Abstract
The cell cycle‐related and expression‐elevated protein in tumor (CREPT) is overexpressed in several human malignancies. However, the clinical relevance of CREPT expression and its biological role in non‐small‐cell lung cancer (NSCLC) remains unclear. In this study, we detected the expression of CREPT in both NSCLC tissues and cell lines by immunohistochemistry, Western blot analysis, and RT‐PCR. The correlation between CREPT expression and clinicopathologic features was analyzed in 271 NSCLC patients. The prognostic value of CREPT expression was evaluated by Kaplan–Meier analysis and Cox regression analysis. CREPT was overexpressed in Calu‐1 cell lines by using plasmid vector and its biological function was explored both in vitro and in vivo. We found that CREPT was significantly overexpressed in NSCLC compared with paired adjacent non‐tumor tissues, and the expression level of CREPT was correlated with tumor differentiation, lymph node metastasis, and clinical stage. Kaplan–Meier analysis showed that the recurrence‐free survival and overall survival of high CREPT expression groups were significantly shorter than those of the low CREPT expression group. Multivariate analysis identified that CREPT might be an independent biomarker for the prediction of NSCLC prognosis. Overexpression of CREPT increased cell proliferation and enhanced the migration and invasion ability of Calu‐1 cells (a human NSCLC cell line with relative low CRPET expression) in vitro. Moreover, CREPT overexpression promoted tumor growth in a nude mice model. These results suggest that CREPT is closely relevant to the proliferation of NSCLC cells and it might be a potential prognostic marker in NSCLC patients.
Keywords: biomarker, cell proliferation, CREPT, non‐small‐cell lung cancer, prognosis
1. INTRODUCTION
Lung cancer is one of the leading causes of cancer related death in the world.1 The two major forms of lung cancer are non‐small‐cell lung cancer (NSCLC, approximately 85% of cases) and small‐cell lung cancer (SCLC, approximately 15% of cases).2 Even though several diagnostic techniques and treatments have been developed,3, 4 approximately 40% of NSCLC patients are first diagnosed at an advanced stage.5 The 5‐year overall survival (OS) rate of these patients was less than 18%.6, 7 Therefore, it is imperative to identify the relevant genes associated with the prognosis of NSCLC and provide individualization treatment.
Cell cycle‐related and expression‐elevated protein in tumor (CREPT) (GenBank NM_021215), a novel gene also called RPRD1B or C20ORF77, is involved in the development and prognosis of various tumors, including liver cancer, breast cancer, retroperitoneal leiomyosarcoma, and endometrial cancer.8, 9, 10, 11, 12 Lu et al10 have shown that CREPT enhances the expression of CyclinD1 by promoting the formation of a chromatin loop, by interacting with RNA polymerase II. Previous studies have shown that CREPT expression is correlated with tumor differentiation, Dukes’ stage, and metastasis in colorectal tumors.12 She et al13 found that CREPT is elevated in retroperitoneal leiomyosarcoma tissues and plays important roles in the progression of retroperitoneal leiomyosarcoma. In our previous study, CREPT silencing significantly inhibited the proliferation and migration of NSCLC cell lines.8 However, the relationship between overexpression of CREPT and prognosis in NSCLC remains unknown. In this study, the expression of CREPT in 271 NSCLC tissues and corresponding adjacent non‐tumor tissues was detected by immunohistochemical staining, and the correlation between CREPT expression and clinicopathologic features were analyzed. Furthermore, CREPT was overexpressed in Calu‐1 cells and its biological function was investigated both in vitro and in vivo.
2. MATERIALS AND METHODS
2.1. Patients and tissue samples
We analyzed 271 NSCLC patients who underwent complete tumor resection with mediastinal lymph node dissection in the Department of Thoracic Surgery, Tangdu Hospital (Xi'an, China) from 2006 to 2010.1, 14 Surgically excised NSCLC tissue samples with matched adjacent non‐cancer lung tissues were embedded in paraffin. Thirty‐five freshly collected paired NSCLC tissues of these samples were stored in liquid nitrogen for further study. Adjacent non‐cancer tissue samples were collected from the same patients and histologically identified to be collagen tissue and bronchial epithelial cells. Patients who received preoperative chemotherapy and radiotherapy were excluded from this study. Clinical information was obtained from the medical records of the enrolled patients. The follow‐up was obtained by telephone interviews to 2014, with a median follow‐up period of 56 months for living patients. The evaluation of histologic classification and differentiation were carried out independently by two pathologists. All tumors were staged according to the pathological TNM classification of the UICC (7th edition). The study protocol was approved by the Regional Ethics Committee for Clinical Research of the Fourth Military Medical University (Xi'an, China). Each patient provided written informed consent for use of their medical records and tissue specimens.
2.2. Cell culture
Human NSCLC cell lines (H520, A549, H838, Spc‐A‐1, and Calu‐1) were purchased from ATCC (Manassas, VA, USA). The cells were cultured in RPMI‐1640 (Gibco, USA) containing 10% FBS (Gibco) and 1% penicillin–streptomycin. Cells were cultured at 37°C in a humidified atmosphere of 5% CO2.
2.3. Immunohistochemistry
The paraffin‐embedded tissues were sliced into 3‐μm sections and deparaffinized, then the slides were boiled in 10 mmol/L citrate buffer for antigen retrieval and blocked with 10% goat serum. The slides were then incubated with primary anti‐CREPT (1:200; GeneTex) or anti‐Ki‐67 (1:200; GeneTex) antibodies overnight. The same concentration of antigen‐specific antibody (Kangwei) was used as negative control. After washing with PBS, the tissue sections were incubated with EnVision HRP (Kangwei, China) as the secondary antibody. Finally, the DAB Elite kit (Zhongshan, China) was used for chemiluminescence analysis. All stained sections were examined by two independent investigators who were blinded to the clinical features and outcomes. The immunohistochemical (IHC) staining scores were based on the following criteria: (i) the percentage of positive cells (0, ≤5%; 1, 6%‐25%; 2, 26%‐50%; 3, 51%‐75%; and 4, >75%); (ii) the staining intensity (0, no color; 1, yellow; 2, brown; and 3, tan); and (iii) the two grades were multiplied together and specimens were assigned to one of four levels: 0, negative (−); 1‐4, weakly positive (+); 5‐8, moderately positive (++); and 9‐12, strongly positive (+++).8, 15 To investigate the correlation of protein expression, (−) and (+) were considered as low expression, and (++) and (+++) were considered as high expression.
2.4. Western blot analysis
Cells and tissue samples were harvested in lysis buffer containing protease inhibitor cocktail. Cell lysates were separated by SDS‐PAGE and transferred onto PVDF membrane. Then the membrane was blocked by 8% non‐fat milk before being incubated with specific antibodies overnight at 4°C (anti‐CREPT, 1:2000; anti‐Ki‐67, 1:3000; β‐actin, 1:2000). After washing with TBST, the membrane was incubated with a HRP‐conjugated secondary antibody (1:3000) at room temperature for 45 minutes. The visualized analysis was carried out using the Millipore chromogenic kit for Western blot analysis (Millipore, Billerica, MA, USA) and quantitatively analyzed using Quantity software (Bio‐Rad, USA).
2.5. Quantitative RT‐PCR
Total RNA from fresh tissues or cells was extracted using TRIzol reagent (Invitrogen, USA) and mRNA was reverse transcribed into cDNA synthesis with a reverse transcription kit (Thermo Fisher Scientific, USA) according to the manufacturer's instructions. The quantitative RT‐PCR (qRT‐PCR) was carried out using a CFX96 Real‐Time PCR system (Bio‐Rad) with SYBR Green reagents (TaKaRa, Japan). Gene expression was normalized to β‐actin. The primers used for real‐time PCR were: CREPT, 5′‐GCTGGAGAAGAAGCTCTCGG‐3′ (forward) and 5′‐ATTTGATTTGGCTTTGCGGAG‐3′ (reverse); and β‐actin, 5′‐TGGCACCCAGCACAATGAA‐3′ (forward) and 5′‐CTAAGTCATAGTCCGCCTAGAAGCA ‐3′ (reverse).
2.6. Plasmid transfection
Plasmid pcDNA3.1 was constructed by PCR amplifying the CREPT gene, using primers 5′‐CGCGATATCATGTCCTCCTTCTCTGAGTC‐3′ (forward) and 5′‐CGCCTCGAGCTAGTCAGTTGAAAACAGGT‐3′ (reverse). The PCR products were the inserted between the EcoRV and XhoI sites in pcDNA3.1 to generate the recombinant plasmid, named pcDNA3.1‐CREPT. Electroporation was used to transfect pcDNA3.1‐CREPT and the empty vector into Calu‐1 cells according to the manufacturer's instructions. To generate stable CREPT overexpressing cell lines, cells were screened by 800 mg/mL G418 after transfection and cells stably expressing CREPT were maintained in medium with 400 mg/mL G418.
2.7. Cell proliferation assay
Cell proliferation was analyzed using CCK‐8 (Beyotime Institute of Biotechnology, Shanghai, China). Cells were cultured in 96‐well plates at a density of 2000 cells/well. Then CCK‐8 solution (10 μL) was added to each well and incubated at 37°C for 2 hours. The optical density of each well was measured at 450 nm and documented every 24 hours. The results were obtained from three independent experiments in triplicate. Growth curves were portrayed based on the optical density value.
2.8. Flow cytometry analysis
Cell cycle progression was quantified using flow cytometry after propidium iodide staining. Cells were collected 48 hours after transfection, then fixed overnight with 70% ice‐cold ethanol and stained with 50 μg/mL propidium iodide (MP Biomedical, USA), and examined using a flow cytometer (FACScan; BD Biosciences, USA) at 488 nm. The results were analyzed by FlowJo 6.1 software.
2.9. Wound healing assay
Cell migration was assessed by a wound healing assay. Briefly, cells (5 × 105 cells/well) stably transfected with CREPT or empty vector were selected using G418 and then cultured in 6‐well plates until confluent. The cell monolayer was scraped in a straight line and washed with PBS, and then images were captured at 0, 12, and 24 hours under a 10× objective (Olympus, Japan). The migration distance was measured and assessed using ImageJ software at six random optical fields on each filter to evaluate cell migration. The percentage change in migration was determined by comparison of the differences in wound width. The experiments were carried out in duplicate.
2.10. Transwell migration and invasion assay
The invasion and migration ability of the cells were examined using Transwell assay with a 24‐well Transwell chamber (8‐μm pore size; Corning Life Sciences, USA) according to the manufacturer's instructions. A total of 1 × 105 serum pre‐starved cells in 400 μL media supplemented were plated in the upper chamber and the bottom chamber contained 600 μL RPMI‐1640 supplemented with 10% FBS. After incubation at 37°C for 48 hours, the non‐migration cells on the top surface of the membrane were completely removed. Meanwhile, the migrated cells on the bottom surface were fixed in dehydrated alcohol for 30 minutes and stained with crystal violet. Cells were counted from five random fields at 400× magnification. Cell invasion assay was carried out in the same conditions except the upper chamber was coated with Matrigel (BD Biosciences).
2.11. In vivo mouse models
All experimental procedures were approved by the Animal Ethics Committee of the Fourth Military Medical University. Nude mice were randomly divided into Calu‐1/pcDNA3.1‐CREPT and Calu‐1/pcDNA3.1 groups. Calu‐1 cells (5 × 106 cells in 0.2 mL PBS) stably transfected with CREPT or empty vector were injected s.c. into 6‐week‐old male BALB/c nude mice. Tumor volume was calculated according to the following formula: tumor volume (mm3) = (short diameter)2 × (long diameter) / 2. Tumor volume was assessed every 5 days. The mice were killed and photographed after 30 days. The tumor tissues were removed and weighed. In addition, IHC staining was carried out to evaluate the expression of CREPT or Ki‐67 in xenograft tissues. Western blot and qRT‐PCR analyses were carried out to assess the expression of Ki‐67.
2.12. Statistical analysis
Data are expressed as values of mean ± SD. Results were evaluated using the t‐test. Associations between two groups were evaluated using the χ 2‐test and Spearman's correlation analysis. Survival curves were examined using the Kaplan–Meier method, and the significance of the difference was evaluated using the log–rank test. Univariate and multivariate survival analyses were undertaken using the Cox hazard regression model. All analyses were carried out with spss 19.0 software (SPSS, Chicago, IL, USA). P < .05 was considered to be statistically significant.
3. RESULTS
3.1. Overexpression of CREPT in NSCLC tissues and cell lines
To compare the expression of CREPT between NSCLC tumor tissues and adjacent non‐tumor tissues, we undertook IHC staining using 271 NSCLC samples and adjacent non‐tumor tissues. The staining intensity was scored on a scale of negative (−), weak (+), moderate (++), and strong (+++) (Figure 1A). The CREPT staining was mainly located in the nucleus and occasionally in cytoplasm. The CREPT was expressed in 262 of 271 (96.7%) NSCLC samples and 114 of 271 (42.1%) corresponding adjacent normal bronchial epithelia (Figure 1B). To confirm the CREPT mRNA and protein levels in NSCLC, 35 pairs of fresh NSCLC tissues and adjacent non‐cancerous lung tissues obtained from patients were used for Western blot analysis and qRT‐PCR. The results showed that higher levels of CREPT were expressed in cancer lesions than in adjacent regions (P < .05; Figure 1D,E). Similarly, CREPT was positively expressed in NSCLC cell lines. The expression of CREPT at the mRNA and protein levels in Calu‐1 cells was the lowest among five NSCLC cell lines (Calu‐1, H520, A549, H838, and SPC‐A‐1) (Figure 1C). The Calu‐1 cell line was thus selected for CREPT overexpression experiments.
Figure 1.
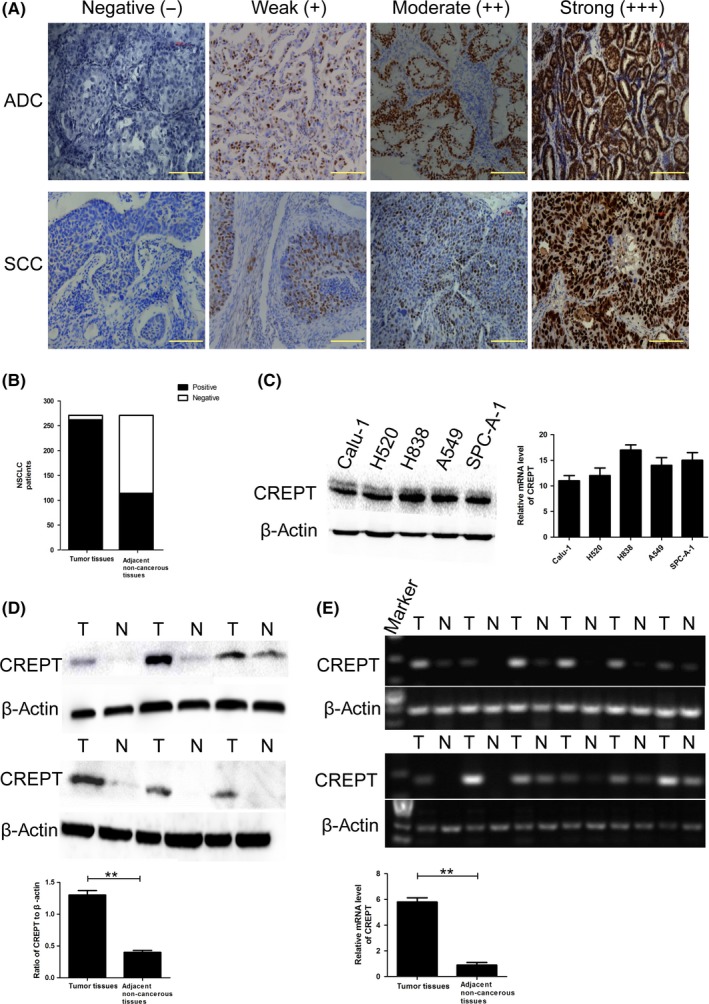
Expression of cell cycle‐related and expression‐elevated protein in tumor (CREPT) in non‐small‐cell lung cancer (NSCLC) tissues and cell lines. A,B, Representative immunohistochemical staining of CREPT in NSCLC tissues (adenocarcinoma [ADC] and squamous cell carcinoma [SCC]) and adjacent non‐tumor lung tissues. C, Protein and mRNA levels of CREPT in different NSCLC cell lines. D,E, Western blot and quantitative RT‐PCR showed that the expression of CREPT in tumor tissue (T) was higher than that in paired adjacent non‐tumor lung tissues (N). Data shown as mean ± SD. **P < .01
3.2. Correlation between CREPT expression and clinicopathologic features
In order to determine the clinical significance of CREPT in NSCLC, the relationship between CREPT expression and clinicopathologic characteristics was investigated (Table 1). The CREPT expression was significantly associated with clinical stage (P = .025), tumor differentiation (P = .000), and lymphatic metastasis (P = .020). There was a strong correlation between CREPT expression and Ki‐67 proliferation index (P = .002). However, no significant relationship was observed between CREPT expression and age (P = .380), gender (P = .627), smoking history (P = .894), tumor size (P = .253), pathology (P = .638), or tumor location (P = .121).
Table 1.
Cell cycle‐related and expression‐elevated protein in tumor (CREPT) expression and clinicpathologic features in 271 patients with non‐small‐cell lung carcinoma
| Variable | Total | CREPT expression | P‐value | |||
|---|---|---|---|---|---|---|
| − | + | ++ | +++ | |||
| Age, y | ||||||
| ≤61 | 139 | 5 | 31 | 87 | 16 | .380 |
| >61 | 132 | 4 | 23 | 89 | 16 | |
| Gender | ||||||
| Male | 204 | 8 | 41 | 131 | 24 | .627 |
| Female | 67 | 1 | 13 | 45 | 8 | |
| Smoking history | ||||||
| Yes | 173 | 7 | 34 | 111 | 21 | .894 |
| No | 98 | 2 | 20 | 65 | 11 | |
| Tumor size, cm | ||||||
| ≤5 | 140 | 3 | 34 | 85 | 18 | .253 |
| >5 | 131 | 6 | 20 | 91 | 14 | |
| Pathology | ||||||
| Adenocarcinoma | 155 | 5 | 30 | 110 | 19 | .638 |
| Squamous carcinoma | 116 | 4 | 24 | 75 | 13 | |
| Clinical stage | ||||||
| I | 69 | 2 | 20 | 43 | 4 | .025a |
| II | 101 | 5 | 25 | 55 | 16 | |
| III | 98 | 2 | 9 | 75 | 12 | |
| IV | 3 | 0 | 0 | 3 | 0 | |
| Tumor differentiation | ||||||
| Well | 15 | 2 | 11 | 2 | 0 | .000b |
| Moderate | 185 | 7 | 41 | 119 | 18 | |
| Poor | 71 | 0 | 2 | 55 | 14 | |
| Lymphatic metastasis | ||||||
| Yes | 150 | 7 | 19 | 100 | 24 | .003b |
| No | 121 | 2 | 35 | 76 | 8 | |
| Location | ||||||
| Central | 177 | 5 | 38 | 111 | 23 | .121 |
| Peripheral | 94 | 4 | 16 | 65 | 9 | |
| Ki67 | ||||||
| Negative | 12 | 2 | 5 | 5 | 0 | .002b |
| Positive | 259 | 7 | 49 | 171 | 32 | |
P ≤ .05.
P ≤ .01.
3.2.1. Overexpression of CREPT promotes cell proliferation by inducing cells to enter G1 phase from S phase
To investigate the role of CREPT on the progression of NSCLC, pcDNA3.1‐CREPT plasmid was adopted to transfect the Calu‐1 cell line and G418 screening to obtain stable cell lines highly expressing CREPT. We confirmed the efficacy of CREPT overexpression using Western blot analyses and qRT‐PCR (Figure 2A). To determine the impact of CREPT on regulating cell growth, CCK‐8 assay was undertaken and results showed that upregulation of CREPT expression in Calu‐1 cells dramatically promotes growth compared with the control group (Figure 2B). To further clarify the mechanism of the promotion effect of tumor cell growth by CREPT, we examined the cell cycle by flow cytometry. The results showed that upregulation of CREPT expression in Calu‐1 cells increased the proportion of cells in G1 phase compared with control cells (40.96 ± 3.21% vs 55.22 ± 4.12%), with concomitant decrease in S phase compared with control cells (33.42 ± 2.11% vs 21.8 ± 2.24%) (Figure 2C). These data strongly indicated that elevated CREPT levels in Calu‐1 cells markedly increased their proliferation through regulating the cell cycle.
Figure 2.
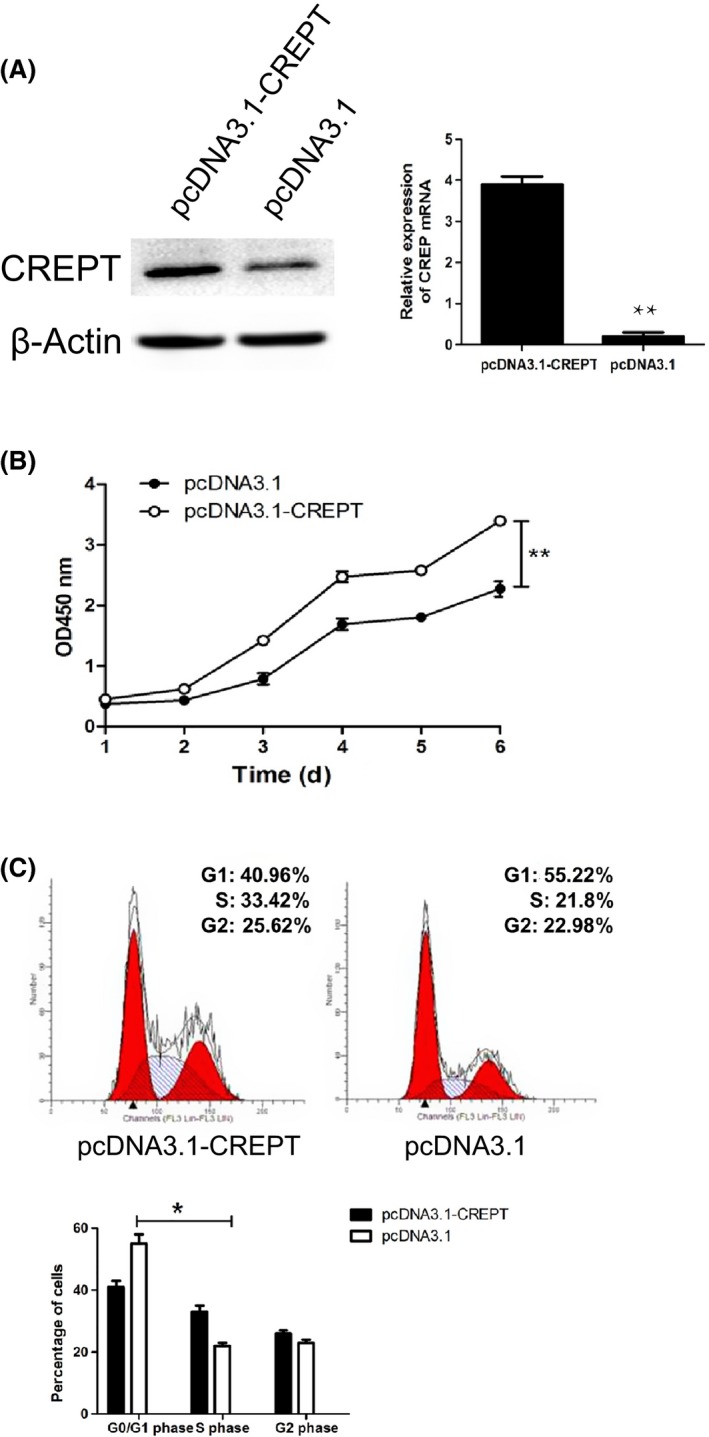
Cell cycle‐related and expression‐elevated protein in tumor (CREPT) overexpression promoted cell proliferation and accelerated the cell cycle in Calu‐1 non‐small‐cell lung cancer cells. A, mRNA and protein levels of CREPT in Calu‐1 cells after transfection of pcDNA3.1‐CREPT and pcDNA3.1 for 48 hours. B, Proliferation of CREPT overexpressed Calu‐1 cells was increased. C, Overexpression of CREPT resulted in G1 phase arrest in Calu‐1 cells (*P < .05, **P < .01)
3.3. Facilitation of Calu‐1 cell migration and invasion in vitro by CREPT
To further explore the potential biological function of CREPT in NSCLC progression, the migration and invasion capacities of CREPT in Calu‐1 cell were assessed using the scratch wound healing assay, Transwell migration assay, and Transwell invasion assay. The scratch wound healing assay showed that pcDNA3.1‐CREPT cells migrated a much longer distance into the wound area than the control cells (P < .05; Figure 3A,B). Transwell migration assay further confirmed that overexpression of CREPT enhanced the migratory ability of Calu‐1 cells (P = .042; Figure 3C,D). In the Transwell invasion assay, more pcDNA3.1‐CREPT cells invaded into the lower chamber than pcDNA3.1 cells (Figure 3E). The quantitative analysis indicated that upregulation of CREPT expression dramatically promoted Calu‐1 cell invasion (P = .033, Figure 3F). These findings suggested that upregulation of CREPT expression in Calu‐1 cells enhances their migration and invasion abilities.
Figure 3.
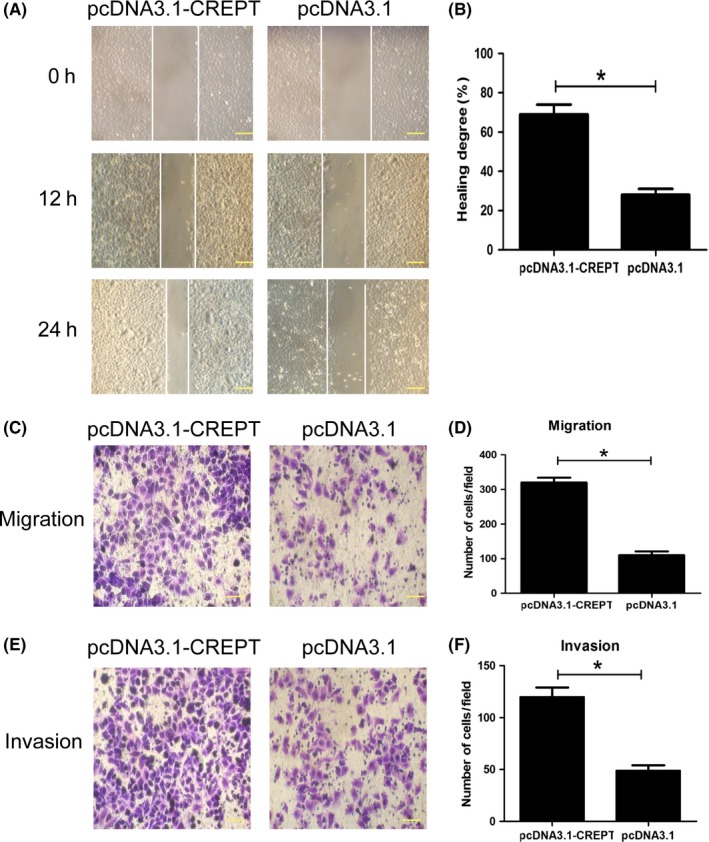
Overexpression of cell cycle‐related and expression‐elevated protein in tumor (CREPT) promotes migration and invasion in Calu‐1 non‐small‐cell lung cancer cells. A, Representative images of wound healing assay for CREPT overexpressed Calu‐1 cells 12 hours and 24 hours after treatment. B, Statistical analysis indicates the overexpression of CREPT in Calu‐1 cells could significantly increase cell motility (P = .029). C, Representative images of Transwell migration assay for CREPT‐overexpressing Calu‐1 cells. D, Statistical analysis of the number of invasion cells between CREPT‐ overexpressing Calu‐1 cells and the control group (P = .039). E Representative images of Transwell invasion assay for CREPT‐overexpressing Calu‐1 cells and control cells. (F) Statistical analysis of the number of invasion cells between CREPT‐overexpressing Calu‐1 cells and the control group (P = .043). *P < .05
3.4. Overexpression of CREPT promotes tumor growth in vivo
We sought to probe whether overexpression of CREPT affects tumor growth, by using a Calu‐1 xenograft model. The same amount of Calu‐1/pcDNA3.1‐CREPT or Calu‐1/pcDNA3.1 cells was s.c. injected into nude mice. After 30 days, the CREPT overexpressing xenograft grew much faster than the control group (Figure 4A). The tumor volumes and weights in the pcDNA3.1‐CREPT group were clearly increased compared to the control group (Figure 4B,C). The expression of CREPT in the pcDNA3.1‐CREPT group was significantly higher than that in tumors treated with pcDNA3.1 (Figure 4D). To determine the effect of CREPT on proliferation, immunohistochemical staining showed that the proliferation marker Ki‐67 was stronger in the pcDNA3.1‐CREPT group (Figure 4D). In addition, the levels of Ki‐67 mRNA and protein in pcDNA3.1‐CREPT tumors were significantly higher than in the control group (Figure 4E). These results confirm that the overexpression of CREPT significantly promotes tumor growth in vivo.
Figure 4.
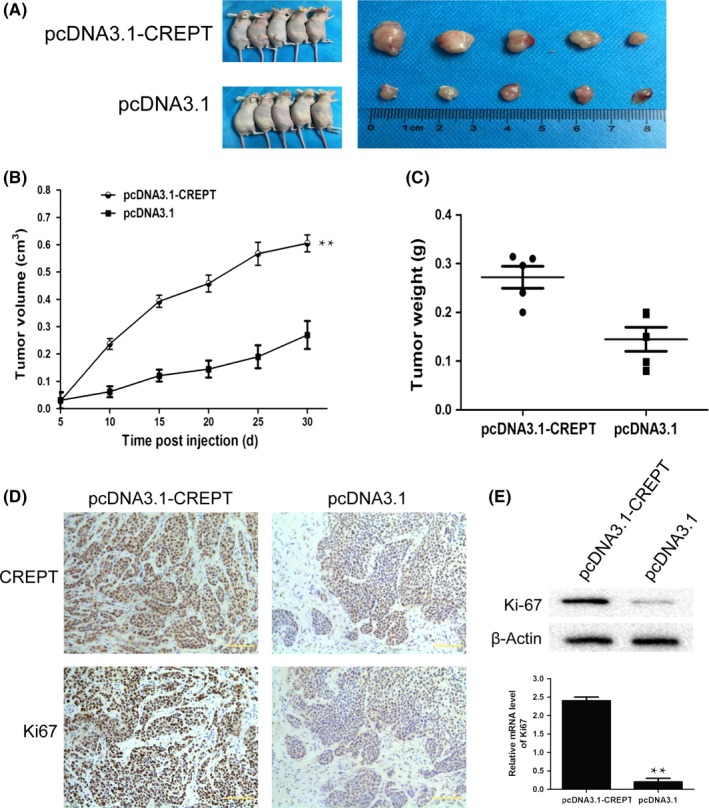
Overexpression of cell cycle‐related and expression‐elevated protein in tumor (CREPT) promotes non‐small‐cell lung tumor growth in vivo. A, Representative images of CREPT‐overexpressing and control Calu‐1 xenografts. B, Tumor volumes of CREPT‐overexpressing and control Calu‐1 xenografts. C, CREPT‐overexpressing Calu‐1 xenografts showed significantly increased tumor weight. D, Representative immunohistochemical staining of CREPT and Ki‐67 in xenograft tissues. E, The mRNA and protein expression level of Ki‐67 in xenograft tissues
3.5. High levels of CREPT predict poor prognosis of NSCLC patients
To explore the relationship between CREPT expression and survival of NSCLC patients, we analyzed the correlation between CREPT expression and clinical outcome in 271 NSCLC patients. Kaplan–Meier analysis showed that patients with higher CREPT expression revealed a significantly shorter OS (log–rank test, P = .01; Figure 5A) and RFS (P = .006; Figure 5B) compared to patients with lower CREPT expression.
Figure 5.
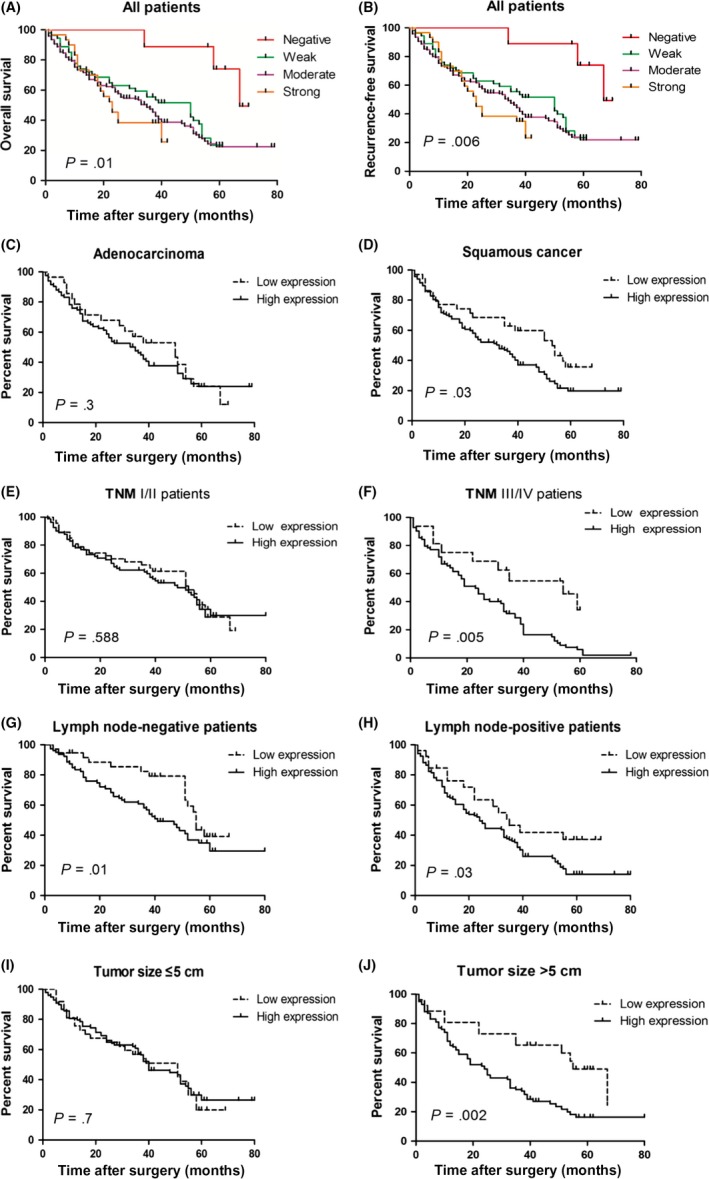
Cell cycle‐related and expression‐elevated protein in tumor (CREPT) overexpression predicts poor prognosis of patients with non‐small‐cell lung cancer. A,B, Patients with lower CREPT expression levels showed better overall survival (P = .01) (A) and better recurrence‐free survival (P = .006) (B). C,D, Kaplan–Meier survival analysis of adenocarcinoma patients (P = .30) (C) and squamous cell carcinoma patients (P = .03) (D), stratified by CREPT expression level. E,F, Kaplan–Meier survival analysis of pTNM I/II patients (P = .588) (E) and pTNM III/IV patients (P = .005) (F), stratified by CREPT expression level. (G,H) Kaplan–Meier survival analysis of lymph node‐negative patients (P = .01) (G) and lymph node‐positive patients (P = .03) (H), stratified by CREPT expression level. I,J, Kaplan–Meier survival analysis of patients with small tumor (diameter ≤5 cm, P = .70) (I) and large tumor (diameter >5 cm, P = .002) (J), stratified by CREPT expression level
In order to investigate the correlation of protein expression and patients’ clinical outcomes, the median CREPT protein expression was used as the cut‐off point to divide high expression and low expression groups for NSCLC patients. In order to assess the relationship between CREPT expression and the clinical outcomes of NSCLC patients according to the different pathology, the patients were divided into the squamous cell carcinoma (SCC) and adenocarcinoma (ADC) groups. The median survival time of SCC patients with high CREPT expression was 31 months (95% confidence interval [CI], 21‐41 months) and low CREPT expression was 53 months (95% CI, 38‐68 months). The result showed that the group with high CREPT expression had a worse prognosis than those in the low CREPT expression group in SCC patients (P = .03; Figure 5). However, there were no obvious significant difference in the survival rate with respect to CREPT expression among ADC patients (P = .3; Figure 5C).
For further analysis, including clinical stages, we divided patients into early (pTNM I/II) stage and advanced stage (pTNM III/IV) to explore the influence of CREPT separately. As shown in Figure 5E, there was a barely detectable statistically significant trend in the survival rate with respect to CREPT expression (P = .588) in pTNM I/II patients. Nevertheless, the low CREPT expression group had a longer survival time compared to the high CREPT expression group in pTNM III/IV patients (P = .005; Figure 5F). Specifically, the median survival time of patients with high CREPT expression was 54 months (95% CI, 28‐80 months), whereas that of patients with low CREPT expression was 19 months (95% CI, 13‐25 months) in pTNM III/IV patients.
Considering the influence of lymph node metastasis on NSCLC patients’ prognosis, we investigated the correlation between the expression of CREPT and prognosis in lymph node‐negative and lymph node‐positive patients. It was clear that patients with high CREPT expression had poor prognosis both in lymph node‐negative and lymph node‐positive patients (P = .01 and.03, respectively; Figure 5G,H).
In the further comparison, CREPT overexpression also correlated with the poor outcomes (P = .002; Figure 5J) in patients with large tumor size (diameter, >5 cm). However, there were no statistical differences in the survival rate between high and low CREPT expression (P = .7; Figure 5I) in the patients with small tumors (diameter ≤5 cm).
To clarify whether CREPT expression represents a prognostic factor in patients with NSCLC, regression analysis using the Cox hazards model was carried out. Based on results of the univariate analysis, factors including CREPT expression, tumor size, lymphatic metastasis, pTNM stage, and tumor differentiation were all significantly associated with OS in NSCLC patients. However, age, gender, smoking history, pathology, and tumor location showed slight variation (all P > .05). The data showed that, besides the TNM stage and tumor differentiation, CREPT expression was associated with poor survival of NSCLC patients (P < .001; Table 2). These results indicated that CREPT might be an independent prognostic marker of NSCLC patients (hazard ratio [HR], 1.223; 95% CI, 1.043‐1.435; P = .013).
Table 2.
Cox proportional hazards model analysis of variables affecting survival in non‐small‐cell lung cancer patients
| Variable | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| P‐value | HR (95% CI) | P‐value | HR (95% CI) | |
| CREPT expression, −/+/++/+++ | <.001a | 1.302 (1.126‐1.506) | .013a | 1.223 (1.043‐1.435) |
| Age, ≤61/>61 years | .289 | 0.956 (0.879‐1.039) | ||
| Gender, male/female | .376 | 0.916 (0. 755‐1.112) | ||
| Smoking, yes/no | .114 | 0.870 (0.732‐1.034), | ||
| Pathology, ADC/SCC | .163 | 1.128 (0.952‐1.336) | ||
| Location, central/peripheral | .644 | 1.042 (0.874‐1.242) | ||
| Tumor size, ≤5 cm/>5 cm | <.001a | 1.138 (1.094‐1.185) | .066 | 1.043 (0.997‐1.090) |
| Lymphatic metastasis, yes/no | <.001a | 1.838 (1.541‐2.192) | .126 | 0.839 (0.679‐1.035) |
| TNM stage, I and II/III and IV | <.001a | 0.421 (0.355‐0.499) | .001a | 0.480 (0.384‐0.599) |
| Tumor differentiation, well/moderate/poor | <.001a | 1.602 (1.360‐1.887) | .109 | 1.163 (0.967‐1.400) |
P ≤ .05.
ADC, adenocarcinoma; CI, confidence interval; CREPT, cell cycle‐related and expression‐elevated protein in tumor; HR, hazard ratio; SCC, squamous carcinoma.
4. DISCUSSION
In this study, we undertook IHC staining to evaluate CREPT expression in 271 NSCLC patients. Immunohistochemical evaluation showed that CREPT was expressed in 96.7% (262/271) NSCLC patients. We also detected the expression of CREPT by using qRT‐PCR and Western blot analysis in the expanded sample quantity. Compared with adjacent non‐tumor tissues, the mRNA and protein levels of CREPT were significantly elevated in NSCLC tissues. Elevated CREPT expression has been reported in 78.9% of retroperitoneal leiomyosarcoma patients and 77.8% of colorectal cancer patients.12, 13 We have further confirmed the elevated expression of CREPT mRNA and protein levels in NSCLC patients by expanding the clinical sample quantity. These findings implied it is easy to detect the expression of CREPT in NSCLC tumor tissues.
A previous study showed that CREPT is associated with clinical stage, lymph node metastasis, and distant metastasis in breast cancer patients.9 She et al. also revealed that the expression of CREPT may be closely related to histological grade in retroperitoneal leiomyosarcoma patients.13 The present study clearly indicated that CREPT expression is significantly correlated with clinical stage, tumor differentiation, and lymphatic metastasis in NSCLC patients. These results indicated that CREPT could play a significant role in NSCLC development and progression.
An earlier study showed that CREPT expression was correlated with poor prognosis in stomach cancer and retroperitoneal leiomyosarcoma patients.10, 13 Our previous study proved that CREPT overexpression was significantly correlated with shorter survival in colorectal cancer patients.12 In the current investigation, the survival analysis revealed that NSCLC patients with higher CREPT expression levels showed significantly shorter survival time compared to those with lower CREPT expression levels. Overexpression of CREPT was related to the malignant behavior of tumors during NSCLC progression. We also evaluated the prognostic significance of CREPT expression in different clinical features of NSCLC patients. Adenocarcinoma is associated with a poorer prognosis and a greater probability of distant recurrence compared with SCC.16 Gene expression profiles are known to differ substantially between ADC and SCC, and each histological subtype has a different etiology, carcinogenesis, and clinicopathologic behavior.17, 18, 19, 20 These studies revealed that the prognostic outcome might be different in SCC and ADC subgroups, which indicated that it is necessary to find an accurate prognostic marker. Based on our analysis, the expression level of CREPT is significantly associated with OS of SCC patients. Specifically, high CREPT expression was a much more significant survival factor for SCC patients than ADC patients. We speculated that CREPT is likely to represent a transformation factor associated with poor prognosis in the SCC development pathway. Considering the limitation of sample size, the differences in the role of CREPT carcinogenesis in SCC and ADC should be validated by further investigations.
We also investigated whether CREPT expression was associated with prognosis in NSCLC patients at different clinical stages. We noticed that the prognostic significance of CREPT expression was apparent in patients with advanced clinical stage, whereas the difference in the survival rate with respect to CREPT expression among patients with early stage was not significant. Further analysis suggested that lymph node metastasis patients with higher CREPT expression had a lower OS compared to those with lower CREPT expression; the difference was significant for lymph node‐negative patients as well as lymph node‐positive patients. A previous study has highlighted the size of the primary tumor as an independent prognostic factor for patients with NSCLC.21 Zhang et al. found that tumor size is an independent prognostic factor, for early stage as well as node‐positive and locally invasive disease.22 We detected that CREPT was a much more significant survival factor for NSCLC patients with larger tumor size (diameter >5 cm) than those with smaller tumor size (diameter ≤5 cm). These findings suggested that the expression of CREPT was specifically more relevant to the survival of advanced staged SCC patients with large tumor size, which can be used as a precise prognostic marker for NSCLC patients.
In the univariate survival analysis, CREPT expression concomitant with TNM stages, tumor differentiation, and lymph node metastasis are all related with NSCLC patients’ prognosis. More importantly, multivariate survival analysis revealed that CREPT expression (HR, 1.223; P = .013) was an independent prognostic factor, along with TNM stage (HR, 0.480; P = .001). We found that CREPT could serve as a potential independent biomarker to predict the prognosis of NSCLC patients.
In a previous study, we explored the biological effect of inhibiting CREPT on NSCLC cell proliferation in vitro.8 These results have motivated us to further investigate the critical role of CREPT in NSCLC cell proliferation. Therefore, we overexpressed CREPT in Calu‐1 cells and investigated its biological function both in vitro and in vivo. According to flow cytometric analysis, we found that upregulated CREPT expression can accelerate Calu‐1 cell proliferation by promoting the transformation of G1 phase to S phase. The CCK‐8 assay suggested that elevated CREPT expression markedly enhanced cell proliferation in Calu‐1 cells. Ki‐67 is recognized as a cell proliferation marker and strictly correlates with cell cycle progression.11 The IHC analysis showed that the expression of CREPT was significantly correlated with the Ki‐67 labelling index in NSCLC specimens. In addition, the tumor volumes and weights in the CREPT overexpression group were clearly increased compared to the control group. We detected the expression of Ki‐67 in xenograft by using IHC, Western blot analysis, and PCR. Compared to the control, much stronger expression of Ki‐67 was detected in xenograft at protein and mRNA levels. Wang et al. reported that overabundant CREPT promoted endometrial tumor growth through regulating the cell cycle.23 The pro‐proliferative role of CREPT was also verified both in vitro and in vivo.
The ability of cancer cells to undergo migration and invasion allows them to spread to lymph nodes and metastasize to distant organs. We undertook Transwell and scratch wound healing assays to investigate the function of CREPT in metastasis and invasion. Our results showed that overexpressed CREPT can promote metastasis and invasion in tumor. Our results provided new evidence supporting the involvement of CREPT in promoting the proliferation, migration, and invasion of NSCLC. Further studies are underway to elucidate the mechanisms underlying CREPT's regulation of NSCLC progression.
Taken together, our results reveal that CREPT was significantly upregulated in NSCLC compared with corresponding non‐tumor tissues. The expression level of CREPT was correlated with tumor differentiation, lymph node metastasis, and clinical stage. More importantly, CREPT overexpression level was associated with poor prognosis of patients. The survival analysis suggested that CREPT could serve as an independent predictor and potential target for NSCLC patients. Moreover, we showed that upregulated CREPT potently promoted NSCLC cell proliferation through regulation of the cell cycle in vitro and in vivo. The molecular mechanisms by which CREPT regulates the progression of tumor are the focus of our future studies.
CONFLICT OF INTEREST
The authors have no conflict of interest.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (No. 81572252). The authors would like to thank Dr. Xiaofei Li (Department of Thoracic Surgery, Tangdu Hospital, Fourth Military Medical University, Xi'an 710038, China) for his kind contributions to the collection of NSCLC tissue samples.
Li W, Zheng G, Xia J, et al. Cell cycle‐related and expression‐elevated protein in tumor overexpression is associated with proliferation behaviors and poor prognosis in non‐small‐cell lung cancer. Cancer Sci. 2018;109:1012–1023. https://doi.org/10.1111/cas.13524
Weimiao Li and Guoxu Zheng equally contributed to this work.
Contributor Information
Zhipei Zhang, Email: zzpzyy@fmmu.edu.cn.
Faguang Jin, Email: jinfag@fmmu.edu.cn.
REFERENCES
- 1. Siegel RL, Miller KD. Jemal A Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7‐30. [DOI] [PubMed] [Google Scholar]
- 2. Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367‐1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mao L. Recent advances in the molecular diagnosis of lung cancer. Oncogene. 2002;21:6960‐6969. [DOI] [PubMed] [Google Scholar]
- 4. Rooney C, Sethi T. Advances in molecular biology of lung disease: aiming for precision therapy in non‐small cell lung cancer. Chest. 2015;148:1063‐1072. [DOI] [PubMed] [Google Scholar]
- 5. Allemani C, Weir HK, Carreira H, et al. Global surveillance of cancer survival 1995‐2009: analysis of individual data for 25,676,887 patients from 279 population‐based registries in 67 countries (Concord‐2). Lancet. 2015;385:977‐1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Douillard JY, Tribodet H, Aubert D, et al. Adjuvant cisplatin and vinorelbine for completely resected non‐small cell lung cancer: subgroup analysis of the lung adjuvant cisplatin evaluation. J Thorac Oncol. 2010;5:220‐228. [DOI] [PubMed] [Google Scholar]
- 7. Kisluk J, Ciborowski M, Niemira M, et al. Proteomics biomarkers for non‐small cell lung cancer. J Pharm Biomed Anal. 2014;101:40‐49. [DOI] [PubMed] [Google Scholar]
- 8. Liu T, Li WM, Wang WP, et al. Inhibiting crept reduces the proliferation and migration of non‐small cell lung cancer cells by down‐regulating cell cycle related protein. Am J Transl Res. 2016;8:2097‐2113. [PMC free article] [PubMed] [Google Scholar]
- 9. Liang Z, Feng Q, Xu L, et al. Crept regulated by mir‐138 promotes breast cancer progression. Biochem Biophys Res Comm. 2017;493:263‐269. [DOI] [PubMed] [Google Scholar]
- 10. Lu D, Wu Y, Wang Y, et al. Crept accelerates tumorigenesis by regulating the transcription of cell‐cycle‐related genes. Cancer Cell. 2012;21:92‐104. [DOI] [PubMed] [Google Scholar]
- 11. Jakobsen JN, Sorensen JB. Clinical impact of Ki‐67 labeling index in non‐small cell lung cancer. Lung cancer 2013;79:1‐7. [DOI] [PubMed] [Google Scholar]
- 12. Zheng G, Li W, Zuo B, et al. High expression of crept promotes tumor growth and is correlated with poor prognosis in colorectal cancer. Biochem Biophys Res Comm. 2016;480:436‐442. [DOI] [PubMed] [Google Scholar]
- 13. She Y, Liang J, Chen L, et al. Crept expression correlates with poor prognosis in patients with retroperitoneal leiomyosarcoma. Int J Clin Exp Pathol. 2014;7:6596‐6605. [PMC free article] [PubMed] [Google Scholar]
- 14. Rami‐Porta R, Wittekind C, Goldstraw P. Complete resection in lung cancer surgery: proposed definition. Lung Cancer. 2005;49:25‐33. [DOI] [PubMed] [Google Scholar]
- 15. Chen P, Li WM, Lu Q, et al. Clinicopathologic features and prognostic implications of Nok/Styk1 protein expression in non‐small cell lung cancer. BMC Cancer. 2014;14:402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jung EJ, Byun JM, Kim YN, et al. Cervical adenocarcinoma has a poorer prognosis and a higher propensity for distant recurrence than squamous cell carcinoma. Int J Gynecol Cancer. 2017;27:1228‐1236. [DOI] [PubMed] [Google Scholar]
- 17. McDoniels‐Silvers AL, Nimri CF, Stoner GD, et al. Differential gene expression in human lung adenocarcinomas and squamous cell carcinomas. Clin Cancer Res. 2002;8:1127‐1138. [PubMed] [Google Scholar]
- 18. Kikuchi T, Daigo Y, Katagiri T, et al. Expression profiles of non‐small cell lung cancers on cdna microarrays: identification of genes for prediction of lymph‐node metastasis and sensitivity to anti‐cancer drugs. Oncogene. 2003;22:2192‐2205. [DOI] [PubMed] [Google Scholar]
- 19. Petty RD, Nicolson MC, Kerr KM, et al. Gene expression profiling in non‐small cell lung cancer: from molecular mechanisms to clinical application. Clin Cancer Res. 2004;10:3237‐3248. [DOI] [PubMed] [Google Scholar]
- 20. Moldvay J, Fabian K, Jackel M, et al. Claudin‐1 protein expression is a good prognostic factor in non‐small cell lung cancer, but only in squamous cell carcinoma cases. Pathol Oncol Res. 2017;23:151‐156. [DOI] [PubMed] [Google Scholar]
- 21. Stojiljkovic D, Santrac N, Stojiljkovic T, et al. Correlation of tumor size as independent factor and disease stage with local recurrence of non‐small cell lung carcinoma and its operability. J BUON. 2015;20:166‐172. [PubMed] [Google Scholar]
- 22. Zhang J, Gold KA, Lin HY, et al. Relationship between tumor size and survival in non‐small‐cell lung cancer (Nsclc): an analysis of the surveillance, epidemiology, and end results (seer) registry. J Thorac Oncol. 2015;10:682‐690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang Y, Qiu H, Hu W, et al. Rprd1b promotes tumor growth by accelerating the cell cycle in endometrial cancer. Oncol Rep. 2014;31:1389‐1395. [DOI] [PubMed] [Google Scholar]


