Abstract
Peripheral T‐ or natural killer (NK)‐cell lymphomas are rare and difficult‐to‐recognize diseases. It remains arduous to distinguish between NK cell‐ and cytotoxic T‐lymphocyte‐derived lymphomas through routine histological evaluation. To clarify the cells of origin, we focused on NK‐cell receptors and examined the expression using immunohistochemistry in 22 cases with T‐ and NK‐cell neoplasms comprising angioimmunoblastic T‐cell lymphoma, anaplastic lymphoma kinase (ALK)‐positive and ‐negative anaplastic large‐cell lymphomas, extranodal NK/T‐cell lymphoma, nasal type, monomorphic epitheliotropic intestinal T‐cell lymphoma, aggressive NK‐cell leukemia, and other peripheral T‐cell lymphomas. Inhibitory receptor leukocyte immunoglobulin‐like receptor subfamily B member 1 (LILRB1) was detected in 14 (64%) cases, whereas activating receptors DNAM1, NKp46, and NKG2D were expressed in 7 (32%), 9 (41%), and 5 (23%) cases, respectively. Although LILRB1 was detected regardless of the disease entity, the activating NK‐cell receptors were expressed predominantly in TIA‐1‐positive neoplasms (DNAM1, 49%; NKp46, 69%; and NKG2D, 38%). In addition, NKp46 and NKG2D were detected only in NK‐cell neoplasms and cytotoxic T‐lymphocyte‐derived lymphomas including monomorphic epitheliotropic intestinal T‐cell lymphoma. One Epstein‐Barr virus‐harboring cytotoxic T‐lymphocyte‐derived lymphoma mimicking extranodal NK/T‐cell lymphoma, nasal type lacked these NK‐cell receptors, indicating different cell origin from NK and innate‐like T cells. Furthermore, NKG2D expression showed a negative impact on survival among the 22 examined cases, which mainly received the standard chemotherapy regimen (log‐rank test, P = .024). We propose that the presence of activating NK‐cell receptors may provide new insights into understanding peripheral T‐cell lymphomas and characterizing them as innate‐like T‐cell neoplasm.
Keywords: cytotoxic molecule, innate‐like T cell, NK‐cell receptor, NKG2D, T/NK‐cell lymphoma
Abbreviations
- AITL
angioimmunoblastic T‐cell lymphoma
- ALK− ALCL
ALK‐negative ALCL
- ALK+ ALCL
ALK‐positive anaplastic large cell lymphoma
- ALK
anaplastic lymphoma kinase
- ANKL
aggressive NK‐cell leukemia
- ATLL
adult T‐cell leukemia/lymphoma
- CI
confidence interval
- CM
cytotoxic molecule
- CTL
cytotoxic T lymphocyte
- EATL
enteropathy‐associated T‐cell lymphoma
- EBV
Epstein‐Barr virus
- ENKL
extranodal NK/T‐cell lymphoma, nasal type
- FFPE
formalin‐fixed, paraffin‐embedded
- GrB
granzyme B
- IHC
immunohistochemistry
- IPI
International Prognostic Index
- LILRB1
leukocyte immunoglobulin‐like receptor subfamily B member 1
- MEITL
monomorphic epitheliotropic intestinal T‐cell lymphoma
- NA
not available
- NK
natural killer
- NKR
NK‐cell receptor
- OS
overall survival
- PD‐1
programmed cell death‐1
- PTCL‐NOS, PTCL
not otherwise specified
- PTCL
peripheral T‐cell lymphoma
- PTNKL
peripheral T‐ or NK‐cell lymphomas
- TCR
T‐cell receptor
- TFH
follicular helper T cell
- WHO
World Health Organization
1. INTRODUCTION
Peripheral T‐ or NK‐cell lymphomas are a group of rare diseases.1 Although they account for 15%‐20% of aggressive lymphomas in Western countries,2 the number is higher in Japan, accounting for 30%‐40% of aggressive lymphomas.2, 3 The differences are explained by the high incidence of HTLV‐I‐associated ATLL and EBV‐associated ENKL.2, 3 Although the current WHO classification includes more than 25 disease entities in PTNKL, making a correct diagnosis per se has a prognostic impact on clinical outcome.1, 2 With increased understanding of PTNKL, subtype‐specific approaches have shown to achieve a promising therapeutic effect, particularly in ENKL.4 Although ATLL is defined as the HTLV‐I‐harboring CD4+ T‐cell neoplasm, ENKL is composed of EBV‐harboring CM‐positive neoplasms of different cell lineages including αβT cells, γδT cells and NK cells.1, 2, 5 Recent observation showed the existence of EBV‐harboring CTL‐type PTCL mimicking ENKL.6 These cases raise concerns regarding differences in the cell of origin between ENKL and EBV‐harboring PTCL‐NOS.
NK‐cell receptors found in mature NK cells contribute to positive or negative signaling during NK‐cell‐mediated cytotoxicity. Activating NKR including DNAM1 (CD226), NKG2D (CD314), and a natural cytotoxicity receptor such as NKp46 (CD335) recognize alterations of cell surface molecules on virus‐infected cells and tumor cells.7 Binding of the activating NKR to ligands induces the activated NK cells to attack their targets.7 In contrast, inhibitory NKR including CD94‐NKG2A and LILRB1 (also known as ILT2/MIR7/CD85j) bind to human leukocyte antigen class I molecules and prevent the NK cells from killing healthy cells.7 However, the expressions of NKR are shown not only in NK cells, but also in unconventional innate‐like T cells comprising γδT cells, invariant NKT cells, mucosal‐associated invariant T cells, and intestinal intraepithelial lymphocytes.8, 9 They play a role in the preservation of tissue integrity and span the innate‐adoptive continuum.8, 9
Previous studies showed that CD94, NKG2A, and NKp46 were usually detected in ENKL, whereas the expression of another group of NKR, the killer cell immunoglobulin‐like receptors seem to be reduced in most ENKL cases.10, 11, 12 The pathological implication of NKR in PTNKL remains to be defined. Therefore, we examined the expressions of LILRB1, DNAM1, NKp46, and NKG2D in several PTNKL entities and carried out a clinicopathological study.
2. MATERIALS AND METHODS
2.1. Case selection and clinical data collection
Selected aggressive PTNKL cases were consecutively diagnosed at St. Marianna University Hospital (Kawasaki, Japan) between January 2011 and October 2016. All cases were diagnosed according to the current WHO classification.1 In this study, PTCL‐NOS were further categorized into 3 groups: i.e, TFH‐type PTCL, CTL‐type PTCL, and others. TFH‐type PTCL was defined as nonspecific T‐cell lymphomas showing CD4+TIA‐1− phenotype and expressing at least two TFH‐associated antigens including PD‐1, CD10, Bcl‐6, and CXCL13.1 In contrast, CTL‐type PTCL was defined as nonspecific T‐cell lymphoma showing TIA‐1+GrB+ phenotype. We also collected clinical data including age, sex, initial sites of involvement, clinical stage according to the Cotswolds‐modified Ann Arbor staging system, IPI, mode of initial therapy, duration of follow up, and status at time of last follow up in each case. Informed consent was obtained from all surviving patients prior to being included in the study. This protocol was approved by the Institutional Review Boards of the St. Marianna University School of Medicine.
2.2. Immunohistochemistry
Immunohistochemistry was carried out on sections of FFPE tissues. In addition to our PTNKL diagnostic panel (the first set: CD3, CD4, CD5, CD8, CM including TIA‐1 and GrB, EBER in situ hybridization; the second set: CD30, CD56, PD‐1, Bcl‐6 and ALK), we assessed the expression profile of TCRβ, TCRδ, LILRB1, DNAM1, NKp46, and NKG2D, which are available for IHC. Heat‐induced antigen retrieval (120°C, 6 min) was carried out using 10 mM citrate buffer, pH 6 (DAKO Japan, Tokyo, Japan). Primary antibodies used were anti‐TCRβ mouse monoclonal antibody (G‐11) (Santa Cruz Biotechnology, Dallas, TX, USA), anti‐TCRδ (H‐41) mouse monoclonal antibody (Santa Cruz Biotechnology), anti‐LILRB1 rabbit monoclonal antibody (Abcam, Cambridge, UK), anti‐CD226 rabbit polyclonal antibody (Sigma‐Aldrich Japan, Tokyo, Japan), anti‐NKP46/NCT1 goat polyclonal antibody (R&D Systems, Minneapolis, MN, USA), and anti‐NKG2D goat polyclonal antibody N‐20 (Santa Cruz Biotechnology). All stains were interpreted as follows: negative, no staining; −/+, equivocal staining but definite positivity in <30% of presumptive neoplastic cells; +/−, definite positivity in 30%‐70% of presumptive neoplastic cells; +, definite positivity in ≥70% of presumptive neoplastic cells.
2.3. PCR‐based TCR gene rearrangement analyses
Genomic DNA was extracted from FFPE tissue using the ReliaPrep™ FFPE gDNA Miniprep System (Promega, Madison, WI, USA). PCR was carried out according to the BIOMED‐2 protocols.11 We initially assessed TCRγ gene (TRG) rearrangement. In cases without monoclonal TRG bands, TCRβ gene (TRB) rearrangements were also analyzed.13
2.4. Statistical analysis
Survival curves were determined using the Kaplan‐Meier method, and the difference was compared using the log‐rank test. All analyses were carried out using EZR (Saitama Medical Center, Jichi Medical University, ver.1.33).14 All P‐values were two‐tailed, and P < .05 was considered significant.
3. RESULTS
3.1. Diagnosis of PTNKL
Between January 2011 and October 2016, a total of 291 cases were diagnosed with malignant lymphomas at our institution. Among them, 22 cases (7.6%) were found to have PTNKL. According to the current WHO classification,1 they were diagnosed with AITL in two, ALK+ ALCL in two, ALK− ALCL in two, ENKL in three, MEITL in three, ANKL in one, and PTCL‐NOS in nine. Nine PTCL‐NOS cases were further regarded as TFH‐type PTCL (5 cases), CTL‐type PTCL (3 cases), and others (true PTCL‐NOS) (1 case). Clinicopathological features of the cases are shown in Table 1. TCRδ was detected in CM‐positive lymphoma cases only including one ALK− ALCL, one EBV‐positive CTL‐type PTCL, and one MEITL case (UPN #10, #18, and #19, respectively) (Table 1). TCRβ was evaluated in CM‐positive lymphoma cases only because of the limited amount of residual specimens. Because the frequencies and intensities of stained lymphoma cells were relatively low, the positive cases included one EBV‐positive CTL‐type PTCL and two MEITL cases (UPN #17, #20, and #21, respectively) (Table 1). Therefore, we considered 3 cases (UPN #10, #18, and #19) as a γδT‐cell lymphoma. Indeed, we detected neither TCRδ nor TCRβ in both ALK+ ALCL cases. Except for 3 cases (UPN #13, #14, and #22), monoclonal TRG bands were detected in the remaining 19 cases. We confirmed that these cases are T‐cell lymphomas. The remaining 3 cases also showed polyclonal TRB bands only, indicating that they are true NK‐cell neoplasms.
Table 1.
Clinicopathological features of 22 examined cases
| UPN | Age/Sex | Diagnosis | Primary site | Clinical stage | IPI score | Immunophenotype | NK receptor | TCRβ | TCRδ | TRG GR | Initial therapy | Survival (days) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CD3 | CD4 | CD5 | CD8 | CD30 | CD56 | PD‐1 | TIA‐1 | GrB | EBER | LILRB1 | DNAM1 | NKp46 | NKG2D | |||||||||||
| 1 | 79/F | AITL | LN | III | 3 | + | + | + | − | − | NA | + | − | − | − | −/+ | − | − | − | NA | − | m | THP‐COP | 1043a |
| 2 | 67/F | AITL | LN | IV | 3 | + | + | + | − | −/+ | NA | + | − | − | − | + | − | − | − | NA | − | m | None | 17 |
| 3 | 61/F | PTCL (TFH) | LN | III | 2 | + | +/− | + | − | − | − | − | − | − | − | +/− | − | − | − | NA | − | m | CHOP | 751a |
| 4 | 63/M | PTCL (TFH) | LN | III | 3 | + | + | + | − | +/− | NA | − | −/+ | − | − | −/+ | − | − | − | NA | − | m | CHOP | 852a |
| 5 | 82/F | PTCL (TFH) | LN | III | 3 | + | + | +/− | − | +/− | NA | + | − | − | − | +/− | − | − | − | NA | − | m | THP‐COP | 1089a |
| 6 | 68/M | PTCL (TFH) | LN | IV | 3 | + | + | + | − | − | NA | + | −/+ | − | − | + | − | − | − | NA | − | m | EPOCH | 570a |
| 7 | 65/M | PTCL (TFH) | LN | III | 3 | + | + | + | − | − | NA | + | − | − | − | +/− | − | − | − | NA | − | m | EPOCH | 933a |
| 8 | 21/M | ALK+ ALCL | LN | II | 0 | − | −/+ | − | − | + | − | NA | + | − | − | − | − | − | − | − | − | m | CHOP | 174a |
| 9 | 62M | ALK+ ALCL | GI tract | IV | 3 | − | +/− | − | − | + | − | NA | + | + | − | + | +/− | − | −/+ | − | − | m | CHOP | 890 |
| 10 | 69/M | ALK− ALCL | GI tract | II | 3 | + | − | + | − | + | − | NA | + | − | − | + | − | − | −/+ | − | + | m | DEX | 107 |
| 11 | 49/M | ALK− ALCL | LN | IV | 2 | + | + | + | − | + | − | NA | − | − | − | − | + | − | − | −/+ | − | m | EPOCH | 574a |
| 12 | 56/M | PTCL‐NOS | GI tract | IV | 2 | + | − | − | − | − | − | NA | − | − | − | − | − | − | −/+ | NA | − | m | CHOP | 330 |
| 13 | 56/M | ENKL | Nasal cavity | I | 1 | − | − | − | − | NA | + | NA | + | + | + | −/+ | − | + | + | − | − | pa | CHOP | 598 |
| 14 | 61/M | ENKL | Nasal cavity | II | 2 | +/− | − | − | +/− | NA | + | NA | + | +/− | + | +/− | + | + | −/+ | − | − | pa | SMILE | 1788a |
| 15 | 64/M | ENKL | Nasal cavity | I | 2 | + | − | − | −/+ | NA | + | NA | + | + | + | + | + | + | −/+ | −/+ | − | m | MILD | 1291a |
| 16 | 68/M | PTCL (CTL) | SC tissue | IV | 4 | + | − | +/− | +/− | − | − | NA | + | + | + | − | − | − | − | −/+ | − | m | DEX | 50 |
| 17 | 72/F | PTCL (CTL) | GI tract | II | 5 | + | − | − | + | − | −− | NA | +/− | + | + | − | − | + | − | +/− | − | m | DEX+l‐Asp | 120 |
| 18 | 73/M | PTCL (CTL) | LN | IV | 4 | + | − | − | + | − | − | NA | + | + | + | + | + | + | + | − | +/− | m | THP‐COP | 35 |
| 19 | 53/M | MEITL | Intestine | II | 2 | + | − | − | − | − | + | NA | + | − | − | + | − | + | + | − | +/− | m | CHOP | 860 |
| 20 | 57/M | MEITL | Intestine | IV | 2 | +/− | − | − | + | − | + | NA | + | + | − | +/− | +/− | + | − | +/− | − | m | NA | 38a |
| 21 | 71/M | MEITL | Intestine | IV | 4 | + | − | − | + | − | − | NA | + | −/+ | − | + | − | + | + | +/− | − | m | None | 134 |
| 22 | 72M | ANKL | BM | IV | 4 | +/− | − | − | − | NA | + | NA | + | +/− | + | + | +/− | + | + | − | − | pb | MILD | 184 |
+, ≥70% positive; +/−, ≥30%~70% positive; −/+, ~30% positive; −, no staining.
AITL, angioimmunoblastic T‐cell lymphoma; ALCL, anaplastic large cell lymphoma; ALK, anaplastic lymphoma kinase; ANKL, aggressive NK‐cell leukemia; BM, bone marrow; CHOP, cyclophosphamide, doxorubicin, vincristine, and prednisolone; DEX, dexamethasone; ENKL, extranodal NK/T‐cell lymphoma, nasal type; EPOCH, etoposide, prednisolone, vincristine, cyclophosphamide, and doxorubicin; GI, gastrointestinal; GR, gene rearrangement; ICE, ifosfamide, carboplatin, and etoposide; IPI, international prognostic index; l‐Asp, l‐asparaginase; LILRB1, leukocyte immunoglobulin‐like receptor subfamily B member 1; LN, lymph node; m, monoclonal; MEITL, monomorphic epitheliotropic intestinal T‐cell lymphoma; MILD, methotrexate, ifosfamide, l‐asparaginase, and dexamethasone; NA, not available; NOS, not otherwise specified; p, polyclonal; PTCL, peripheral T‐cell lymphoma; SC, subcutaneous; SMILE, dexamethasone, methotrexate, ifosfamide, l‐asparaginase, and etoposide; TFH, follicular helper T cell; THP‐COP, pirarubicin, cyclophosphamide, vincristine, and prednisolone; UPN, unique patient number.
Alive at the last observation point.
TRB gene rearranged band was also undetected.
3.2. NKR expression in PTNKL
Representative cases are presented in Figure 1. A total of 14 cases (64%) were positive for LILRB1 (Table 1). This molecule was expressed regardless of the disease entity. The expression was shown in ANKL (1/1, 100%), ENKL (2/3, 67%), CTL‐type PTCL (1/3, 33%), MEITL (3/3, 100%), ALK+ ALCL (1/2, 50%), ALK− ALCL (1/2, 50%), TFH‐type PTCL (4/5, 80%), and AITL (1/2, 50%). In contrast, activating NKR, DNAM1, NKp46, and NKG2D were detected mainly in TIA‐1‐positive neoplasms (46%, 69%, and 38%, respectively). Expression of DNAM1 was shown in ANKL (1/1, 100%), ENKL (2/3, 67%), CTL‐type PTCL (1/3, 33%), MEITL (1/3, 33%), ALK+ ALCL (1/2, 50%), and ALK− ALCL (1/2, 50%). This molecule was also detected in the reticuloendothelial cells surrounding neoplastic cells (Figure 2). Expression of NKp46 was shown in ANKL (1/1, 100%), ENKL (3/3, 100%), CTL‐type PTCL (2/3, 67%), and MEITL (3/3, 100%). Furthermore, NKG2D was also expressed in ANKL (1/1, 100%), ENKL (1/3, 33%), CTL‐type PTCL (1/3, 33%) and MEITL (2/3, 67%). Compared with the staining pattern of DNAM1, these molecules were detected exclusively in neoplastic cells. Although the expression trend in NKG2D was similar to that in NKp46, the positive rate was lower than that of NKp46. One EBV‐harboring CTL‐type PTCL case (UPN #16) lacked the expression of all examined NKR in spite of the extranodal disease presentation (Figure 1F).
Figure 1.
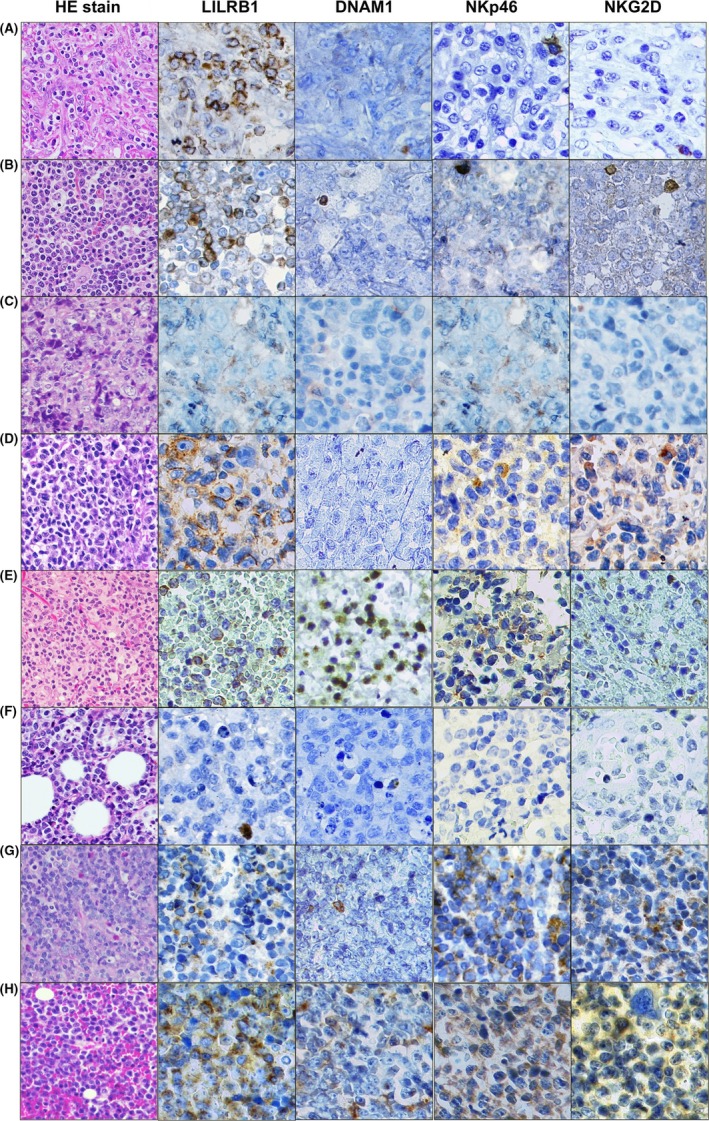
Expression of natural killer (NK) cell receptors (NKR) in peripheral T‐ or NK‐cell lymphomas. Each biopsy specimen was morphologically assessed using hematoxylin and eosin (HE) staining and immunohistochemistry. A, Angioimmunoblastic T‐cell lymphoma (AITL) case (unique patient number [UPN] #2). This case showed expression of inhibitory NKR leukocyte immunoglobulin‐like receptor subfamily B member 1 (LILRB1). B, Follicular helper T‐cell (TFH)‐type peripheral T‐cell lymphoma (PTCL) case (UPN #4). This case also expressed LILRB1. C, Anaplastic lymphoma kinase (ALK)‐positive anaplastic large cell lymphoma (ALCL) case (UPN #8). This case was negative for any NKR whereas NKp46 was detected in the bystander cells. D, ALK‐negative ALCL case (UPN #10). This case expressed only LILRB1. E, Extranodal NK/T‐cell lymphoma, nasal type (ENKL) case (UPN #14). This case was an NK‐cell‐derived lymphoma and showed expression of LILRB1 and activating NKR DNAM1 and NKp46, whereas NKG2D was not detected in the lymphoma cells. F, CTL‐type PTCL case (UPN #16). Although this case mimics ENKL because of the positivity of Epstein‐Barr virus‐encoded small RNAs, it lacked expression of all examined NKR. G, Monomorphic epitheliotropic intestinal T‐cell lymphoma case (UPN #19). This case showed expression of LILRB1, NKp46 and NKG2D, but not DNAM1. H, Aggressive NK‐cell leukemia case (UPN #22). This case was positive for all examined NKR
Figure 2.
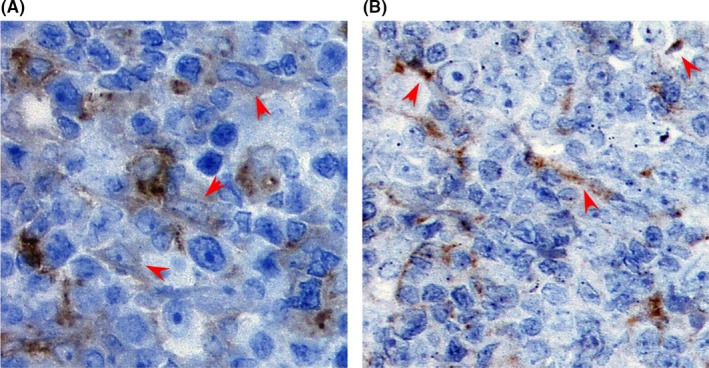
Dendritic cells and endothelial cells in follicular helper T‐cell‐type peripheral T‐cell lymphoma cases expressed DNAM1. A, Although the lymphoma cells were negative for DNAM1, dendritic cells and endothelial cells were positive for this molecule as shown by red arrowheads (unique patient number [UPN] #3). DNAM1 was more weakly stained in dendritic cells than in endothelial cells. B, As shown by red arrowheads, DNAM1 was clearly detected in endothelial cells in DNAM1‐negative case (UPN #7)
3.3. Clinical features and outcomes of examined cases
Median age at diagnosis was 64.5 years (range, 21‐82), and the male/female ratio was 3.5. As an initial involved site, nodal and extranodal presentations were observed in 10 (45%) and 12 (55%) cases, respectively. Six cases (27%) showed limited‐stage disease. According to the IPI score, low, low‐intermediate, high‐intermediate, and high‐risk categories were determined in 2 (9%), 7 (32%), 8 (36%), and 5 cases (23%), respectively. Thirteen of 19 treated cases (68%) received CHOP‐like chemotherapy regimen. Median follow‐up duration was 20.4 months (range, 0.6‐63.9), and the median OS for the entire group was 28.3 months (95% CI 4.4, NA) (Figure 3A). Although there was no significant difference in the OS by age ≥60 years (log‐rank test, P = .796), IPI high‐risk group (log‐rank test, P < .001) had a negative impact on the OS (Figure 3B). Expressions of LILRB1 (log‐rank test, P = .796), DNAM1 (log‐rank test, P = .655), and NKp46 (log‐rank test, P = .508) showed no adverse effect on the OS (Figure 3C‐E), whereas the expression of NKG2D (log‐rank test, P = .024) had a negative impact on the OS (Figure 3F).
Figure 3.
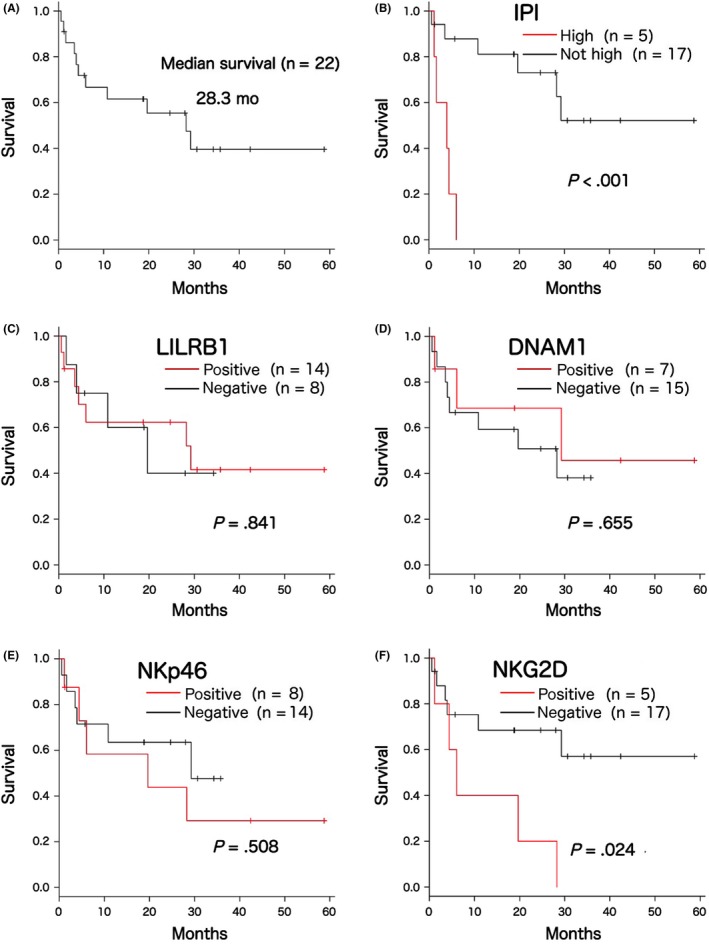
Clinical outcomes of the examined 22 cases. A, Overall survival (OS) of the 22 examined cases. Median survival duration was estimated as 28.3 months. B, Although age ≥60 years was not associated with OS, high‐risk group of international prognostic index (IPI) showed a negative impact on the OS (log‐rank test, P < .001). C‐F, Expression of natural killer (NK) cell receptors (NKR) and OS. Although the expression of (C) leukocyte immunoglobulin‐like receptor subfamily B member 1 (LILRB1), (D) DNAM1, and (E) NKp46 had no impact on the OS, expression of (F) NKG2D showed an adverse effect on the OS (log‐rank test, P = .024)
4. DISCUSSION
In the present study, we confirmed the expression of LILRB1, DNAM1, NKp46 and NKG2D in some PTNKL entities. Although LILRB1 expression had not yet been evaluated in NK‐cell malignancies, we verified the presence in ANKL and ENKL, and further found that the expression pattern was not disease‐specific. Previous reports showed that LILRB1 was expressed not only in Sézary cells, but also in B‐cell lines.15, 16 In addition, the stimulation was shown to suppress the proliferation in spite of the different cell lineage.15, 16 The frequent expression of LILRB1 suggests that this molecule may have a potential to mediate growth‐inhibitory signals in PTNKL cells. Our observation raises the possibility that LILRB1 might be a novel therapeutic target for PTNKL.
Compared with LILRB1, the expression of activating NKR was relatively restricted to CM‐positive neoplasms. Although NKp46 and NKG2D were detected mainly in neoplastic cells, DNAM1 was also expressed in the surrounding reticuloendothelial cells. Increased expression levels of DNAM1 were observed in inflammatory monocytes and activated endothelial cells.17, 18 The enhanced expression of DNAM1 likely leads to acceleration of cell adhesion in the expressing cells.17, 18 This molecule may function in cell‐cell interactions in addition to its cytotoxic effects in the tumor environment. Recent study showed that NKp46 was detected in ALK+ ALCL cells but not in EATL cells.12 Nevertheless, we observed that NKp46 was detected in some of the surrounding bystander cells but not in ALK+ ALCL cells (Figure 1C). Although MEITL had been included in EATL in the previous WHO classification, our three MEITL cases (two αβT‐cell and one γδT‐cell types) were all positive for NKp46. Our observations show that the expression of NKp46 and NKG2D is a hallmark of lymphomas derived from NK cells and unconventional innate‐like T cells. Although aberrant expression might occur in neoplastic cells, the presence of the activating NKR on non‐γδT‐cell lymphoma cells may mirror the acquisition of its natural cytotoxic function. Therefore, even though the lymphoma cells are infected with EBV, non‐γδT‐cell type CTL‐derived lymphomas lacking the expression of activating NKR should be distinguished from ENKL.
Although several problems such as sensitivity of IHC and loss of expression remain, NKG2D was not so frequently expressed in CM‐positive neoplasms. Expression levels of NKG2D in NK cells are likely increased through STAT3 activation, which is considered one of the important alterations in PTNKL.19, 20 Previous study indicated that approximately 30% of cases of ENKL presented STAT3 phosphorylation.21 In addition, the activation of STAT3 seems to be associated with a poor clinical outcome in ENKL.22 Our result may imply that the detection of NKG2D had selected higher risk cases among the patients with relatively poor prognostic diseases such as ANKL, ENKL, and MEITL. Although it is difficult to draw a conclusion as a result of the small numbers of cases, the innate lymphoid cell‐associated marker might show an impact on clinical outcome when such standard regimens are adopted.
In summary, we showed, for the first time, that DNAM1 and NKG2D are expressed in aggressive PTNKL. Although we failed to discriminate between T‐ and NK‐cell neoplasms by recognition of the expression profile of NKR in PTNKL, the detection of activating NKR including DNAM1, NKp46, and NKG2D characterizes CM‐positive PTNKL as neoplasms derived from NK cells and unconventional innate‐like T cells. We propose that the presence of the activating NKR may provide new insights into the understanding of ENKL‐like T‐cell lymphomas. Further studies are needed to determine whether the expression of activating NKR delineates the biologically and clinically distinct groups of PTNKL and to relate them to the pathogenesis.
CONFLICTS OF INTEREST
Authors declare no conflicts of interest for this article.
Uemura Y, Isobe Y, Uchida A, et al. Expression of activating natural killer‐cell receptors is a hallmark of the innate‐like T‐cell neoplasm in peripheral T‐cell lymphomas. Cancer Sci. 2018;109:1254–1262. https://doi.org/10.1111/cas.13512
Y Uemura and Y Isobe contributed equally to this work.
REFERENCES
- 1. Swerdlow SH, Campo E, Pileri SA, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127:2375‐2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. International T‐cell lymphoma project . International peripheral T‐cell and natural killer/T‐cell lymphoma study: pathology findings and clinical outcomes. J Clin Oncol. 2008;25:4124‐4130. [DOI] [PubMed] [Google Scholar]
- 3. Aoki R, Karube K, Sugita Y, et al. Distribution of malignant lymphoma in Japan: analysis of 2260 cases, 2001‐2006. Pathol Int. 2008;58:174‐182. [DOI] [PubMed] [Google Scholar]
- 4. Yamaguchi M, Kwong Y‐K, Kim WS, et al. Phase II study of SMILE chemotherapy for newly diagnosed stage IV, relapsed, or refractory extranodal natural killer (NK)/T‐cell lymphoma, nasal type: the NK‐cell Tumor Study Group Study. J Clin Oncol. 2011;29:4410‐4416. [DOI] [PubMed] [Google Scholar]
- 5. Pongpruttipan T, Sukpanichnant S, Assanasen T, et al. Extranodal NK/T‐cell lymphoma, nasal type, includes cases of natural killer cell and αβ, γδ, and αβ/γδ T‐cell origin: a comprehensive clinicopathologic and phenotypic study. Am J Surg Pathol. 2012;36:481‐499. [DOI] [PubMed] [Google Scholar]
- 6. Kato S, Asano N, Miyata‐Takata T, et al. T‐cell receptor (TCR) phenotype of nodal Epstein‐Barr virus (EBV)‐positive cytotoxic T‐cell lymphoma (CTL). A clinicopathologic study of 39 cases. Am J Surg Pathol. 2015;39:462‐471. [DOI] [PubMed] [Google Scholar]
- 7. Campbell KS, Hasegawa J. Natural killer cell biology: an update and future directions. J Allergy Clin Immunol. 2013;132:536‐544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fan X, Rudensky AY. Hallmarks of tissue‐resident lymphocytes. Cell. 2016;164:1198‐1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Seyda M, Elkhal A, Quante M, et al. T cells going innate. Trends Immunol. 2016;37:546‐556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Haedicke W, Ho FCS, Chott A, et al. Expression of CD94/NKG2A and killer immunoglobulin‐like receptors in NK cells and a subset of extranodal cytotoxic T‐cell lymphomas. Blood. 2000;95:3628‐3630. [PubMed] [Google Scholar]
- 11. Dukers DF, Vermeer MH, Jaspars LH, et al. Expression of killer cell inhibitory receptors is restricted to true NK cell lymphomas and a subset of intestinal enteropathy‐type T cell lymphomas with a cytotoxic phenotype. J Clin Pathol. 2001;54:224‐228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Freud AG, Zhao S, Wei S, et al. Expression of the activating receptor, NKp46 (CD335), in human natural killer and T‐cell neoplasia. Am J Clin Pathol. 2013;140:853‐866. [DOI] [PubMed] [Google Scholar]
- 13. van Dongen JJM, Langerak AW, Brüggemann M, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T‐cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED‐2 concerted action BMH4‐CT98‐3936. Leukemia. 2003;17:2257‐2317. [DOI] [PubMed] [Google Scholar]
- 14. Kanda Y. Investigation of the freely available easy‐to‐use software ‘EZR’ for medical statistics. Bone Marrow Transplant. 2013;48:452‐458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nikolova M, Musette P, Bagot M, et al. Engagement of ILT2/CD85j in Sézary syndrome cells inhibits their CD3/TCR signaling. Blood. 2001;103:1796‐1798. [DOI] [PubMed] [Google Scholar]
- 16. Naji A, Menier C, Maki G, et al. Neoplastic B‐cell growth is impaired by HLA‐G/ILT2 interaction. Leukemia. 2012;26:1889‐1892. [DOI] [PubMed] [Google Scholar]
- 17. Vo AV, Takenaka E, Shibuya A, et al. Expression of DNAM‐1 (CD226) on inflammatory monocytes. Mol Immunol. 2016;69:70‐76. [DOI] [PubMed] [Google Scholar]
- 18. Chen L, Xie X, Zhang X, et al. The expression, regulation and adhesion function of a novel CD molecule, CD226, on human endothelial cells. Life Sci. 2003;73:2373‐2382. [DOI] [PubMed] [Google Scholar]
- 19. Zhu S, Phatarpekar PV, Denman CJ, et al. Transcription of the activating receptor NKG2D in natural killer cells is regulated by STAT3 tyrosine phosphorylation. Blood. 2014;124:403‐411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Inghirami G, Chan WC, Pileri S, the AIRC 5xMILE consortium ‘genetics‐driven targeted management of lymphoid malignancies’ . Peripheral T‐cell and NK cell lymphoproliferative disorders: cell of origin, clinical and pathological implications. Immunol Rev. 2015;263:124‐159. [DOI] [PubMed] [Google Scholar]
- 21. Tsutsui M, Yasuda H, Suto H, et al. Frequent STAT3 activation is associated with Mcl‐1 expression in nasal NK‐cell lymphoma. Int J Lab Hematol. 2010;32:419‐426. [DOI] [PubMed] [Google Scholar]
- 22. Chen Y‐W, Guo T, Shen L, et al. Receptor‐type tyrosine‐protein phosphatase κ directly targets STAT3 activation for tumor suppression in nasal NK/T‐cell lymphoma. Blood. 2015;125:1589‐1600. [DOI] [PubMed] [Google Scholar]


