Abstract
Hypoxia plays a critical role in the metastasis of gastric cancer (GC), yet the underlying mechanism remains largely unclear. It is also not known whether long, noncoding RNAs (lncRNAs) are involved in the contribution of hypoxia to GC metastasis. In the present study, we found that lncRNA BC005927 can be induced by hypoxia in GC cells and mediates hypoxia‐induced GC cell metastasis. Furthermore, BC005927 is frequently upregulated in GC samples and increased BC005927 expression was correlated with a higher tumor‐node‐metastasis stage. GC patients with higher BC005927 expression had poorer prognoses than those with lower expression. Additional experiments showed that BC005927 expression is induced by hypoxia inducible factor‐1 alpha (HIF‐1α); ChIP assay and luciferase reporter assays confirmed that this lncRNA is a direct transcriptional target of HIF‐1α. Next, we found that EPHB4, a metastasis‐related gene, is regulated by BC005927 and that the expression of EPHB4 was positively correlated with that of BC005927 in the clinical GC samples assessed. Intriguingly, EPHB4 expression was also increased under hypoxia, and its upregulation by BC005927 resulted in hypoxia‐induced GC cell metastasis. These results advance the current understanding of the role of BC005927 in the regulation of hypoxia signaling and offer new avenues for the development of therapeutic interventions against cancer progression.
Keywords: EPHB4, gastric cancer, hypoxia, lncRNA, metastasis
1. INTRODUCTION
Gastric cancer (GC) is one of the most common human cancers: it is the third most common human cancer in the world, and the second most common cancer in China.1 Despite recent advances in clinical and experimental oncology, the prognosis of GC remains poor, with the 5‐year survival rate of patients with this type of cancer remaining relatively low.2 As the major cause of death is metastasis, which greatly hinders treatment success,3 better understanding of the mechanisms underlying GC metastasis is crucial for improving the diagnosis and treatment of GC patients. A hypoxic microenvironment plays a critical role in the metastasis and recurrence of certain cancer types, including GC, hepatocellular carcinoma (HCC) and colorectal cancer.4, 5, 6, 7, 8, 9 Indeed, it is well established that hypoxia contributes to metastasis by regulating the expression of various protein‐coding genes,10, 11 and our previous study also found that hypoxia could prompt gastric cancer metastasis and hypoxia inducible factor‐1 (HIF‐1) was involved in this process12, 13 Further study explored that HIF‐1 regulated protein‐coding genes by binding to hypoxia response elements (HRE) in gene promoters in hypoxic GC.14
However, only 2% of the human transcriptome is composed of protein‐coding mRNAs; most of the transcriptome comprises noncoding transcripts that yield long, noncoding RNAs (lncRNAs), which are defined as transcripts longer than 200 nucleotides (nt).15, 16, 17 Thus far, more than 10 000 lncRNAs have been reported as playing crucial roles in diverse cancer processes by various mechanisms, such as chromatin remodeling18, 19 and transcriptional regulation, in addition to aiding in the repair of DNA damage.20, 21 Although a number of lncRNAs have been reported to play critical roles in diverse processes, only a few examples of lncRNA‐mediated transcriptional regulation in hypoxia‐related cancer have been described. For example, Yang et al22 have shown that a hypoxic microenvironment suppresses lncRNA low expression in tumor (lncRNA‐LET), which is associated with HCC metastasis. In another study, Fan Yang et al23 found that lincRNA‐p21 is a hypoxia‐responsive lncRNA that is essential for hypoxia‐enhanced glycolysis. Therefore, an understanding of the roles and specific regulatory mechanisms of lncRNAs in hypoxia‐induced GC metastasis is urgently needed.
Using microarrays, we previously identified a small number of lncRNAs that are aberrantly expressed in GC under hypoxia compared with normoxia.24 In the present study, we describe our further analysis of one such lncRNA, BC005927, an 817‐bp, unspliced, polyadenylated transcript transcribed from the intron of the SERPINE1 gene on chromosome 7 that is upregulated by hypoxia, showing increased expression in hypoxia‐induced GC cells and GC tissues. We found that BC005927 promotes GC metastasis, whereas downregulation of BC005927 inhibits GC invasion in vitro and in vivo under hypoxic conditions. We further show that BC005927 is a direct transcriptional target of HIF‐1α through the interaction of its promoter region with a putative HIF‐1α response element. Moreover, we show that alteration of BC005927 expression influences the expression of EPHB4, which is located 300 kb upstream of BC005927, indicating that BC005927 affects GC cell invasion and metastasis partially through EPHB4. This study advances the current understanding of the roles of lncRNAs, which include regulation of the pathogenesis of hypoxia‐related GC, thus facilitating the development of lncRNA‐directed diagnostics and therapeutics.
2. MATERIALS AND METHODS
2.1. Ethics statement
For tissue specimens, signed informed consent was obtained from the patients who contributed dissected tissues. All GC cases were clinically and pathologically confirmed. Experimental procedures were approved by the Institutional Review Board of the Fourth Military Medical University. All animal experiments were carried out in accordance with national guidelines for the care and use of laboratory animals and with the approval of the Institutional Committee for Animal Research.
2.2. RNA extraction and quantitative real‐time PCR
Total RNA was extracted from cultured cells or tissues using TRIzol reagent. cDNA was synthesized using a PrimeScript RT Reagent Kit (TaKaRa, Dalian, China), and real‐time PCR was carried out using SYBR Premix Ex Taq II (TaKaRa) and measured using a CFX96TM Manager (Bio‐Rad, Hercules, CA, USA). PCR primers were purchased from TaKaRa. The primers are listed in Table S1. PCR conditions were as follows: 95°C for 5 seconds, 55°C for 30 seconds, and 72°C for 30 seconds for 45 cycles. β‐Actin expression was used as an internal control, the 2−ΔΔCt method was used for relative quantitation of gene expression levels, and all RT‐PCR were carried out in triplicate.
2.3. Clinical samples
Samples from 95 GC patients who had undergone gastrectomy with lymph node dissection for GC at Xijing Hospital between November 2007 and April 2009 were included in the present study (Table S2). None of the patients received preoperative chemotherapy. The resected specimens were histologically examined by hematoxylin‐eosin (HE) staining. Primary tumor samples and corresponding non‐tumor mucosa were collected from each patient immediately after the surgical process and were snap‐frozen in liquid nitrogen until further use. Total RNA from the frozen tissues was isolated with TRIzol (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions.
2.4. Western blotting
Cellular proteins were extracted using lysis buffer at 4°C on ice. Lysates were centrifuged at 9500 g for 10 minutes at 4°C, and the supernatants were collected. Western blotting was then done according to standard procedures.25 Rabbit anti‐EPHB4 monoclonal antibody used was obtained from Abcam (Cambridge, MA, USA) and mouse anti‐β‐actin monoclonal antibody from Sigma‐Aldrich (St Louis, MO, USA). Bands were detected using an ECL system (Santa Cruz Biotechnology, Santa Cruz, CA, USA), and β‐actin expression was used as an internal control.
2.5. Construct design and cell transfection
On the basis of the BC005927 sequence, 4 shRNAs were designed using small interfering RNA Target Finder (InvivoGen, San Diego, CA, USA) as follows: 5′‐CCACCAGTTACCTGCAATA‐3′, 5′‐GGAACAAAGATGGTTTCTA‐3′, 5′‐CCAAGACAAACACACTCAT‐3′, and 5′‐GATGAGCAGTGGTTTGAAA‐3′. Four shRNAs for HIF1α were designed using small interfering RNA Target Finder (InvivoGen) as follows: 5′‐CTGGGAATGACCGACATGT‐3′, 5′‐GCTCAGACCAACAATTTCA‐3′, 5′‐GCTGACAACAGGAGGAGAA‐3′, and 5′‐CCAGATTCATCATCAATGA‐3′. Lentiviral vectors encoding shRNAs or a nonsilencing control were generated using a GV248 vector (GeneChem Co., Ltd, Shanghai, China). Stable transfectants overexpressing BC005927 or EPHB4 were generated by lentiviral transduction using a GV166 vector (GeneChem Co., Ltd). An empty vector was used as a negative control. Stably transfected cells were selected with puromycin (Sigma‐Aldrich) and confirmed through fluorescence microscopy and RT‐PCR.
2.6. Chromatin immunoprecipitation assay
HIF‐1 binding to uc003uxs promoter was analyzed by ChIP on gastric cancer cells. SGC7901 cells exposed to hypoxic conditions (1% O2, 24 hours) were fixed with 1% paraformaldehyde, and chromatin derived from isolated nuclei was sheared by using a F550 microtip cell sonicator (Fisher Scientific). After centrifugation, supernatants containing sheared chromatin were incubated with an anti‐HIF‐1α antibody or control IgG. Protein A sepharose was then added, incubation was continued overnight, and immune complexes were subsequently eluted. Complexes were next treated with RNase and proteinase K and were extracted with phenol/chloroform and then with chloroform. DNA was precipitated, washed, dried, resuspended in water and analyzed by PCR. The primers were as follows: site 1 (sense, 5′‐CCCCGCTATTCCTCTATTTTCTTT‐3′ and antisense, 5′‐ACCATCCTCCCTGCTCTCCT‐3′) or site 2 (sense, 5′‐CTTCTTCCGCTCGACTTTC‐3′ and antisense, 5′‐TGACCGGCTTTCATCACTA‐3′).
2.7. In vitro migration and invasion assays
For transwell migration assays, 5 × 104 cells in serum‐free RPMI 1640 medium were added to the upper chamber of each insert (BD Biosciences, Franklin Lakes, NJ, USA). For invasion assays, the chamber inserts were coated with 50 mg/L Matrigel (BD Biosciences, San Jose, CA, USA). After 4 hours of incubation at 37°C, 1 × 105 cells in serum‐free RPMI 1640 medium were added to the upper chamber. For both assays, medium supplemented with serum was used as a chemoattractant in the lower chamber. After incubation in a normoxic (37°C and 5% CO2) or hypoxic (37°C, 1% O2, 5% CO2, and 94% N2) chamber for 24 or 48 hours, cells on the upper surface of the membrane were removed. The cells on the lower surface were fixed in 100% methanol for 15 minutes, air dried, stained with 0.1% crystal violet, and counted under a microscope (Olympus Corp., Tokyo, Japan) to calculate relative numbers. Nine random fields were analyzed per insert. Each experiment was conducted in triplicate in 3 independent experiments.
2.8. High‐content screening assay
Briefly, 5 × 103 cells were plated into each well of a 96‐well plate and incubated at 37°C. After 24 hours, the culture medium was replaced with serum‐free RPMI 1640 medium, and cells were cultured for an additional 24 hours. The cells were then washed twice with ice‐cold PBS and stained with Hoechst 33342 for 15 minutes in an incubator. The cells were subsequently washed twice with ice‐cold PBS, and culture medium was added to each well. Cell motility was detected with a Cellomics ArrayScan VTI HCS (Thermo Scientific, USA) according to the manufacturer's instructions (5 replicate wells per group).
2.9. In vivo metastasis assays
Nude mice were purchased from the Experimental Animal Center of the Fourth Military Medical University. For in vivo metastasis assays, 2 × 106 SGC7901 infected with a lentivirus containing BC005927 siRNA, MKN28 with a lenti‐BC005927 or corresponding negative control were suspended in 0.2 mL PBS and injected into the tail vein of each mouse. After 8 weeks, the mice were killed, and their tumor nodules were counted under a stereomicroscope (Olympus Corp.). The tumor tissues derived from various organs were then dissected and histologically examined. Each tumor cell line was injected into 10 mice.
2.10. Luciferase assay
To investigate whether BC005927 is transcriptionally regulated by HIF‐1α, SGC7901 cells expressing either control shRNA or HIF‐1α shRNA were transfected with a pGL3‐based construct containing the BC005927 promoter plus a Renilla luciferase plasmid. Twenty‐four hours later, the cells were cultured under normoxic or hypoxic conditions for another 24 hours. Reporter activity was measured using a luciferase assay kit (Promega, Madison, WI, USA) and plotted after normalization with respect to Renilla luciferase activity (mean ± SD).
2.11. Bisulfite sequencing PCR analyses
Genomic DNA was extracted from GC cells with the QIAamp DNA Mini Kit (Qiagen, Germantown, MD, USA) and subjected to bisulfite modification using an EpiTect® Bisulfite kit (Qiagen) according to the manufacturer's protocol. PCR was carried out and the PCR product was ligated into T Vector. After transformation, individual colonies were selected, and the insert was sequenced and analyzed by BiQ_Analyzer.
2.12. Statistical analysis
The SPSS 12.0 program (SPSS Inc., Chicago, IL, USA) was used for statistical analysis. Data are presented as mean ± standard error for at least 3 independent experiments. Differences between groups were analyzed using Student's t test when comparing only 2 groups or a one‐way analysis of variance when comparing more than 2 groups. The chi‐squared test was used to analyze the relationships between EPHB4 expression and various clinicopathological characteristics. BC005927 and EPHB4 expression levels in clinical GC tissues and corresponding adjacent non‐tumorous tissues were compared using Wilcoxon signed‐rank test, and correlations between BC005927 and EPHB4 expression in the tissue specimens were explored using Pearson's correlation. P < .05 was considered statistically significant.
3. RESULTS
3.1. Aberrant expression of BC005927 lncRNA in hypoxia‐induced GC cells and GC tissues
In our previous study, we found that lncRNA BC005927, which is an 817‐bp, exon sense‐overlapping lncRNA on chromosome 7, was upregulated by hypoxia comparing hypoxia‐induced GC to normoxia conditions using microarrays.24 Bioinformatics analysis showed promoter regions of lnc‐BC005927 containing HRE. We then carried out real‐time RT‐PCR to examine expression levels of this lncRNA in GC cells under both normoxic and hypoxic conditions (after 8, 16, 24, and 48 hours in 1% O2). We found, compared to expression under normoxic conditions, BC005927 was strongly induced by hypoxia in SGC7901 cells, MKN45 cells and MKN28 cells in a time‐dependent method (Figure 1A1, A2, A3). These data indicate the specificity of BC005927 in hypoxic induction. The results suggest that BC005927 can, indeed, be regulated by hypoxia in GC cells.
Figure 1.
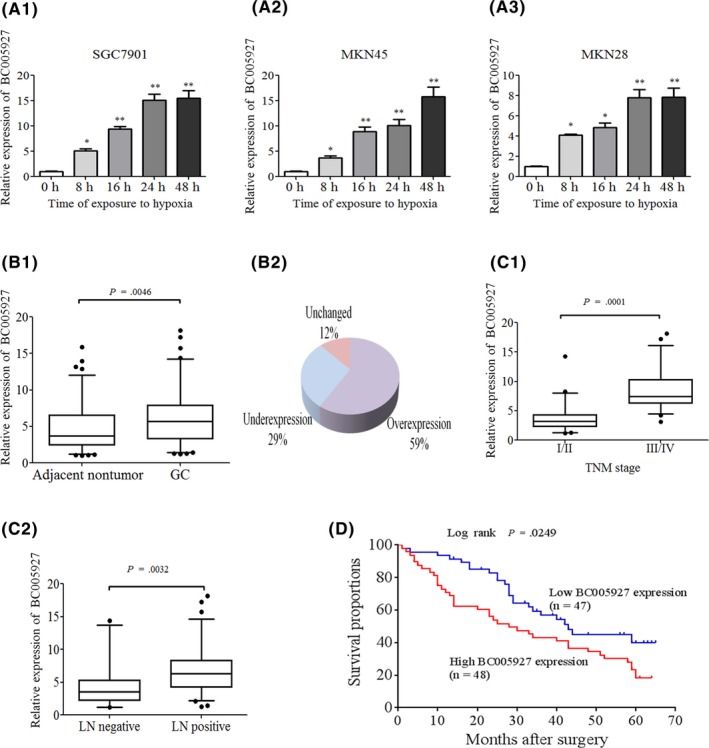
Aberrant expression of BC005927 long, non‐coding RNA (lncRNA) in hypoxia‐induced gastric cancer (GC) cells and GC tissues. A, lncRNA‐BC005927 is upregulated under hypoxic culture conditions (1% O2, 5% CO 2, 94% N2) in (A1) SGC7901 cells, (A2) MKN45 cells and (A3) MKN28 cells. B1, B2, C1, C2, lncRNA‐BC005927 expression was assessed by real‐time PCR in 95 pairs of human GC and adjacent tissues. β‐Actin was used as an internal control. Significant differences between samples were analyzed using the Wilcoxon signed‐rank test (P = .002, n = 95). (D) Kaplan ‐Meier's analysis of the correlation between BC005927 expression and overall survival (OS) of GC patients. In all panels, results are representative of at least 3 independent experiments *P < .05, **P < .01
To further study BC005927 expression in tumor tissues in vivo, we examined a panel of paired tumor and normal primary tissue specimens that were collected from patients with GC (n = 95). BC005927 transcripts were found to be expressed at higher levels in tumor tissues compared to non‐tumor tissues obtained from the same donor after normalization to β‐actin expression (P = .0046, Figure 1B1). Overall expression level of BC005927 increased in 56 GC samples (58.95%), was unchanged in 11 samples and was downregulated in 28 samples (Figure 1B2), indicating that upregulation of BC005927 is a frequent event in GC. We next examined the relationship between BC005927 expression levels and the clinicopathological characteristics of 95 tumor tissue samples. As shown in Figure 1C1 and C2, BC005927 overexpression was correlated with advanced clinical tumor‐node‐metastasis stage (P = .0001) and lymph node metastasis (P = .0032). BC005927 expression in GC patients was not found to be correlated with age, gender, tumor size or cell differentiation (data not shown). Moreover, we examined whether the level of BC005927 expression was associated with survival in GC patients. Patients were subsequently divided into high expression (n = 48) and low expression groups (n = 47) based on BC005927 expression levels greater (equal) or less than the median expression (5.676). Kaplan‐Meier survival analyses showed that patients with high BC005927 expression had a shorter survival time than those with low BC005927 expression (Figure 1D). Therefore, the high expression of this lncRNA might be associated with GC metastasis.
3.2. BC005927 is induced by HIF‐1α, and HIF‐1α interacts with the HIF‐1α response element in the BC005927 promoter region
To determine whether BC005927 functions through regulating HIF‐1 transcriptional activity by binding to the HIF‐1 response element, we analyzed the promoter region of BC005927 and inspected the genomic sequence upstream of the gene encoding this lncRNA by bioinformatics analysis (Genomatix Software Suite for sequence analysis, MatInspector). Two putative HRE (5‐RCGTG‐3) were found within the promoter region (Figure 2A), so we first speculated whether HIF‐1α could modulate BC005927 expression at the transcriptional level. RT‐PCR results showed that knockdown of HIF‐1α in SGC7901 and MKN45 cells greatly attenuated hypoxia‐induced BC005927 upregulation (Figure 2B, C1, C2). Conversely, ectopic expression of HIF‐1α significantly increased BC005927 expression in MKN28 cells (Figure 2D1, D2). These data indicate that HIF‐1α might be responsible for hypoxia‐induced BC005927 expression.
Figure 2.
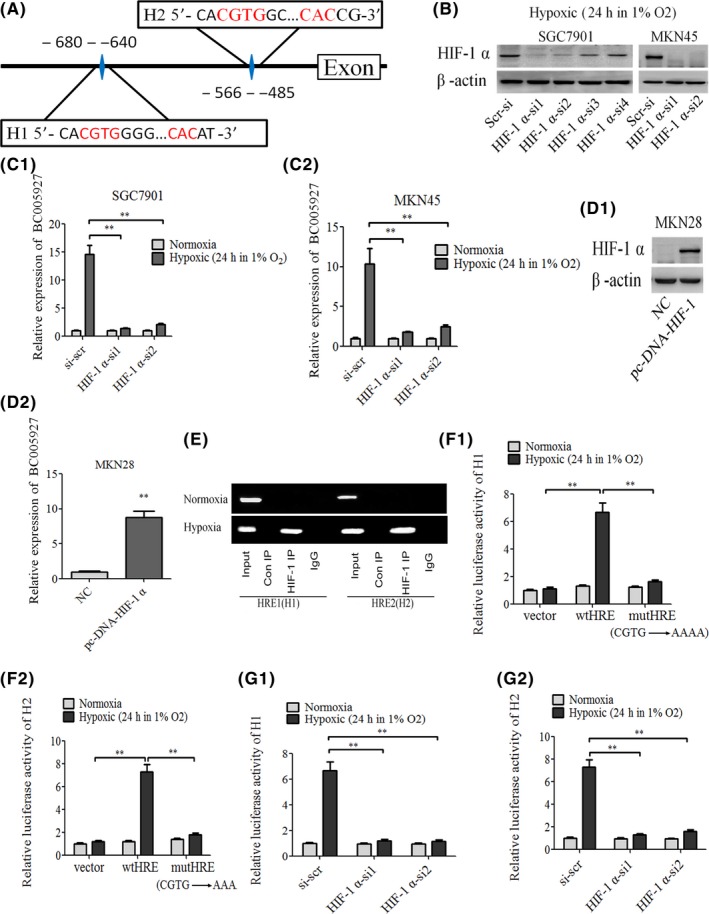
BC005927 is induced by hypoxia inducible factor‐1 alpha (HIF‐1α), and HIF‐1α interacts with the HIF‐1α response element in the BC005927 promoter region. A, Schematic illustration of consensus HIF‐1α response element in BC005927 gene promoter. B, C1, C2, SGC7901, MKN45 cells expressing control shRNA, HIF‐1α shRNAs were cultured under hypoxic conditions for 24 hours. Cell lysates and total RNA were subjected to western blot and real‐time RT‐PCR analyses, respectively. Data shown are mean ± SD (n = 3). D1, D2, Cell lysates and total RNA of MKN28 cells expressing NC (negative control) or pc‐DNA‐HIF‐1α were subjected to western blot and real‐time RT‐PCR analyses, respectively. Data shown are mean ± SD (n = 3). E, SGC7901 cells were cultured under normoxic or hypoxic conditions for 24 hours. Lysates were then subjected to ChIP assay. ChIP products were amplified by PCR reaction. F1, F2, SGC7901 cells were cotransfected with the indicated reporter constructs and Renilla luciferase plasmid. Twenty‐four hours after transfection, cells were cultured under normoxic or hypoxic conditions for 24 hours. Reporter activity was then measured and plotted after normalizing with respect to Renilla luciferase activity (mean ± SD). G1, G2, SGC7901 cells expressing control shRNA, HIF‐1α‐si1, or HIF‐1α‐si2 were cotransfected with the hypoxia response element (HRE)‐WT reporter constructs and Renilla luciferase plasmid. Twenty‐four hours after transfection, cells were cultured under normoxic or hypoxic conditions for 24 hours. Reporter activity was then measured and plotted after normalizing with respect to Renilla luciferase activity (mean ± SD). In all panels, results are representative of at least 3 independent experiments. **P < .01
Next, ChIP assays verified the association of HIF‐1α and the chromatin fragments corresponding to the two HRE within the BC005927 gene (Figure 2E). In addition, we used the HRE‐luciferase reporter system to examine how lnc affects HIF‐1‐regulated luciferase expression. DNA fragments containing wild‐type or mutant HRE were inserted into the promoter region of a firefly luciferase reporter plasmid. As expected, luciferase expression from the wild type, but not the mutant reporter, was markedly induced by hypoxia (Figure 2F1, F2). This hypoxia‐induced luciferase expression, however, was greatly inhibited by knockdown of HIF‐1α (Figure 2G1, G2). These data show that BC005927 is transcriptionally upregulated by HIF‐1α.
3.3. BC005927 is involved in cell invasion of GC cells and mediates hypoxia‐induced invasion of GC cells
To investigate the role of BC005927 in GC metastasis and invasion, the GC cell lines SGC7901 and MKN45 were transfected with BC005927‐specific shRNAs or a control shRNA. Four different shRNAs targeting BC005927 were tested, and the two that most effectively knocked down its expression (BC005927‐si1 and BC005927‐si2) were selected for subsequent studies. The MKN28 cell line was chosen for experiments assessing the upregulation of BC005927 expression. MKN28 cells were infected with a recombinant lentivirus expressing BC005927 (LV‐BC005927) or a control lentivirus (LV‐NC), and stable clones, MKN28‐BC005927 and MKN28‐NC, were established. Upregulation and knockdown of BC005927 expression was confirmed by qRT‐PCR (Figure 3A1, B1, C1).
Figure 3.
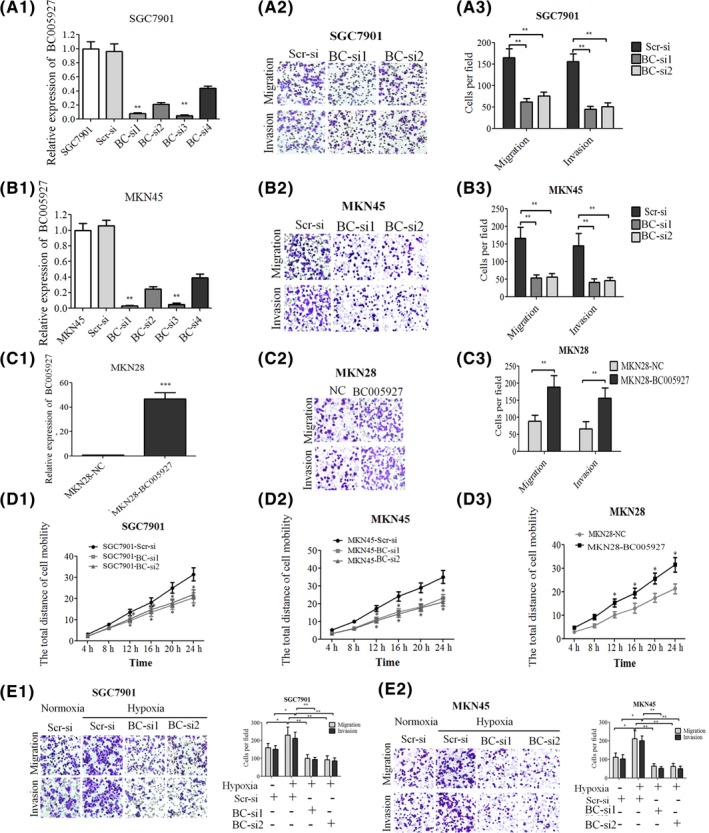
BC005927 is involved in cell invasion of gastric cancer (GC) cells and mediates hypoxia‐induced invasion of GC cells. SGC7901 and MKN45 cells transfected with lenti/si‐BC005927. A1, B1, MKN28 cells transfected with lenti‐BC005927 or (C1) NC were confirmed by real‐time PCR. Transwell migration and invasion assays of (A2, A3) SGC7901 and (B2, B3) MKN45 cells were carried out after transfection with (C2, C3) BC005927 BC‐si, MKN28 cells after transfection with BC005927. D1, D2, D3, Cell mobility was examined with a Cellomics ArrayScan VTI 1700 Plus. In all panels, results are representative of at least 3 independent experiments. E1, E2, Representative images and number of metastatic or invasive BC005927‐downregulated SGC7901 and MKN45 cells under normoxic or hypoxic conditions. In all panels, results are representative of at least 3 independent experiments
Given the intronic position of BC005927 within SERPINE1, we next aimed to determine the effect of BC005927 knockdown/upregulation on SERPINE1 expression in GC cells. As a result, we found that the level of SERPINE1 expression was not significantly altered following BC005927 knockdown or following overexpression of BC005927 relative to the negative control (Figure S1). This result confirms that the knockdown/knockup strategy did not appreciably affect the levels of the host SERPINE1 transcript and that the phenotypic effects observed following knockdown/knockup of BC005927 were driven directly by BC005927.
We then determined whether BC005927 could affect normoxic GC cell migration and invasion. Transwell assays with or without Matrigel indicated that SGC7901 and MKN45 cells with stable BC005927 knockdown showed significantly decreased migration and invasion compared with control cells (Figure 3A2, A3, B2, B3). Moreover, high‐content screening results showed that cell motility was reduced in the BC005927 knockdown cells (Figure 3D1, D2). Conversely, upregulation of BC005927 expression in MKN28 cells significantly enhanced cell migration, invasion and motility (Figure 3C2, C3, D3). We found that expression of BC005927 in MKN28 cells did not induce tumor cell growth, and knockdown of BC005927 expression in SGC7901 and MKN45 cells did not significantly reduce cell viability (data not shown).
Given that BC005927 can be upregulated by hypoxia, we hypothesized that BC005927 might have a role in hypoxia‐induced migration and invasion. To test this hypothesis, we first determined the migration and invasion ability of GC cells under hypoxic conditions and found that hypoxia significantly increased the metastasis and invasion potentials of SGC7901 and MKN45 cells. Upon the knockdown of BC005927, SGC7901 and MKN45 metastasis and invasion abilities induced by hypoxia were dramatically diminished (Figure 3E1, E2).
3.4. BC005927 promotes the metastasis and invasion of GC cancer cells in vivo
To further explore the roles of BC005927, we injected MKN28‐BC005927, SGC7901‐BC005927 si1, SGC7901‐BC005927 si2 or the corresponding control cells into nude mice through the lateral tail vein. Histological analyses (Figure 4C1, C2) confirmed that the incidence of lung and liver metastases was dramatically increased in the mice injected with MKN28‐BC005927 and dramatically decreased in those injected with SGC7901‐BC005927 si1 and SGC7901‐BC005927 si2 compared with the control group. Number of metastatic lung and liver nodules was significantly increased in the MKN28‐BC005927 groups and significantly decreased in the SGC7901‐BC005927 si1 and SGC7901‐BC005927 si2 groups compared with the control group (Figure 4A, B1, B2). Taken together, these observations suggest that BC005927 is a positive metastatic regulator of GC.
Figure 4.
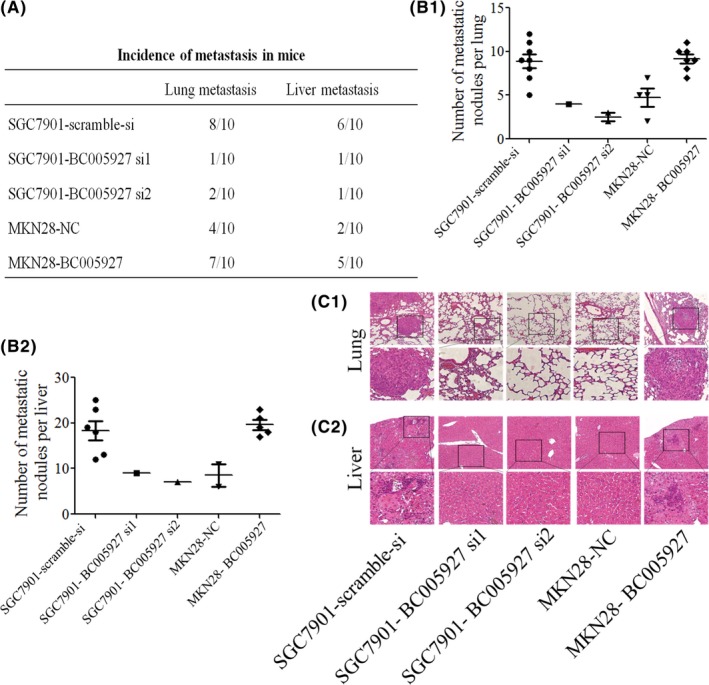
BC005927 promotes the metastasis and invasion of gastric cancer (GC) cells in vivo. SGC7901 cells transfected with BC005927‐si1, BC005927‐si2 or a negative control, MKN28 cells transfected with the LV‐BC005927‐expression vector or the control vector were injected into nude mice by the tail vein for an in vivo metastasis assay. Animals were killed 6 weeks after injection. A, Incidence of lung and liver metastasis in each group of nude mice. B1, Number of metastatic lung foci observed in each group. B2, Number of metastatic liver foci observed in each group. C1, Images showing representative hematoxylin and eosin (HE) staining of lung tissue samples from the different experimental groups. C2, Images showing representative HE staining of liver tissue samples from the different experimental groups. In all panels, the results are representative of at least 3 independent experiments
3.5. Association of BC005927 with EPHB4
To explore the mechanism by which BC005927 promotes GC cell migration and invasion, we attempted to identify the potential target genes of lncRNA. Recently, several studies have found that many lncRNAs are involved in molecular regulation pathways through interactions with neighboring proteins. Thus, we sought to identify proteins associated with this lncRNA using a bioinformatics analysis. We retrieved genomic locus information from the UCSC genome browser (http://genome.ucsc.edu/) and found that the tumor oncogene EPHB4, a metastasis‐associated gene, is located 300 kb upstream of BC005927 (both of them located within chr7 q22.1) (Figure 5A), and we also found another tumor‐related gene TRIP6 nearby. To further validate the association between lncRNA BC005927 and EPHB4 or TRIP6, we carried out RT‐PCR and western blotting using SGC7901 and MKN45 cells with downregulated BC005927 and MKN28‐BC005927 cells. TRIP6 expression was unchanged (data not shown), whereas we observed significantly decreased EPHB4 expression in the BC005927 shRNA‐transfected cells and increased expression in BC005927‐overexpression cells compared with the control cells (Figure 5B1, B2, C1, C2).
Figure 5.
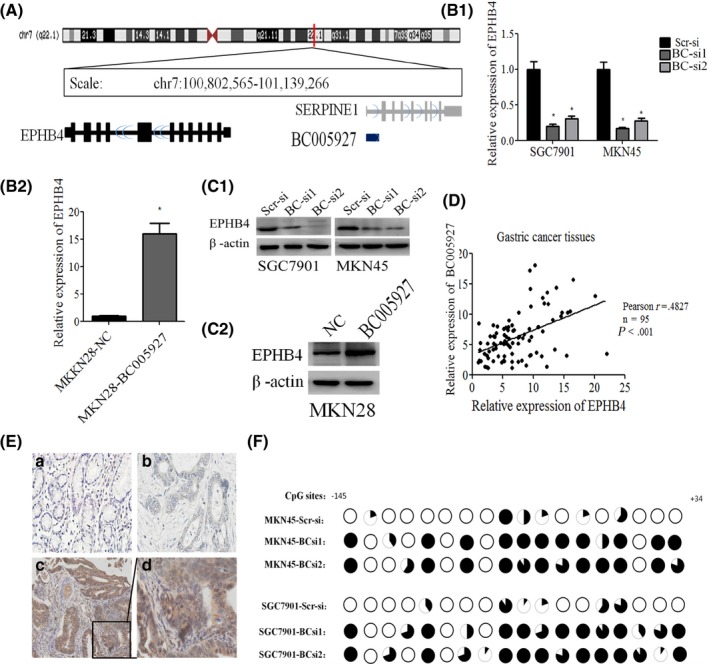
Association of BC005927 with EPHB4. A, Diagram of genes located around BC005927. B1, B2, C1, C2, mRNA and protein levels of EPHB4 were assessed by RT‐PCR and western blotting in SGC7901 and MKN45 cells transfected with an BC005927 siRNA‐lentivirus, MKN28 cells transfected with BC005927 lentivirus or a negative control (NC). D, Correlation between relative mRNA levels of BC005927 and EPHB4 in 95 gastric cancer (GC) tissue specimens. E, Immunohistochemical analysis of EPHB4 in metastatic and nonmetastatic GC: (a) noncancerous region of GC, (b) primary site of nonmetastatic GC, and (c,d) primary site of metastatic GC. F, Methylation mapping of 18 CpG island in EPHB4 promoter region obtained from bisulfite sequencing in scrambled siRNA control MKN45 or SGC7901 cells, and MKN45 or SGC7901cells infected with BC005927 siRNA. Each circle in the figure represents a single CpG site. For each cell line, percentage methylation at a single CpG site is calculated from the sequencing results of 10 independent clones. Black circles, 100% methylated; white circles, 0% methylation. In all panels, results are representative of at least 3 independent experiments.*P < .05
Moreover, we found that the expression of EPHB4 was positively correlated with BC005927 expression in GC tissues (Figure 5D). Immunohistochemistry results showed strong staining of EPHB4 at primary sites in the samples obtained from patients with metastatic GC compared with those from patients with nonmetastatic GC (Figure 5E). In addition, increased EPHB4 expression in the GC tissues was significantly correlated with TNM stage and lymph node metastasis, but was not associated with other parameters, such as gender, age or GC cell differentiation (Table S3).
3.6. Upregulation of EPHB4 is potentially involved in the oncogenic function of BC005927
As a potential downstream target of BC005927, we next set out to validate whether EPHB4 could also affect GC cell metastasis and invasion. GC cell lines SGC7901 and MKN45 were transfected with EPHB4‐specific shRNAs or a control shRNA. MKN28 cells were infected with a recombinant lentivirus expressing EPHB4 (LV‐EPHB4) or a control lentivirus (LV‐NC), and the stable clones MKN28‐EPHB4 and MKN28‐NC were established. Upregulation and knockdown of EPHB4 expression were confirmed by qRT‐PCR and western blot (Figure 6A1, A2, B1). Transwell assays showed that EPHB4 downregulation significantly impaired GC cell migration and invasion (Figure 6C1, C2), whereas its upregulation increased GC cell migration and invasion (Figure 6C3).
Figure 6.
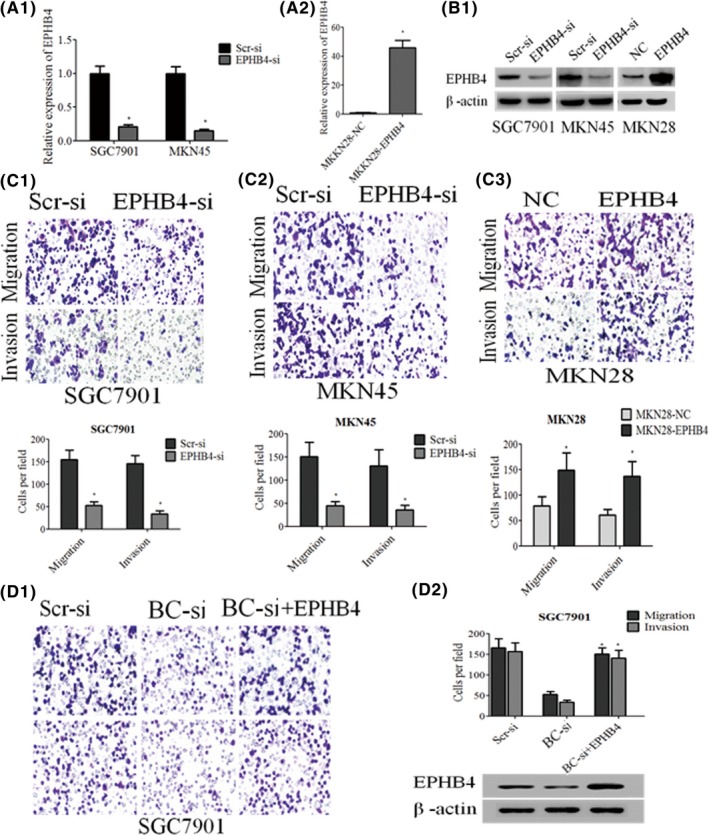
Upregulation of EPHB4 is potentially involved in the oncogenic function of BC005927. A1, A2, B, EPHB4 mRNA and protein levels were detected by qRT‐ PCR and western blot analysis. C1, C2, C3, Transwell assays were conducted to determine the migratory and invasive abilities of si‐BC005927‐transfected SGC7901 and MKN45 cells, BC005927‐overexpression MKN28 cells. D1, D2, Transwell assays were conducted to determine the migratory and invasive abilities of si‐NC, si‐BC005927, or both si‐BC005927 and EPHB4‐transfected SGC7901 cells. D2, Western blot analysis of EPHB4 expression. In all panels, results are representative of at least 3 independent experiments.*P < .05
We next carried out rescue experiments to investigate whether EPHB4 is involved in the BC005927‐induced increase in GC cell metastasis and invasion. SGC7901 cells were cotransfected with si‐BC005927 and EPHB4‐expressing lentivirus, and this was shown to rescue the decreased expression of EPHB4 induced by the knockdown of BC005927 (Figure 6D2, lower panel). Moreover, a transwell assay indicated that this cotransfection could partially rescue si‐BC005927‐impaired migration and invasion in SGC7901 cells (Figure 6D1, D2, upper panel). These data indicate that BC005927 promotes GC cell metastasis and invasion through upregulation of EPHB4 expression.
Next, to investigate the potential mechanism by which BC005927 induces the expression of EPHB4 in GC cells, we focused on DNA methylation of EPHB4, because lncRNAs are known to be involved in epigenetic regulation. Then, we carried out bisulfite sequencing of cloned alleles over the region of −145 to +34 of the EPHB4 promoter encompassing 18 CpG sites.26 Results showed that EPHB4 CpG islands were densely hypomethylated in control SGC7901‐si‐Scr and MKN45‐si‐Scr cells, but GC cells transduced with BC005927 siRNA showed more methylated CpG dinucleotides (Figure 5F).
Taken together, these results show that knockdown of BC005927 can downregulate the expression of EPHB4 in GC cells by regulating EPHB4 DNA methylation, which illustrates that EPHB4 may act as a downstream target of BC005927 in GC cells.
3.7. Upregulation of EPHB4 by BC005927 mediates hypoxia‐induced GC metastasis
To determine whether EPHB4 is involved in BC005927‐induced GC cell metastasis under hypoxia upregulated by lncRNA BC005927 in vitro, we measured its expression levels in BC005927‐downregulated GC cells compared with control cells under hypoxia. Western blot results showed that EPHB4 expression increased under hypoxia compared with normoxia. This increasing trend was abrogated following BC005927 knockdown (Figure 7A1, A2), indicating that overexpression of EPHB4 is mediated by BC005927 upregulation under hypoxia.
Figure 7.
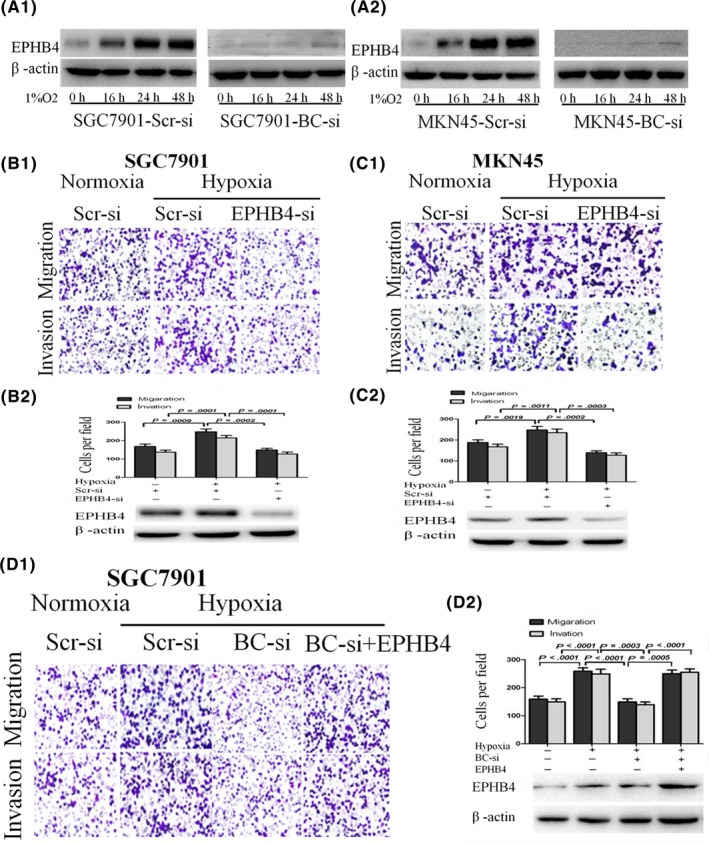
Upregulation of EPHB4 by BC005927 mediates hypoxia‐induced gastric cancer (GC) metastasis. A1, A2, Protein levels of EPHB4 in SGC7901 and MKN45 cells. The cells were transfected with a BC005927 siRNA‐lentivirus or negative control (NC) and then exposed to hypoxia after transfection. The cells were then collected and subjected to western blot analysis at specific time points, as indicated. β‐Actin served as an internal control. B1, C1, Transwell migration and invasion assays of SGC7901‐NC and SGC7901‐si‐EPHB4 cells and MKN45‐NC and MKN28‐si‐EPHB4 cells were carried out under normoxic or hypoxic conditions. B2, C2, Protein levels of EPHB4 were determined by western blot assays. D1, D2, Transwell migration and invasion assays of SGC7901 cells were carried out after transfection with a BC005927 siRNA‐lentivirus, EPHB4 expression vector, or negative control (hypoxia) under normoxic or hypoxic conditions. Protein levels of EPHB4 were determined by western blot assays. In all panels, results are representative of at least 3 independent experiments
Moreover, we conducted transwell experiments to investigate the role of EPHB4 in hypoxic GC. Results showed that the metastasis and invasion abilities of GC cells induced by hypoxia were dramatically diminished following the downregulation of EPHB4 expression (Figure 7B1, B2, C1, C2). To further examine the function of EPHB4 in BC005927‐induced GC cell metastasis and invasion, SGC7901 were cotransfected with EPHB4‐expressing vector and si‐BC005927, and transwell assays showed that impairment of the effects of siRNA‐mediated BC005927 on hypoxia‐induced GC cell migration and invasion were partially relieved by EPHB4 but not by the negative control (Figure 7D1, D2). Taken together, these results indicate that EPHB4 expression is increased under hypoxia and that EPHB4 upregulation by BC005927 mediates hypoxia‐induced GC metastasis.
4. DISCUSSION
It has been well established that hypoxia contributes to GC metastasis and invasion by regulating the expression of various protein‐coding genes.12, 13 However, approximately 97%‐98% of transcriptional gene products do not encode proteins; these ncRNAs have biological functions in cancer development, fetal development, and cell growth.27, 28 Of all the studied non‐coding genes, lncRNAs have been garnering increasing interest. Recently, several studies have indicated functional roles of lncRNAs in tumorigenesis.29, 30, 31, 32, 33 However, hypoxic lncRNAs are still an emerging research topic, and only a handful of lncRNAs are known to be involved in the development of disease under hypoxia. For example, Yang et al22 showed that a hypoxic microenvironment suppresses lncRNA‐LET, which is associated with HCC metastasis. In another study, Yang et al23 found that lincRNA‐p21 is a hypoxia‐responsive lncRNA that is essential for hypoxia‐enhanced glycolysis. However, whether lncRNA is also involved in hypoxic GC has remained unknown to date.
In a previous study, we carried out a microarray analysis and identified a small number of lncRNAs that are aberrantly expressed in human hypoxia‐related GC cells compared to normoxia‐related GC cells.24 In this study, among the upregulated lncRNAs, we found that lncRNA BC005927, an 817‐bp, unspliced, polyadenylated transcript transcribed from the intron of the SERPINE1 gene on chromosome 7, is overexpressed in hypoxia‐induced GC cells. Further examination showed that the average level of BC005927 in GC tissues is significantly higher than that in corresponding non‐tumor tissues. The high expression level of BC005927 in GC patients was also found to be associated with advanced tumor TNM stage and lymph node metastasis. These results indicate that BC005927 may play vital roles in the metastasis and invasion of GC and may have important functions in GC development and progression. However, the mechanism underlying the high expression of BC005927 under hypoxia is unknown. Through bioinformatics analysis, we found that the BC005927 promoter contains a conserved HIF‐1α‐binding site. In addition, our results showed that BC005927 is a direct transcriptional target of HIF‐1α. Our results indicate that the absence of HIF‐1α expression may contribute to the downregulation of BC005927 in GC.
Applying loss‐of‐function and gain‐of‐function approaches, we further identified that lncRNA BC005927 plays a key role in GC cell metastasis and invasion under hypoxia, indicating that it is a hypoxia‐inducible lncRNA that is essential for the hypoxia‐enhanced metastasis and invasion of GC.
Recent studies have indicated that lncRNAs can function by regulating neighboring protein‐coding genes. For example, Li et al34 showed that linc‐POU3F3 promotes cell viability and proliferation in ESCC cells by regulating the expression of the POU3F3 gene, which is located 4 kb downstream of linc‐POU3F3. In another study, Du et al35 found that lncRNA‐HULC can downregulate its neighboring gene p18 and promote hepatoma cell proliferation. In the present study, using a bioinformatics analysis, we found that EPHB4 is located 300 kb upstream of lncRNA BC005927. EPHB4 is a member of the Eph receptors, the largest subfamily of receptor tyrosine kinases (RTK), and was initially isolated from a human hepatocellular carcinoma cell line Hep3Ba.36 Recent studies have shown that expression of EPHB4 is increased in many types of cancer, including prostate,37 ovarian,38, 39 glioma,40 bladder,41 prostate,42 breast,43 colon,44, 45 and pancreatic cancer.46 Various studies exploring the role of EPHB4 overexpressed in cancer cells have suggested that EPHB4 can promote tumor development by stimulating angiogenesis,47, 48 increasing cancer cell survival,43, 49, 50 and facilitating invasion and migration.51, 52 Overexpression of EPHB4 in the mammary epithelium of MMTV‐neuT transgenic mice accelerated the onset of tumor formation and increased metastasis to the lungs.53 In xenograft models of breast and prostate cancer, knockdown of EPHB4 significantly inhibits tumor growth indicating that EPHB4 directly contributes to tumor progression.42, 43 Similarly, targeting EPHB4 using a monoclonal antibody also significantly reduced tumor growth in vivo.39 EPHB4 overexpression also plays an important role in gastric carcinogenesis in Chinese patients. However, the specific function of EPHB4 in gastric carcinogenesis requires further study to be completely understood.54
In our study, we found that EPHB4 was highly expressed at primary sites in patients with metastatic GC compared to those with nonmetastatic GC; increased EPHB4 expression in GC tissues was also significantly correlated with TNM stage and lymph node metastasis. Further experiments showed that knockdown of EPHB4 inhibited GC cell metastasis and invasion, whereas its upregulation impaired cell metastasis and invasion. These results indicate that the EPHB4 gene is a metastasis‐associated gene in GC.
Moreover, we found that expression of EPHB4 is positively correlated with BC005927 expression in GC tissues. Accordingly, we also found that BC005927 knockdown could downregulate EPHB4 expression at the mRNA and protein levels, and upregulation of BC005927 enhanced EPHB4 expression. More importantly, the upregulation of EPHB4 was able to reverse the inhibition of GC cell migration and invasion as a result of the knockdown of BC005927 expression. To further explore the relationship between EPHB4 and BC005927 under hypoxia, we investigated the expression of EPHB4 in GC cells with reduced BC005927 expression and control cells under hypoxia. Results showed that EPHB4 expression was increased under hypoxia, although this trend of upregulation disappeared following the knockdown of BC005927. Moreover, transwell experiments showed that EPHB4 mediates GC cell metastasis and invasion under hypoxia, with an increase in EPHB4 expression reversing the impairment of GC cell migration and invasion mediated by reduced BC005927 expression. Therefore, these data confirm that BC005927 is a key regulatory factor of GC that regulates EPHB4 expression.
Recently, increasing evidence confirmed that lncRNA can regulate DNA methylation of protein‐coding genes during the development of disease,55, 56, 57, 58 and several studies reported that EPHB4 expression is activated by demethylation of the EPHB4 CpG island in the development of a variety of cancers.59 Interestingly, using BSP assays, we observed a robust increase in levels of DNA methylation of EPHB4 following depletion of BC005927.
Expression of EPHB4 was significantly increased in hypoxic GC cells and tissues, which was at least partially mediated by BC005927, suggesting that its upregulation may be a key factor for the development of GC and may indicate a higher risk of metastasis. We show here that BC005927 possibly regulates the invasive and metastatic abilities of GC cells under hypoxia, partially through its regulation of EPHB4. Further studies are needed to investigate the exact mechanism of EPHB4 DNA demethylation that is induced by BC005927. Our findings further the understanding of GC pathogenesis and will facilitate the development of lncRNA‐directed diagnostics and therapeutics for the treatment of cancer.
CONFLICTS OF INTEREST
Authors declare no conflicts of interest for this article.
Supporting information
ACKNOWLEDGMENTS
The authors would like to thank Xijing Hospital (Xi'an, China) for specimens. The study was funded by National Natural Science Foundation of China (No. 81401918, 81602170, 81472301, 81472300, 81272349, 81502102, 81372608 and 81302164), the Science and Technology Plan Project of Guangdong Province (No. 2013B021800049), the Natural Science Foundation of Guangdong Province (No. 2014A030313595), and the Science and Technology Plan Project of Guang Zhou City (No. 201607010077).
Liu X, Wang Y, Sun L, et al. Long noncoding RNA BC005927 upregulates EPHB4 and promotes gastric cancer metastasis under hypoxia. Cancer Sci. 2018;109:988–1000. https://doi.org/10.1111/cas.13519
Funding information
The Science and Technology Plan Project of Guang Zhou City (Grant/Award Number: ‘201607010077’), the Science and Technology Plan Project of Guangdong province (Grant/Award Number: ‘2013B021800049’), the Natural Science Foundation of Guangdong province (Grant/Award Number: ‘2014A030313595’), National Natural Science Foundation of China (Grant/Award Number: ‘81401918, 81602170, 81472301, 81472300, 81272349, 81502102, 81372608, 81302164’).
Xiangqiang Liu, Yafang Wang, and Li Sun contributed equally to this work.
Contributor Information
Yongan Zhou, Email: zhou.yongan@163.com.
Lili Liu, Email: lily123fmmu@163.com.
REFERENCES
- 1. Pogorzelski M, Ting S, Gauler TC, et al. Impact of human papilloma virus infection on the response of head and neck cancers to anti‐epidermal growth factor receptor antibody therapy. Cell Death Dis. 2014;5:e1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277‐300. [DOI] [PubMed] [Google Scholar]
- 3. Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127:679‐695. [DOI] [PubMed] [Google Scholar]
- 4. Wang JR, Gan WJ, Li XM, et al. Orphan nuclear receptor Nur77 promotes colorectal cancer invasion and metastasis by regulating MMP‐9 and E‐cadherin. Carcinogenesis. 2014;35:2474‐2484. [DOI] [PubMed] [Google Scholar]
- 5. Fu J, Chen Y, Cao J, et al. p28GANK overexpression accelerates hepatocellular carcinoma invasiveness and metastasis via phosphoinositol 3‐kinase/AKT/hypoxia‐inducible factor‐1alpha pathways. Hepatology. 2011;53:181‐192. [DOI] [PubMed] [Google Scholar]
- 6. Sun HX, Xu Y, Yang XR, et al. Hypoxia inducible factor 2 alpha inhibits hepatocellular carcinoma growth through the transcription factor dimerization partner 3/ E2F transcription factor 1‐dependent apoptotic pathway. Hepatology. 2013;57:1088‐1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Miyake S, Kitajima Y, Nakamura J, et al. HIF‐1alpha is a crucial factor in the development of peritoneal dissemination via natural metastatic routes in scirrhous gastric cancer. Int J Oncol. 2013;43:1431‐1440. [DOI] [PubMed] [Google Scholar]
- 8. Tanaka T, Nakamura J, Kitajima Y, et al. Loss of trefoil factor 1 is regulated by DNA methylation and is an independent predictive factor for poor survival in advanced gastric cancer. Int J Oncol. 2013;42:894‐902. [DOI] [PubMed] [Google Scholar]
- 9. Tong WW, Tong GH, Chen XX, Zheng HC, Wang YZ. HIF2alpha is associated with poor prognosis and affects the expression levels of survivin and cyclin D1 in gastric carcinoma. Int J Oncol. 2014;46:233‐242. [DOI] [PubMed] [Google Scholar]
- 10. Tsai YP, Wu KJ. Hypoxia‐regulated target genes implicated in tumor metastasis. J Biomed Sci. 2012;19:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kapusta A, Kronenberg Z, Lynch VJ, et al. Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Gen. 2013;9:e1003470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu L, Sun L, Zhao P, et al. Hypoxia promotes metastasis in human gastric cancer by up‐regulating the 67‐kDa laminin receptor. Cancer Sci. 2010;101:1653‐1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang Y, Li Z, Zhang H, et al. HIF‐1alpha and HIF‐2alpha correlate with migration and invasion in gastric cancer. Cancer Biol Ther. 2010;10:376‐382. [DOI] [PubMed] [Google Scholar]
- 14. Liu L, Sun L, Zhang H, et al. Hypoxia‐mediated up‐regulation of MGr1‐Ag/37LRP in gastric cancers occurs via hypoxia‐inducible‐factor 1‐dependent mechanism and contributes to drug resistance. Int J Cancer. 2009;124:1707‐1715. [DOI] [PubMed] [Google Scholar]
- 15. Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298‐1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Qiao HP, Gao WS, Huo JX, Yang ZS. Long non‐coding RNA GAS5 functions as a tumor suppressor in renal cell carcinoma. Asian Pac J Cancer Prev. 2013;14:1077‐1082. [DOI] [PubMed] [Google Scholar]
- 17. Yang H, Zhong Y, Xie H, et al. Induction of the liver cancer‐down‐regulated long noncoding RNA uc002mbe.2 mediates trichostatin‐induced apoptosis of liver cancer cells. Biochem Pharmacol. 2013;85:1761‐1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gupta RA, Shah N, Wang KC, et al. Long non‐coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071‐1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang F, Zhang L, Huo XS, et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54:1679‐1689. [DOI] [PubMed] [Google Scholar]
- 20. Wang X, Arai S, Song X, et al. Induced ncRNAs allosterically modify RNA‐binding proteins in cis to inhibit transcription. Nature. 2008;454:126‐130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Berghoff EG, Clark MF, Chen S, Cajigas I, Leib DE, Kohtz JD. Evf2 (Dlx6as) lncRNA regulates ultraconserved enhancer methylation and the differential transcriptional control of adjacent genes. Development. 2013;140:4407‐4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang F, Huo XS, Yuan SX, et al. Repression of the long noncoding RNA‐LET by histone deacetylase 3 contributes to hypoxia‐mediated metastasis. Mol Cell. 2013;49:1083‐1096. [DOI] [PubMed] [Google Scholar]
- 23. Yang F, Zhang H, Mei Y, Wu M. Reciprocal regulation of HIF‐1alpha and lincRNA‐p21 modulates the Warburg effect. Mol Cell. 2014;53:88‐100. [DOI] [PubMed] [Google Scholar]
- 24. Wang Y, Liu X, Zhang H, et al. Hypoxia‐inducible lncRNA‐AK058003 promotes gastric cancer metastasis by targeting gamma‐synuclein. Neoplasia. 2014;16:1094‐1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang Y, Li H, Hou S, Hu B, Liu J, Wang J. The noncoding RNA expression profile and the effect of lncRNA AK126698 on cisplatin resistance in non‐small‐cell lung cancer cell. PLoS ONE. 2013;8:e65309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kuang SQ, Bai H, Fang ZH, et al. Aberrant DNA methylation and epigenetic inactivation of Eph receptor tyrosine kinases and ephrin ligands in acute lymphoblastic leukemia. Blood. 2010;115:2412‐2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maamar H, Cabili MN, Rinn J, Raj A. linc‐HOXA1 is a noncoding RNA that represses Hoxa1 transcription in cis. Genes Dev. 2013;27:1260‐1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kornienko AE, Guenzl PM, Barlow DP, Pauler FM. Gene regulation by the act of long non‐coding RNA transcription. BMC Biol. 2013;11:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Reis EM, Verjovski‐Almeida S. Perspectives of long non‐coding RNAs in cancer diagnostics. Front Genet. 2012;3:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Flockhart RJ, Webster DE, Qu K, et al. #BRAFV600E remodels the melanocyte transcriptome and induces BANCR to regulate melanoma cell migration. Genome Res. 2012;22:1006‐1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ramos AD, Diaz A, Nellore A, et al. Integration of genome‐wide approaches identifies lncRNAs of adult neural stem cells and their progeny in vivo. Cell Stem Cell. 2013;12:616‐628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xu D, Yang F, Yuan JH, et al. LncRNA‐LALR1 accelerates hepatocyte proliferation during liver regeneration by activating Wnt/beta‐Catenin signaling. Hepatology. 2013;58:739‐751. [DOI] [PubMed] [Google Scholar]
- 33. Kim K, Jutooru I, Chadalapaka G, et al. HOTAIR is a negative prognostic factor and exhibits pro‐oncogenic activity in pancreatic cancer. Oncogene. 2013;32:1616‐1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li W, Zheng J, Deng J, et al. Increased levels of the long intergenic non‐protein coding RNA POU3F3 promote DNA methylation in esophageal squamous cell carcinoma cells. Gastroenterology. 2014;146:1714‐1726 e5. [DOI] [PubMed] [Google Scholar]
- 35. Du Y, Kong G, You X, et al. Elevation of highly up‐regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down‐regulating p18. J Biol Chem. 2012;287:26302‐26311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu Q, Suo Z, Kristensen GB, Baekelandt M, Nesland JM. The prognostic impact of EphB2/B4 expression on patients with advanced ovarian carcinoma. Gynecol Oncol. 2006;102:15‐21. [DOI] [PubMed] [Google Scholar]
- 37. Astin JW, Batson J, Kadir S, et al. Competition amongst Eph receptors regulates contact inhibition of locomotion and invasiveness in prostate cancer cells. Nat Cell Biol. 2010;12:1194‐U175. [DOI] [PubMed] [Google Scholar]
- 38. Alam SM, Fujimoto J, Jahan I, Sato E, Tamaya T. Coexpression of EphB4 and ephrinB2 in tumour advancement of ovarian cancers. Br J Cancer. 2008;98:845‐851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Spannuth WA, Mangala LS, Stone RL, et al. Converging evidence for efficacy from parallel EphB4‐targeted approaches in ovarian carcinoma. Mol Cancer Ther. 2010;9:2377‐2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nakada M, Niska JA, Tran NL, McDonough WS, Berens ME. EphB2/R‐Ras signaling regulates glioma cell adhesion, growth, and invasion. Am J Pathol. 2005;167:565‐576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xia G, Kumar SR, Stein JP, et al. EphB4 receptor tyrosine kinase is expressed in bladder cancer and provides signals for cell survival. Oncogene. 2006;25:769‐780. [DOI] [PubMed] [Google Scholar]
- 42. Xia G, Kumar SR, Masood R, et al. EphB4 expression and biological significance in prostate cancer. Cancer Res. 2005;65:4623‐4632. [DOI] [PubMed] [Google Scholar]
- 43. Kumar SR, Singh J, Xia G, et al. Receptor tyrosine kinase EphB4 is a survival factor in breast cancer. Am J Pathol. 2006;169:279‐293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dopeso H, Mateo‐Lozano S, Mazzolini R, et al. The receptor tyrosine kinase EPHB4 has tumor suppressor activities in intestinal tumorigenesis. Cancer Res. 2009;69:7430‐7438. [DOI] [PubMed] [Google Scholar]
- 45. Davalos V, Dopeso H, Castano J, et al. EPHB4 and survival of colorectal cancer patients. Cancer Res. 2006;66:8943‐8948. [DOI] [PubMed] [Google Scholar]
- 46. Li M, Zhao ZW. Clinical implications of EphB4 receptor expression in pancreatic cancer. Mol Biol Rep. 2013;40:1735‐1741. [DOI] [PubMed] [Google Scholar]
- 47. Noren NK, Lu M, Freeman AL, Koolpe M, Pasquale EB. Interplay between EphB4 on tumor cells and vascular ephrin‐B2 regulates tumor growth. Proc Natl Acad Sci USA. 2004;101:5583‐5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Martiny‐Baron G, Korff T, Schaffner F, et al. Inhibition of tumor growth and angiogenesis by soluble EphB4. Neoplasia. 2004;6:248‐257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kumar SR, Masood R, Spannuth WA, et al. The receptor tyrosine kinase EphB4 is overexpressed in ovarian cancer, provides survival signals and predicts poor outcome. Br J Cancer. 2007;96:1083‐1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kumar SR, Scehnet JS, Ley EJ, et al. Preferential induction of EphB4 over EphB2 and its implication in colorectal cancer progression. Cancer Res. 2009;69:3736‐3745. [DOI] [PubMed] [Google Scholar]
- 51. Yang NY, Pasquale EB, Owen LB, Ethell IM. The EphB4 receptor‐tyrosine kinase promotes the migration of melanoma cells through Rho‐mediated actin cytoskeleton reorganization. J Biol Chem. 2006;281:32574‐32586. [DOI] [PubMed] [Google Scholar]
- 52. Wimmer‐Kleikamp SH, Lackmann M. Eph‐modulated cell morphology, adhesion and motility in carcinogenesis. IUBMB Life. 2005;57:421‐431. [DOI] [PubMed] [Google Scholar]
- 53. Munarini N, Jager R, Abderhalden S, et al. Altered mammary epithelial development, pattern formation and involution in transgenic mice expressing the EphB4 receptor tyrosine kinase. J Cell Sci. 2002;115:25‐37. [DOI] [PubMed] [Google Scholar]
- 54. Li M, Zhao ZW, Zhang Y, Xin Y. Over‐expression of Ephb4 is associated with carcinogenesis of gastric cancer. Digest Dis Sci. 2011;56:698‐706. [DOI] [PubMed] [Google Scholar]
- 55. Li D, Feng J, Wu T, et al. Long intergenic noncoding RNA HOTAIR is overexpressed and regulates PTEN methylation in laryngeal squamous cell carcinoma. Am J Pathol. 2013;182:64‐70. [DOI] [PubMed] [Google Scholar]
- 56. Szafranski P, Dharmadhikari AV, Brosens E, et al. Small noncoding differentially methylated copy‐number variants, including lncRNA genes, cause a lethal lung developmental disorder. Genome Res. 2013;23:23‐33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Di Ruscio A, Ebralidze AK, Benoukraf T, et al. DNMT1‐interacting RNAs block gene‐specific DNA methylation. Nature. 2013;503:371‐376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lai F, Shiekhattar R. Where long noncoding RNAs meet DNA methylation. Cell Res. 2014;24:263‐264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wu QH, Lind GE, Aasheim HC, et al. The EPH receptor Bs (EPHBs) promoters are unmethylated in colon and ovarian cancers. Epigenetics. 2007;2:237‐243. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


