowest mean condition as well as the full protein name are given in the table.
(XLSX)
Accession, peptide count, unique peptides, confidence score, ANOVA p-value, maximum fold change and highest and lowest mean condition as well as the full protein name are given in the table.
(XLSX)
Accession, peptide count, unique peptides, confidence score, ANOVA p-value, maximum fold change and highest and lowest mean condition as well as the full protein name are given in the table.
(XLSX)
Pathway name, source, number of overlapping genes, number of all pathway genes and P and Q values of enrichment are listed.
(XLSX)
Only proteins passing the cut-off of 0.05 for ANOVA were used.
(XLS)
Pathway name, source, number of overlapping genes, number of all pathway genes and P and Q values of enrichment are listed.
(XLSX)
Only proteins passing the cut-off of 0.05 for ANOVA were used.
(XLS)
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD008583 (http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD008583).