Abstract
Degree of histological differentiation is an important characteristic of cancers and may be associated with malignant potential. However, in squamous cell carcinomas, a key transcriptional factor regulating tumor differentiation is largely unknown. Chemoradiotherapy (CRT) is a standard treatment for locally advanced esophageal squamous cell carcinoma; however, the survival rate is still below 40%. From microarray data, single‐minded 2 (SIM2) was overexpressed in the epithelial subtype. Here, we investigated the correlation between SIM2 expression and its clinical implication, and in vitro and in vivo functions of SIM2 in tumor differentiation and in CRT sensitivity. Although SIM2 was suppressed in cancerous tissues, SIM2‐high ESCC showed a favorable prognosis in CRT. Transient SIM2 expression followed by 3D culture induced expression of differentiation markers and suppressed epithelial‐mesenchymal transition‐ and basal‐cell markers. Levels of PDPN‐high tumor basal cells and of expression of genes for DNA repair and antioxidant enzymes were reduced in stable transfectants, and they showed high CDDP and H2O2 sensitivities, and their xenografts showed a well‐differentiated histology. Reduction of tumor basal cells was restored by knockdown of aryl hydrocarbon receptor nuclear translocator (ARNT) that interacted with SIM2. Together, SIM2 increases CRT sensitivity through tumor differentiation by cooperation with ARNT.
Keywords: ARNT, chemoradiotherapy, differentiation, esophageal squamous cell carcinoma, SIM2
1. INTRODUCTION
Esophageal cancer is the sixth most common cause of cancer deaths worldwide.1 Neoadjuvant chemotherapy (CT) or neoadjuvant chemoradiotherapy (CRT) followed by esophagectomy, or definitive CRT have been standard initial treatments for locally advanced esophageal squamous cell carcinoma (ESCC) in Asia and Western countries. Although improvement in neoadjuvant CT and definitive CRT has been achieved, the 5‐year survival rate of locally advanced ESCC is still 37%‐55% as a result of local recurrence, lymph node metastasis, and distant metastasis.2, 3 Therefore, further investigation into the progression and treatment resistance of ESCC is needed. Degree of tumor differentiation is the major histopathological classification factor in squamous cell carcinoma such as ESCC, head and neck squamous cell carcinoma (HNSCC) and uterine cervical squamous cell carcinoma and has many similarities in therapeutic strategy. In squamous cell carcinoma, the differentiation degree has been thought to influence treatment sensitivity and prognosis, as is shown in several reports on their relationship. Broders classified genito‐urinary cancer including squamous cell carcinoma by the ratio of differentiated cells and undifferentiated cells, and the undifferentiated histology presented a poor prognosis.4 Histological grade including keratinization of HNSCC was reported to influence prognosis.5 Additionally, histological differentiation was reported to be correlated with local recurrence in squamous cell carcinoma of the skin, ear and lip.6 However, it is unknown whether this factor is associated with the response to neoadjuvant therapy, because study using pretreatment biopsy samples has been limited. Furthermore, no key transcription factor involved in the differentiation of squamous cell carcinoma has been reported. By comparing gene expression profiles among pre‐ and post‐treatment biopsy specimens of 30 ESCC patients and 121 pretreatment ESCC biopsy specimens, we recently discovered a good responder subtype of ESCC with cytotoxic T‐lymphocyte signatures activated by CRT.7 In the complete response (CR) cases, 999 overexpressed genes including at least 234 tumor‐specific CTL activation‐associated genes such as IFNG, PRF1, and GZMB, were found in post‐treatment biopsy specimens. Clustering analysis using expression profiles of these 234 genes in 121 pretreatment ESCC allowed us to distinguish the immune‐activated cases, designating them as I‐type, from other cases. Further comparative study identified a series of epithelial‐mesenchymal transition (EMT)‐related genes overexpressed in early relapse cases. Clinical outcome of CDH2‐negative epithelial cases in the I‐type was significantly better than that of CDH2‐positive mesenchymal cases in the I‐type (64% vs 12% in 5‐year overall survival). Interestingly, SIM2 was found to be overexpressed in CDH2‐negative epithelial cases in the I‐type as shown in Table S7 of our previous paper.7
Single‐minded 2 (SIM2) is located in a minimum region of chromosome 21 often implicated in Down syndrome called Down syndrome chromosomal region, and is a member of the basic HLH (helix‐loop‐helix)‐PER‐ARNT‐SIM (bHLH‐PAS) family.8 SIM2 is comparable with other bHLH‐PAS family members, hypoxia inducible factor alpha (HIF1α) and aryl hydrocarbon receptor (AHR), for binding to the partner, aryl hydrocarbon receptor nuclear translocator (ARNT) or ARNT2. SIM2‐ARNT dimer binds to central midline elements (CME) in the regulatory regions of target genes and actively represses gene expression through the carboxy‐terminal transrepression domain of SIM2.9, 10, 11 In addition, SIM2‐ARNT dimer is capable of binding not only to CME but also to hypoxia‐response elements that is normally bound by HIF‐1α.12 There are two different spliced isoforms of human SIM2, SIM2‐long (SIM2) and SIM2‐short (SIM2s), which differ in their 3′ ends.8 In mice, Sim2s has been reported to exert a less repressive effect on hypoxia‐induced gene expression than does Sim2, and Sim2s binds to CME and activates expression of the CME‐controlled reporter gene through an Arnt transactivation domain‐dependent mechanism.13 However, their differential functions in humans are yet unknown.
Substantial misregulation of SIM2 expression has been reported in several cancer types.14, 15, 16, 17 In breast cancer, SIM2s directly downregulates SNAI2 expression and inhibits EMT, and represses tumor growth and invasion.15, 18, 19 In addition, Sim2s increases the expression of genes that are associated with mammary lactogenic differentiation in mice.20 Conversely, knockdown of SIM2s causes growth inhibition and increases cell death through apoptosis in cultured colon carcinoma and pancreatic carcinoma cell lines,14, 16, 21 and decreases growth of colon carcinoma‐derived xenograft.8 Increased expression of SIM2s and SIM2 is notably associated with the development and progression of prostate tumor.17, 22, 23 Thus, the expression and the role of SIM2 and SIM2s are dependent on the tumor type. In this study, we showed the functional role of SIM2 and its clinical implications in squamous cell carcinoma, particularly in ESCC.
2. MATERIALS AND METHODS
2.1. Clinical samples
Sixty pairs of ESCC tissues and their matched non‐cancerous tissues were provided from patients who underwent esophagectomy at the National Cancer Center Hospital (Tokyo, Japan), and 85 biopsy samples of stage II/III ESCC before CRT were provided by the National Cancer Center Hospital East (Kashiwa, Japan) after obtaining written informed consent from each patient and approval by the Center's Ethics Committee (Nos.17‐031 and 19‐014). All experiments were carried out in accordance with the guidelines and regulations of the Committee.
2.2. Cell culture
Esophageal cancer cell lines (TE1, TE3, TE5, TE6, TE8, TE10, KYSE510, and T.Tn), were purchased from the Japanese Collection of Research Bioresources Cell Bank. Esophageal epithelial cells (HEEpiC) were purchased and cultured by the supplier's protocol (ScienCell, San Diego, CA, USA). TE1, TE3, TE5, TE6, TE8, TE10, and KYSE510 were routinely propagated in RPMI 1640 (Wako, Tokyo, Japan) supplemented with 10% FBS, penicillin and streptomycin. T.Tn was propagated in DMEM/Ham's F‐12 (Wako) supplemented with 10% FBS, penicillin and streptomycin. All cell lines were maintained at 37°C, 5% CO2 and 95% humidified air. We used 3.5‐cm NanoCulture Plate (SCIVAX, Kawasaki, Japan) for 3D culture.
2.3. RT‐PCR and quantitative real‐time PCR
Total RNA was isolated by suspending the cells in an ISOGEN lysis buffer (Nippon Gene, Toyama, Japan) followed by precipitation with isopropanol. Reverse transcription was carried out by SuperScript III First‐Stand Synthesis System (Invitrogen, Carlsbad, CA, USA). PCR was carried out by AccuPrime Taq DNA Polymerase System (Invitrogen) within the linear range of amplification, typically 19‐30 cycles, for all splicing isoforms of SIM2, long isoform of SIM2, short isoform of SIM2 (SIM2s), ARNT, ARNT2, FN1, VIM, SNAI2, TWIST1, PDPN, SPRR1A, FLG and ACTB. Quantitative real‐time PCR was carried out for long isoform of SIM2, ARNT, ARNT2, VIM, PDPN, SPRR1A, FLG, FANCD2, BRCA1, BARD1, XRCC5, SOD2 and ACTB by a Bio‐Rad iCycler with iQ Syber Green Supermix (Bio‐Rad, Hercules, CA, USA). Results are presented as linearized Ct values normalized to the housekeeping ACTB and the indicated reference value (2−ΔΔCt). Primers used for the study are listed in Table S1.
2.4. 5‐Azacytidine treatment
Cells were plated at 2 × 106 cells per 10‐cm dish. One day after plating, the cells were treated with 5‐azacytidine (AzaC, 2 μmol/L; Focus Biomolecules, Plymouth Meeting, PA, USA) for 48 hours.
2.5. Bisulfite sequence
Bisulfite modification of DNA isolated from 10 pairs of esophageal cancer tissues and their matched non‐cancerous tissues was conducted by using MethylEasy Xceed (Human Genetic Signatures, Sydney, Australia) according to the manufacturer's protocol. PCR for bisulfite‐treated DNA was done by carrying out 40 cycles using EpiTaq HS (TaKaRa, Ohtsu, Japan) with primers for the SIM2 promoter. Sequencing was carried out by Eurofins Genomics Inc. (Tokyo, Japan). Primers used for the study are listed in Table S2.
2.6. Plasmid construction and transfection
pCMV6‐AC‐GFP containing the long isoform of the SIM2 cDNA and pCMV6‐neo containing SIM2s cDNA were purchased from OriGene Technologies (Rockville, MD, USA). Cells were plated at 2 × 106 per 10‐cm dish, and transfected with either pCMV6‐AC‐GFP‐SIM2 or pCMV6‐SIM2s or no insert of pCMV6‐neo (OriGene Technologies) by using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's protocol. Forty‐eight hours later, cells were selected with G418 (0.4 mg/mL) for 1 month. Colonies were transferred to larger plates, and expression of different splicing isoforms of SIM2 mRNAs was examined by quantitative real‐time PCR.
2.7. siRNA transfection
ARNT siRNA purchased from Ambion (Austin, TX, USA) was introduced to SIM2‐transfectants using Thermo Scientific DharmaFECT Transfection Reagents (Thermo Fisher Scientific, Waltham, MA, USA). Quantitative real‐time PCR and flow cytometry were carried out after siRNA treatment of the SIM2‐transfectants.
2.8. Flow cytometry
Cells (5 × 105 cells) were incubated with anti‐PDPN antibody (1:100; Abcam, Cambridge, UK) or control mouse IgG at room temperature for 30 minutes, then incubated with Alexa488‐conjugated antimouse IgG antibody (1:500; Invitrogen) on ice for 30 minutes. Dead cells were labeled with propidium iodide and excluded from the analysis. Flow cytometry was carried out using FACSCalibur (Becton, Dickinson and Co., Franklin Lakes, NJ, USA), and analyzed by Cell Quest software (Becton, Dickinson and Co.).
2.9. Immunocytochemistry
Specimens fixed in formalin and embedded in paraffin were cut into 4‐μm sections, dewaxed, and dehydrated. Sections were blocked by 10% FBS in PBS, and reacted with primary antibodies against PDPN (1:40; Abnova, Taipei, Taiwan) at 4°C overnight, followed by incubation with EnVision + Dual Linl System‐HRP (Dako, Carpinteria, CA, USA). Subsequently, the sections were subjected to DAB reaction (Dako) for 5 minutes, and counterstained with hematoxylin.
2.10. Animal experiment
The protocol for the animal experiments was approved by the committee for Ethics of Animal Experimentation and was in accordance with the Guideline for Animal Experiments at the National Cancer Center. SIM2 overexpressing cells (5 × 106 cells) were transplanted into subcutaneous spaces at three sites in the backs of 6‐week‐old female C.B17/Icr‐scid (scid/scid) mice. Tumor growth was observed for 3‐11 weeks. Two mice were used for each group.
2.11. Immunofluorescence analysis
Cells were grown on glass chamber slides, fixed with 4% paraformaldehyde, permeabilized with −20°C methanol and 0.5% Triton X‐100/PBS, and blocked with 0.1 mol/L NH4Cl, 10% FBS and 3% BSA in PBS. Cells were incubated with anti‐green fluorescent protein (GFP) antibody (1:500; OriGene Technologies) and anti‐ARNT antibody (1:50; OriGene Technologies) at 4°C overnight, then incubated with Alexa488‐conjugated antimouse IgG antibody (1:1000; Invitrogen) and Alexa488‐conjugated antigoat IgG antibody (1:1000; Invitrogen) at room temperature for 30 minutes and stained with DAPI.
2.12. Immunoprecipitation
TE8 cells were plated at 2.5 × 106 per 10‐cm dish and transiently transfected with either pCMV6‐AC‐GFP‐SIM2 or no insert of pCMV6‐neo (OriGene Technologies) by using Lipofectamine 2000 reagent (Invitrogen). Cells were treated with 5 μmol/L proteasome inhibitor, MG‐132 (Sigma‐Aldrich, St Louis, MO, USA) for 18 hours and, 48 hours after transfection, cells were collected and lysed in lysis buffer (20 mmol/L Tris‐HCl, 20% glycerol, 300 mmol/L KCl, 5 mmol/L 2‐mercaptoethanol, 1 mmol/L Pefabloc (Roche, Basel, Switzerland), and 25 μmol/L MG‐132. Immunoprecipitation was conducted using Immunoprecipitation Kit‐Dynabeads Protein G (Life Technologies, Rockville, MD, USA). Cell lysates were incubated with anti‐GFP antibody (OriGene Technologies) or control mouse IgG at 4°C overnight, and then incubated with 50 μL Dynabeads at 4°C for 4 hours. The Dynabeads‐antibody‐antigen complex was washed and resuspended in SDS sample buffer, then incubated at 95°C for 5 minutes. Eluted proteins were fractionated by SDS‐PAGE and detected by western blot.
2.13. Western blot
Samples were separated using SDS‐PAGE (7.5% acrylamide). Proteins were transferred to a nitrocellulose membrane and blocked with 5% membrane blocking agent (GE Healthcare, Buckinghamshire, UK) in PBS, and probed with anti‐SIM2 antibody (1:250; Abcam, Cambridge, UK), anti‐ARNT antibody (1:200; Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti‐SOD2 antibody (1:5000; Abcam), or anti‐β‐actin antibody (1:1000; Cell Signaling Technology, Danvers, MA, USA) at 4°C overnight, or anti‐α tubulin antibody (1:1000; Santa Cruz Biotechnology) at room temperature for 2 hours, then washed and incubated with HRP‐conjugated antigoat immunoglobulin (Dako, Carpinteria, CA, USA) or HRP‐conjugated antimouse immunoglobulin (Dako) at room temperature for 2 hours. Immunoreactive protein bands were identified with Pierce ECL Plus Western Blotting Substrate (Thermo Scientific).
2.14. CDDP treatment
Mock‐ or SIM2‐transfectants were plated at 2 × 105 cells per 3.5‐cm NanoCulture Dish (SCIVAX). One day after plating, the cells were treated with CDDP (0, 5 μmol/L) every 2 days for 14 days. Number of viable cells was counted by Trypan blue‐exclusion test.
2.15. H2O2 or fluorouracil (5‐FU) treatment
Mock‐ or SIM2‐transfectants were plated at 1 × 104 cells per 96‐well NanoCulture Plate (SCIVAX). Three day after plating, those cells were treated with H2O2 or 5‐FU for 1 day or 3 days. Number of viable cells was counted by CellTiter‐Glo Luminescent Cell Viability Assay (Promega, Madison, WI, USA).
2.16. Irradiation experiment
SIM2 overexpressing cells were plated at 2 × 105 cells per 6‐cm dish. One day after plating, the cells were irradiated with γ‐rays (0, 1, 5, 10 Gy), then incubated for 7 days. Number of viable cells was counted by using a Trypan blue‐exclusion test.
2.17. Statistical analysis
All data are expressed as the mean + SE, and analyzed using the unpaired t test. In clinical outcome data, P‐values were calculated by log‐rank analysis. P‐values <.05 were considered significant. Ekuseru‐Toukei 2010 (Social Survey Research Information Co., Ltd, Tokyo, Japan) was used for all statistical analyses.
3. RESULTS
3.1. SIM2 is frequently underexpressed in ESCC by promoter methylation
First, by quantitative real‐time PCR, we compared the mRNA expression levels of SIM2, ARNT and ARNT2 between cancerous and non‐cancerous tissues of 60 ESCC patients who underwent esophagectomy. In 90% of them, SIM2 was suppressed in the cancerous tissues as compared with the non‐cancerous tissues (Figure 1A, upper). In contrast, no or quite low SIM2s mRNA was detected in both the cancerous and non‐cancerous tissues by RT‐PCR (data not shown). Both ARNT and ARNT2 mRNAs slightly increased in the cancerous tissues, but ARNT2 mRNA levels were quite low in both the cancerous and non‐cancerous tissues (Figure 1A, lower; Figure S1). Next, we examined SIM2 expression in 85 ESCC biopsy specimens before definitive CRT and in 4 normal specimens. In 93% of 85 ESCC, SIM2 was underexpressed compared with the average of the 4 normal specimens (Figure 1B). Importantly, the upper 30% of patients in the SIM2 expression level showed a favorable prognosis compared to the lower 30% of patients (Figure 1C). To address the relationship between SIM2 expression and promoter methylation, we examined the expression of SIM2, SIM2s, ARNT, and ARNT2 in 8 ESCC cell lines using RT‐PCR. In accordance with the above results from the clinical samples, no or quite low SIM2s and ARNT2 mRNA and high ARNT mRNA were detected in all of the 8 cell lines and normal esophageal epithelial cells (Figure 2A; Figure S2). SIM2 mRNA level was quite low in TE10 only (Figure 2A). Transcription start site of SIM2 is localized at 1.2 kb upstream of the translation initiation site, and the SIM2 promoter region is reported to be GC‐rich.24 To examine whether promoter methylation is involved in the suppression of SIM2, we treated TE10 with AzaC. SIM2 mRNA significantly increased with treatment of AzaC (Figure 2B). Bisulfite sequencing suggested that methylation of CpG sites in the SIM2 promoter was inversely correlated with SIM2 mRNA level in 5 surgical samples with high‐end expression level and in another 5 samples with low‐end expression level of which the RNA and DNA were available (Figure 2C). Hypermethylation was preferentially observed in the 5 surgical samples with low SIM2 expression. Taken together, promoter methylation is thought to be one of the causes of suppression of SIM2 expression in ESCC.
Figure 1.
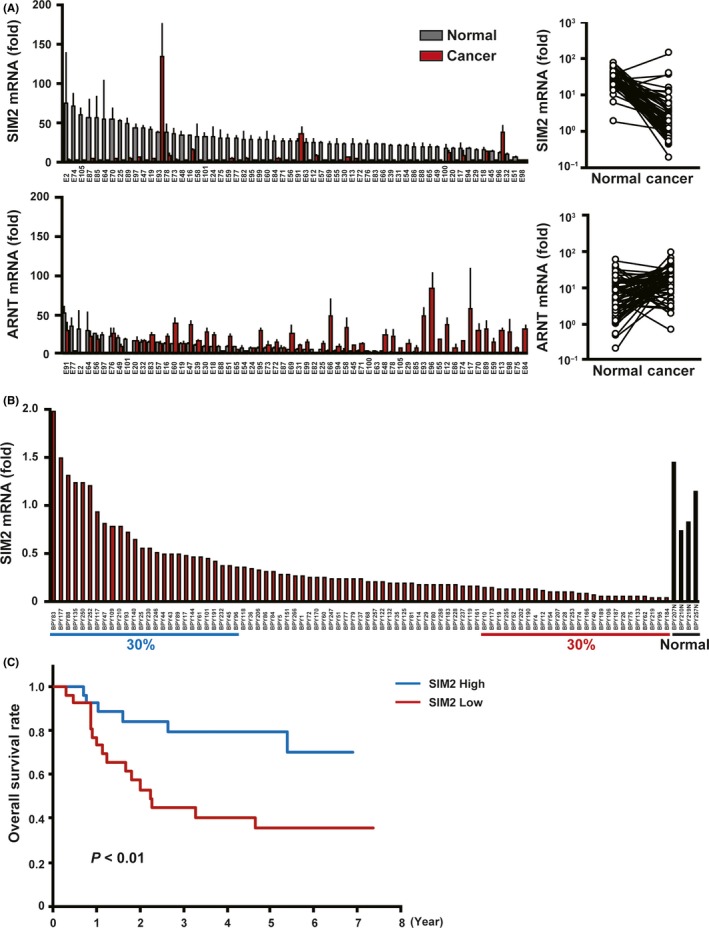
Esophageal squamous cell carcinoma (ESCC) with SIM2 expression is chemoradiotherapy (CRT) sensitive. A, Real time RT‐PCR of SIM2 and ARNT in 60 pairs of surgically resected cancerous tissue (red column) and non‐cancerous tissue (gray column) (mean + SE for a triplicate analysis). Plot graphs (right) represent means of SIM2 and ARNT mRNA expression in cancerous tissue and non‐cancerous tissue of each patient. B, Real‐time RT‐PCR of SIM2 in 85 cancerous tissues and 4 non‐cancerous tissues of stages II‐III ESCC before CRT. C, Overall survival of ESCC who were treated with definitive CRT. Blue line represents 30% of patients with the highest SIM2 expression, and red line represents 30% of patients with the lowest SIM2 expression. P‐values were calculated by log‐rank analysis
Figure 2.
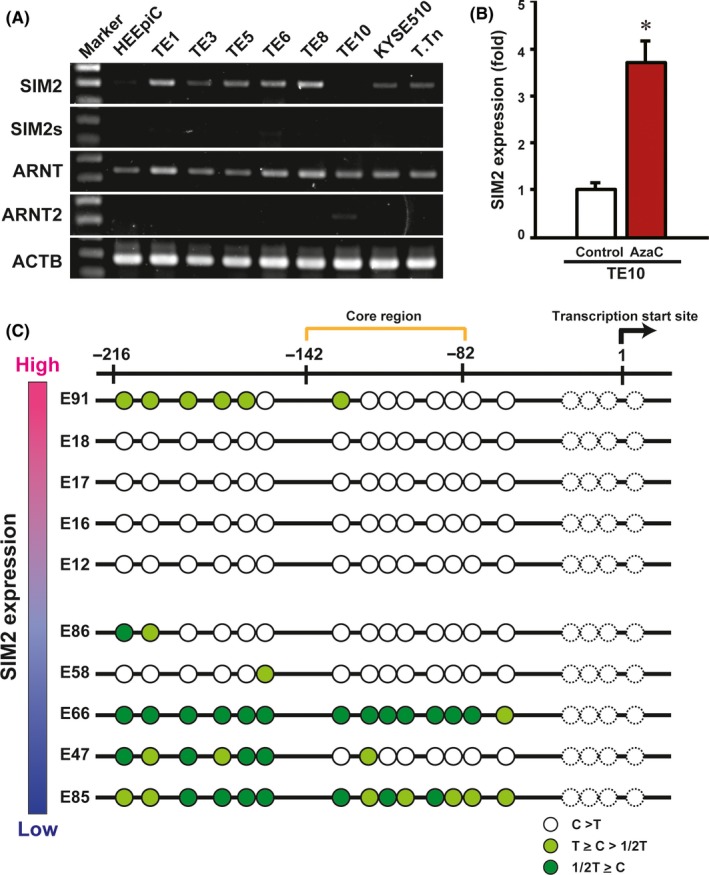
Induction of SIM2 by 5‐azacytidine (AzaC) treatment and promoter methylation. A, Semiquantitative RT‐PCR of SIM2, SIM2s, ARNT, ARNT2, and ACTB in normal esophageal epithelial cells (HEEpiC) and 8 esophageal squamous cell carcinoma‐derived cell lines TE1, TE3, TE5, TE6, TE8, TE10, KYSE510, and T.Tn. B, Real‐time RT‐PCR of SIM2 in TE10 with (red column) and without (white column) AzaC treatment (n = 3, mean + SE; *P < .05). C, Bisulfite sequencing of the SIM2 promoter in 5 cancerous tissues with high SIM2 expression and 5 cancerous tissues with low SIM2 expression (Figure 1). Each circle represents cytosine of CpG sites after bisulfite treatment. White circle signifies that the cytosine signal is higher than the thymidine signal. Light green circle signifies that the cytosine signal is higher than half of the thymidine signal but lower than the thymidine signal. Green circle signifies that the cytosine signal is lower than half of the thymidine signal. Dotted circle, not informative
3.2. SIM2 promotes differentiation of squamous cell carcinoma in 3D culture
First, we transiently transfected SIM2 or SIM2s cDNA to TE8 cells, cultured in an ordinary high‐adherent tissue culture plate, and then examined the expression of EMT regulator genes (SNAI2 and TWIST1), EMT markers (FN1 and VIM), and a basal cell marker (PDPN).25, 26, 27, 28 Overexpression of SIM2s did not affect expression of all of the genes, whereas overexpression of SIM2 repressed SNAI2 and TWIST1 expression, but did not affect FN1, VIM or PDPN (Figure 3A).
Figure 3.
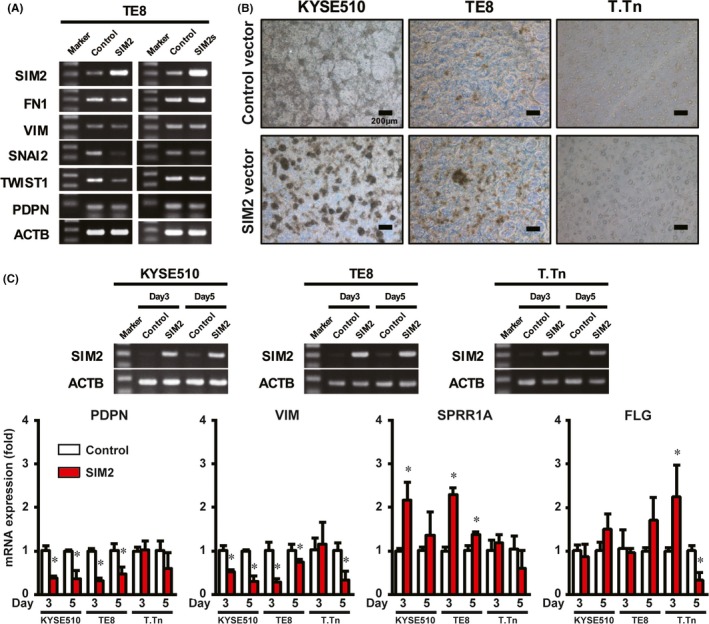
SIM2 is involved in differentiation of esophageal squamous cell carcinoma (ESCC). A, Semiquantitative RT‐PCR of SIM2, FN1, VIM, SNAI2, TWIST1, PDPN, and ACTB in 2D cultured TE8 cells 3 days after transfection of pCMV6‐neo or pCMV‐AC‐GFP‐SIM2 (left) or pCMV6‐SIM2s (right). B, Photographic images of 3D cultured KYSE510, TE8, and T.Tn cells 3 days after transfection of pCMV6‐neo (upper) or pCMV‐AC‐GFP‐SIM2 (lower). C, Real‐time RT‐PCR of PDPN, VIM, SPRR1A, and FLG in 3D cultured KYSE510, TE8, and T.Tn cells 3 or 5 days after transfection of pCMV6‐neo (Control, white column) or pCMV‐AC‐GFP‐SIM2 (SIM2, red column) (n = 3, mean + SE; *P < .05)
To examine the effect of SIM2 on cell differentiation, we next used a 3D culture system which was reported to induce differentiation of squamous cell carcinoma through adhesion restriction.29 Before the SIM2 cDNA transfection experiment, we confirmed both an increase and a decrease of mRNA of differentiation markers (CEA, FLG, IVL, KRT1, LOR, and SPRR1A) and EMT/basal cell markers (VIM and PDPN), respectively,25, 26, 27, 28 in KYSE510, TE8, and T.Tn by the 3D culture (Figure S3). Overexpression of SIM2 in KYSE510, TE8 and T.Tn followed by the 3D culture appeared to increase spheroid formation more effectively than did the control (Figure 3B). Overexpression of SIM2 in KYSE510 and TE8 significantly increased SPRR1A mRNA and decreased VIM and PDPN mRNA at day 3 or 5, and that in T.Tn significantly induced FLG and repressed VIM (Figure 3C). No effect of 3D culture in SIM2 mRNA level was observed in these three cell lines (Figure S3B). These results of in vitro 3D cultures suggest that SIM2 has an important role as an ESCC differentiation inducer.
3.3. SIM2 inhibits self‐renewal of PDPN‐positive tumor basal cells
To investigate in vitro long term and the in vivo effect of SIM2 in ESCC cells, we established transfectants stably expressing SIM2, KYSE510‐SIM2‐27, KYSE510‐SIM2‐37, TE8‐SIM2‐2, TE8‐SIM2‐3, T.Tn‐SIM2‐9, and T.Tn‐SIM2‐23 (Figure 4A), and their mock‐transfected counterparts, KYSE510‐Mock, TE8‐Mock, T.Tn‐Mock. Given that PDPN is a basal cell marker in normal esophageal mucosa26, 27 and is also a marker of tumor basal cells with high tumor‐initiating ability in squamous cell carcinoma,28 we compared the proportion of PDPN‐positive cells in the SIM2‐transfectants to that in the mock transfectants by flow cytometry using anti‐PDPN antibody. We first confirmed that there was no difference between the PDPN‐positive tumor basal cell ratios of mock‐ and SIM2‐transfectants stained with control IgG (data not shown). The PDPN‐positive tumor basal cell ratio in KYSE510‐Mock was 50.6%, whereas that in KYSE510‐SIM2‐27 and KYSE510‐SIM2‐37 was markedly reduced to 2.6% and 2.7%, respectively. In T.Tn‐SIM2‐9 and T.Tn‐SIM2‐23, PDPN‐positive tumor basal cell ratios were 58.6% and 42.0%, respectively, and were reduced clearly from 90.3% in T.Tn‐Mock. Although 98.5% in TE8‐Mock was PDPN‐positive, the ratio was reduced to 80.8% and 74.0% in TE8‐SIM2‐2 and TE8‐SIM2‐3, respectively (Figure 4B). We further investigated the histology of the spheroids formed by 3D culture. T.Tn‐Mock cells produced many small spheroids that were composed mainly of PDPN‐positive cells. However, T.Tn‐SIM2‐9 cells produced larger spheroids whose cores were composed of PDPN‐negative cells (Figure 4C, yellow arrow). These data suggest that SIM2 inhibits self‐renewal of PDPN‐positive tumor basal cells and produces differentiated daughter cells.
Figure 4.
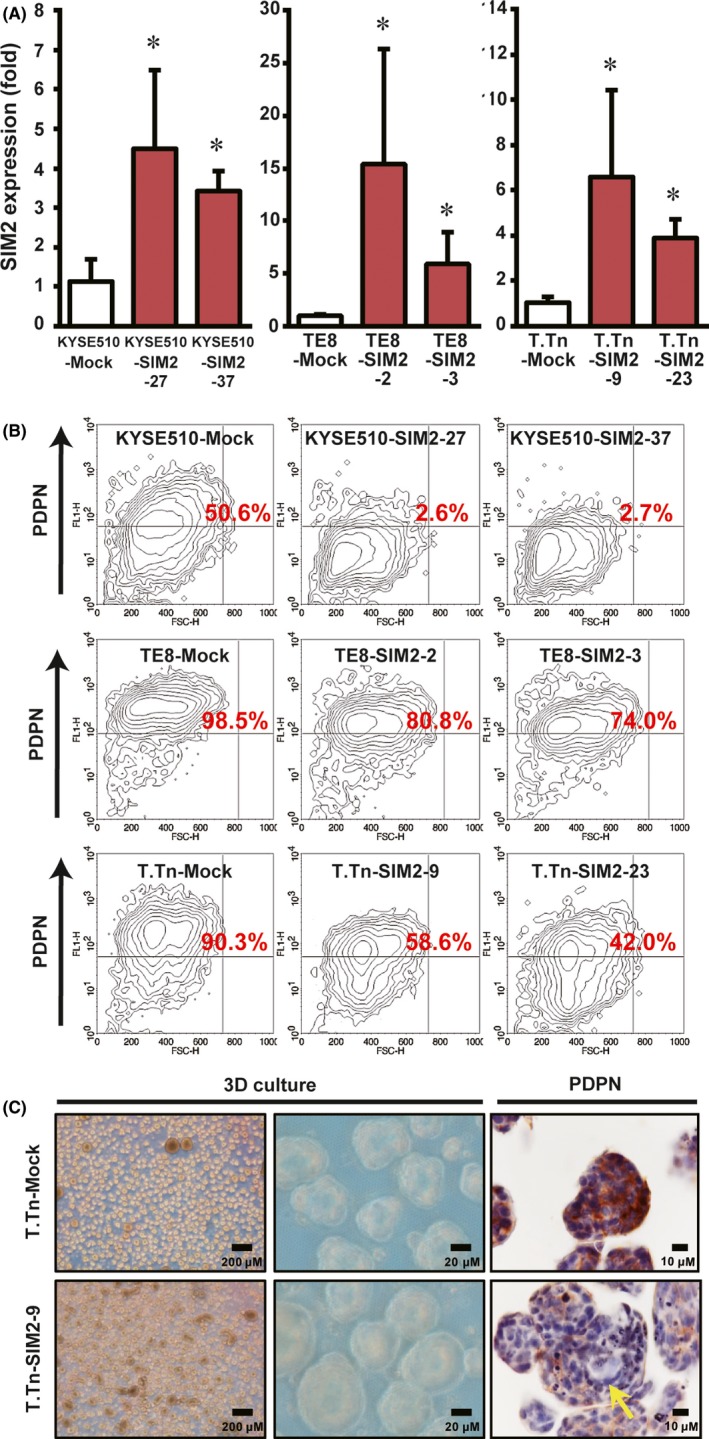
SIM2 decreases PDPN‐positive tumor basal cell ratio. A, Real‐time RT‐PCR of SIM2 in mock‐ (white column) and SIM2‐transfectants (red column) of KYSE510, TE8, and T.Tn (n = 6, mean + SE; *P < .05). B, Flow cytometric analysis of PDPN in mock‐ and SIM2‐transfectans of KYSE510, TE8, and T.Tn. Numbers in red represent PDPN‐positive cell ratio. C, Immunostaining for PDPN in 3D cultured T.Tn‐Mock (upper) and T.Tn‐SIM2‐9 (lower) cells. PDPN‐negative cells are indicated by yellow arrow
3.4. SIM2 induces well‐differentiated histology
To investigate the effect of SIM2 on tumor formation, we transplanted mock‐ and SIM2‐transfectants of KYSE510 and T.Tn into scid/scid mice subcutaneously, because TE8 had quite low tumor formation ability. KYSE510‐Mock developed larger tumors than did KYSE510‐SIM2‐27 and KYSE510‐SIM2‐37 in 3‐7 weeks. Although T.Tn‐Mock developed tumors in 8‐11 weeks, T.Tn‐SIM2‐9 formed only one small tumor by the 11th week and T.Tn‐SIM2‐23 formed no tumor (Figure 5A). Xenografts of KYSE510‐Mock showed moderate‐differentiated histology, and KYSE510‐SIM2‐27 showed a well‐differentiated histology with many cancer pearls (orbicular keratinized regions), which often appear at the central part of the tumor nest in well‐differentiated squamous cell carcinomas (Figure 5B,C). In this well‐differentiated tumor of KYSE‐SIM2‐27, a single or thin layer of the basal cells on the outer edge of the tumor nest was PDPN‐positive, and any regions inside the basal cell layer were PDPN‐negative. Very impressively, although the tumor nest of T.Tn‐Mock showed a typical poor‐differentiated histology and was composed mainly of PDPN‐positive basal cells, that of T.Tn‐SIM2‐9 showed a well‐differentiated histology with many large cancer pearls and was composed of both PDPN‐positive thin basal cell layers and PDPN‐negative thick differentiated cell layers (Figure 5B,C). These in vivo results suggest that SIM2 also has an important role in histological differentiation of ESCC.
Figure 5.
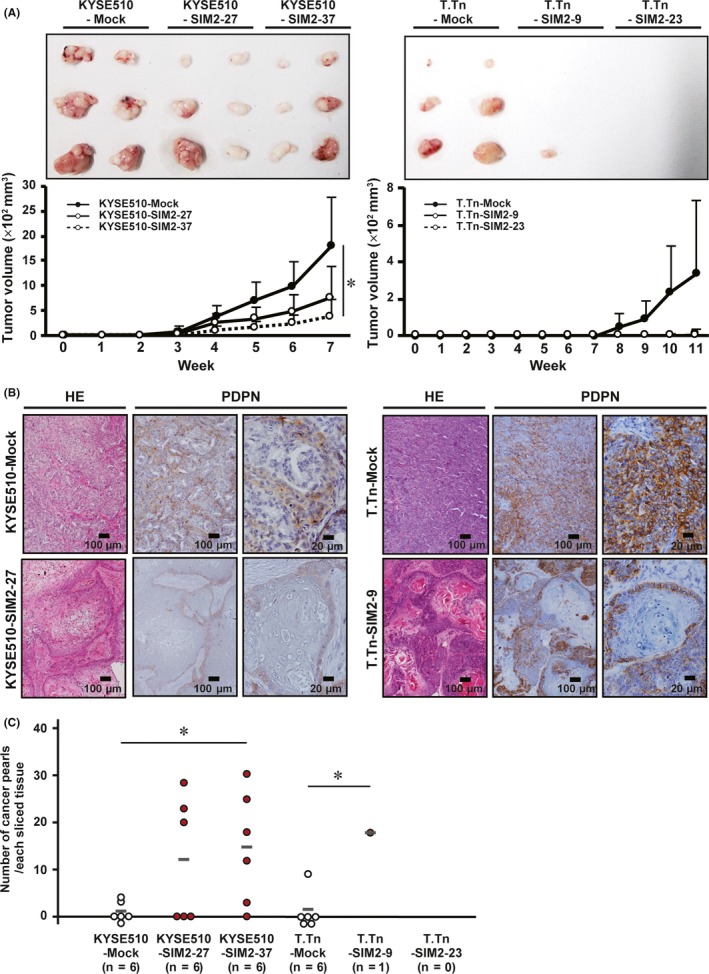
SIM2 induces well‐differentiated histology and in vivo growth suppression of KYSE510 and T.Tn cells. A, Xenograft tumor growth curves of mock‐ and SIM2‐transfectants of KYSE510 and T.Tn in 3 sites of 2 mice (n = 6, mean + SE). B, Hematoxylin and eosin staining and immunocytochemistry for PDPN in xenografts of KYSE510‐Mock, KYSE510‐SIM2‐27, T.Tn‐Mock, and T.Tn‐SIM2‐9. C, Number of cancer pearls per slice in the xenograft of KYSE510‐Mock, KYSE510‐SIM2‐27, KYSE510‐SIM2‐37, T.Tn‐Mock, T.Tn‐SIM2‐23, and T.Tn‐SIM2‐9. Gray bars represent average number of cancer pearls. Each number of cancer pearls between the Mock‐ and SIM2‐transfectants was discriminated by white and red (n = 6, *P < .05)
3.5. SIM2 and ARNT cooperatively inhibit self‐renewal of PDPN‐positive tumor basal cells
SIM2 has a nuclear localization signal, and the SIM2‐ARNT complex actively represses gene expression.30 Although SIM2 and ARNT were expressed in a good prognostic subset of ESCC cases, SIM2s and ARNT2 were rarely expressed (Figure 2A). Thus, it was thought that SIM2 and ARNT play an important role in ESCC cells. To investigate the subcellular localization of SIM2 and ARNT, SIM2‐GFP transfected cells (TE8‐SIM2‐2) were stained with anti‐GFP antibody and anti‐ARNT antibody. SIM2‐GFP fusion and ARNT were colocalized mainly in the nuclei of TE8‐SIM2‐2 cells (Figure 6A). To confirm that SIM2 binds to ARNT in the nuclei of ESCC cells, we subsequently immunoprecipitated the SIM2‐ARNT complex in both transient and stable TE8 transfectants by anti‐GFP antibody. Immunoprecipitation experiments of SIM2‐GFP and ARNT showed that SIM2‐GFP interacted with ARNT (Figure 6B; Figure S4). Furthermore, we knocked down ARNT or SIM2 expression in KYSE510 cells by small interfering RNA (siRNA) (Figure 6C,D). The PDPN‐positive tumor basal cell ratio was increased significantly by ARNT or SIM2 siRNA compared with the control siRNA. The experiments were carried out by using two kinds of ARNT or SIM2 siRNAs, and the decrement of both the mRNA and protein was confirmed (Figure S5). We also carried out ARNT‐knockdown experiments in the corresponding SIM2‐overexpressed cells (KYSE510‐SIM2‐27), and showed a significant increase of PDPN‐positive cells (Figure S6). These data suggest that SIM2 induces differentiation of PDPN‐positive tumor basal cells by cooperation with ARNT.
Figure 6.
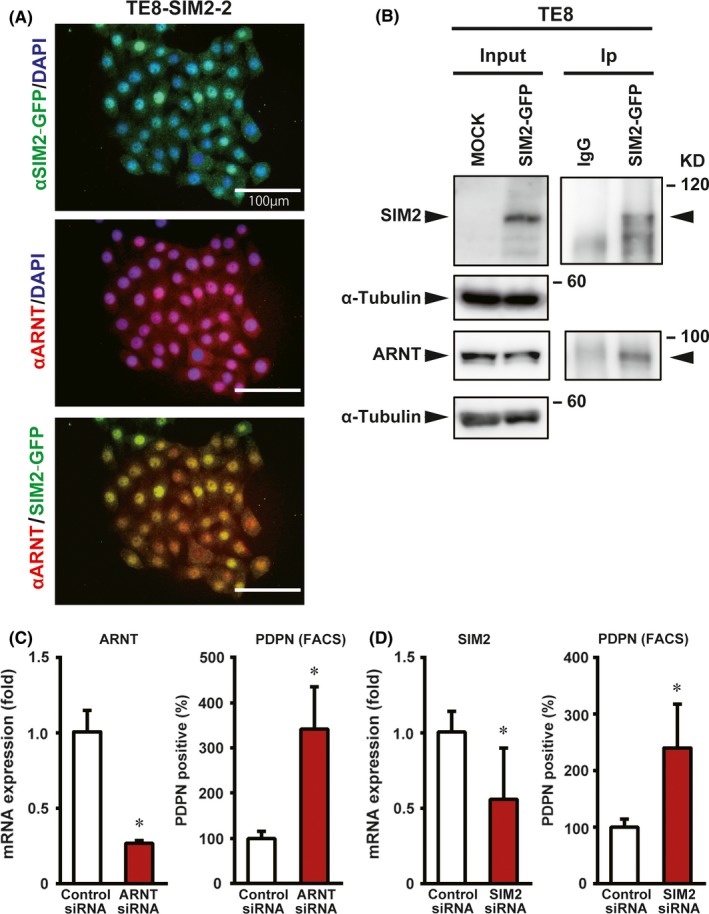
SIM2 and ARNT cooperatively decrease PDPN‐positive tumor basal cells. A, Immunofluorescence showing colocalization of SIM2‐GFP and ARNT in the nuclei of TE8‐SIM2‐2 cells. B, Immunoprecipitation of SIM2‐GFP with anti‐green fluorescent protein (GFP) antibody 1 day after transient SIM2‐GFP transfection to TE8 cells followed by western blot with anti‐SIM2, ‐ ARNT, or‐α tubulin antibody. Input, unprecipitated extracts. IgG, control IP. C, Real‐time RT‐PCR of ARNT (left) and percentage of PDPN‐positive cells (right) in KYSE510 cells that were transfected with control siRNA (white column) or ARNT siRNA (red column) (n = 3, mean + SE; *P < .05). D, Real‐time RT‐PCR of SIM2 (left) and percentage of PDPN‐positive cells (right) in KYSE510 that were transfected with control siRNA (white column) or SIM2 siRNA (red column) (n = 3, mean + SE; *P < .05)
3.6. SIM2 improves CDDP and H2O2 sensitivity of ESCC cells
Survival analyses of the subsets of 85 ESCC patients showed that prognosis is better if SIM2 mRNA expression is high in pretreatment biopsy samples, suggesting that SIM2 is involved in CRT sensitivity (Figure 1). To examine the hypothesis of SIM2‐regulated chemosensitivity of ESCC cells, we continuously treated T.Tn‐Mock, T.Tn‐SIM2‐9, and T.Tn‐SIM2‐23 with CDDP, which is a key drug in the treatment of ESCC, on alternate days for 14 days using 3D culture (Figure 7A, left). The viable cell ratio of T.Tn‐SIM2‐9 and T.Tn‐SIM2‐23 was significantly decreased compared to T.Tn‐Mock (Figure 7A, right). Subsequently, we treated TE8‐Mock and TE8‐SIM2‐2 with H2O2, which is a superoxide precursor produced by irradiation, for 24 hours using 3D culture. The viable cell ratio of TE8‐SIM2‐2 was significantly decreased compared with TE8‐Mock (Figure 7B). IC50 of TE8‐SIM2‐2, TE8‐SIM2‐3, T.Tn‐SIM2‐9 and T.Tn‐SIM2‐23 on γ‐ray irradiation was also significantly decreased compared with TE8‐Mock and T.Tn‐Mock, respectively (Figure S7). In addition, we confirmed independence between SIM2 and ARNT expression by reciprocal siRNA treatments (Figure S8A) and showed a slight increase of viable cells in SIM2 or ARNT siRNA‐transfected cells 24 hours after H2O2 treatment (Figure S8B).
Figure 7.
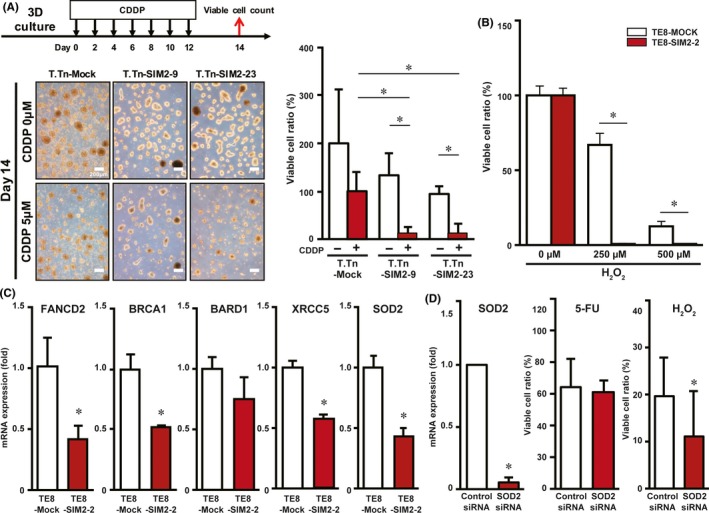
Sensitivity of SIM2‐transfectants to CDDP and H2O2 is associated with decreased expression of DNA repair and antioxidant enzymes. A, Viable cell ratios of T.Tn‐Mock, T.Tn‐SIM2‐9 and T.Tn‐SIM2‐23 cells that were treated with alternate day administration of CDDP in 3D culture for 14 days (n = 3, mean + SE; *P < .05). B, Viable cell ratios of TE8‐Mock (white column) and TE8‐SIM2‐2 cells (red column) that were treated with H2O2 in 3D culture for a day (n = 3, mean + SE; *P < .05). C, Real‐time RT‐PCR of FANCD2, BRAC1, BARD1, XRCC5, and SOD2 in 3D cultured TE8‐Mock (white column) and TE8‐SIM2‐2 (red column) cells (n = 3, mean + SE; *P < .05). D, Real‐time RT‐PCR of SOD2 (left) and viable cell ratios in fluorouracil (5‐FU) (middle) or H2O2 (right) treatment in TE8 that were transfected with control siRNA (white column) or SOD2 siRNA (red column) (n = 3, mean + SE; *P < .05)
Among SIM2‐target genes identified by microarray of TE8‐Mock and TE8‐SIM2‐2 cells in 3D culture (data not shown), we focused DNA repair and antioxidant enzymes (FANCD2, BRCA1, BARD1, XRCC5, and SOD2) as candidate SIM2‐target genes responsible for the increased sensitivity to CRT because they have been reported to be involved in chemo‐ or radio‐resistances, and carried out quantitative real‐time RT‐PCR on TE8‐Mock and TE8‐SIM2‐2 cells in 3D culture. TE8‐SIM2‐2 showed significant repression of FANCD2, BRCA1, XRCC5, and SOD2 compared to TE8‐Mock (Figure 7C). To this end, we treated untransfected TE8 cells with each siRNA of these 4 genes and investigated cell viability after 5‐FU or H2O2 treatment. Among the 4 siRNAs, a single SOD2 siRNA transfection decreased the viable cell ratio only in the H2O2‐treated cells (Figure 7D). The experiments were carried out by using two kinds of SOD2 siRNAs, and the decrement of both the mRNA and protein was confirmed (Figure S9).
4. DISCUSSION
Epigenetic changes in DNA without concomitant changes in the underlying genetic code are known to occur in human cancers.31 Promoter methylation resulting in repression of tumor suppressor genes may drive tumorigenesis. We found that SIM2 expression is repressed in cancerous tissues compared with non‐cancerous tissues in 90% of 60 ESCC patients (Figure 1A,B). Promoter hypomethylation of SIM2 was found in 4 of 5 ESCC cases with high SIM2 expression, whereas hypermethylation was found in 3 of 5 cases with low SIM2 expression (Figure 2C). Therefore, promoter methylation is thought to be one of the causes that represses SIM2 in ESCC.
We showed that overexpression of the long isoform of SIM2 decreased the expression of the major EMT regulators, SNAI2 and TWIST1, whereas the short splicing isoform SIM2s did not affect their expression (Figure 3A). Therefore, the SIM2 long isoform is thought to reduce mesenchymal characteristics in ESCC cells. Further 3D culture experiments of ESCC cells showed that overexpression of SIM2 decreased the expression of VIM and PDPN and increased the expression of differentiation markers SPRR1A and FLG (Figure 3B,C). Together, our results suggest that SIM2 plays a key role in the regulation of differentiation in squamous cell carcinoma in vitro. Interestingly, PDPN has been reported to not only be a candidate marker of tumor‐initiating cells but also a metastasis‐promoting factor in squamous cell carcinoma. When PDPN expression was knocked down, ESCC cells have shown defective invasion and tumorigenic activities.32 In our study, stable SIM2‐transfectants showed decreased ratios of PDPN‐positive tumor basal cells (Figure 4B). Tumors derived from SIM2‐transfected KYSE510 and T.Tn cells were small and showed a well‐differentiated histology with PDPN‐positive tumor basal cells located at the outer edge of the tumor nests (Figure 5B). In Figure 4B, SIM2 transfection strongly reduced PDPN‐positive cells in KYSE510 and T.Tn cells, but the reduction was limited in TE8 cells. TE8 cells are thought to have a high self‐renewal ability of PDPN‐positive tumor basal cells, because TE8 contained 98.5% PDPN‐positive cells. In this case, differentiation ability or asymmetrical cell division ability was not able to occur by SIM2 only. SIM2‐overexpressed cells form spheroids and proliferate more than control cells (Figures 3B and 4C). This seems not to be matched with the result of xenograft tumor growth (Figure 5A), although the differentiation phenotype is consistent. In xenograft of SIM2‐overexpressed cells, complete cell differentiation occurs, resulting in keratinization and/or cell death; however, only incomplete cell differentiation occurs under 3D culture. Therefore, only in vivo tumor growth of SIM2‐overexpressed cells may be delayed or suppressed. These results suggest that SIM2 has an important role not only in histological differentiation but also in tumorigenesis, at least in the subset of ESCC.
In addition, immunofluorescence and immunoprecipitation of transient or stable SIM2‐transfectants showed that SIM2 is localized in the nuclei and interacted with ARNT (Figure 6A,B; S4). Furthermore, knockdown of ARNT expression in the transient or stable SIM2‐transfectants resulted in an increment of PDPN‐positive tumor basal cells, suggesting that SIM2 and ARNT cooperatively induce differentiation of PDPN‐positive tumor basal cells (Figure 6C,D; S6).
High SIM2 expression in cancerous tissue did not significantly affect the prognosis of ESCC patients who underwent esophagectomy alone (data not shown); however, in patients who were treated with definitive CRT, high SIM2 expression was associated with a good prognosis (Figure 1C). Knockdown of PDPN expression has been reported to reduce resistance to anticancer drugs, 5‐FU and CDDP.32 Our findings were congruous with the previous report that SIM2‐transfectants reduced the PDPN‐positive basal cell ratio and improved sensitivity to CDDP (Figure 4B,C, 7A). Taken together, it is plausible that the basal cell reduction by induction of differentiation is one of the mechanisms of CRT sensitivity of ESCC with high SIM2 expression. FANCD2, BRCA1, BARD1, and XRCC5 have been reported to play pivotal roles in the DNA repair pathway, and FANCD2 and BARD1 interact with BRCA1 in the repair of DNA interstrand cross‐links and DNA double‐strand breaks, respectively.33, 34, 35 SOD2 is known to efficiently catalyze the dismutation of reactive oxygen species,36 which are induced by irradiation. In line with the facts, FANCD2, BRCA1, XRCC5, and SOD2 were downregulated in the 3D cultured SIM2‐transfectant (Figure 7C). Among the 4 siRNAs of these 4 genes, only a single SOD2 siRNA transfection decreased the mRNA and the viable cell ratio in the H2O2‐treated cells (Figure 7D). These results suggest that SIM2 increases chemo‐ and radiosensitivity by simultaneous repression of multiple DNA repair enzymes and repression of a single antioxidant enzyme.
Microvessel density in squamous cell carcinoma has been reported to be associated with radiosensitivity.37 Such microvessels were often observed in the thick stroma in well‐differentiated ESCC (Figure S10A) and also in the xenograft with a well‐differentiated histology of KYSE510 cells (Figure S10B). Interestingly, the xenograft of the SIM2‐transfectant T.Tn‐SIM2‐9 also showed increased angiogenesis (Figure S10C). Tumor angiogenesis is another complex phenotype at the crossroads of multiple intra‐ and extracellular signaling, and it is possible that SIM2 stimulates angiogenesis indirectly but contributes to chemo‐ and radiosensitivity through delivery of CDDP and oxygen to tumor basal cells in vivo.
We summarized possible SIM2 roles in good (left) and poor (right) responders to CRT for ESCC (Figure S11). In the left panel, ESCC tumors with high SIM2 expression may maintain squamous differentiation potential of the esophageal mucosa and restrain the expansion of the PDPN‐positive tumor basal cell layer. Downregulation of DNA repair and antioxidant enzymes may also contribute to the enhanced sensitivity to CRT. The upstream signals of SIM2 may become drug targets for differentiation therapy. However, the downstream signals, which should be identified less in well‐differentiated cancers (Figure S11, left panel), may also be therapeutic targets to poor prognostic ESCC with low SIM2 expression (Figure S11, right panel). By contrast, the right panel shows the loss of the SIM2‐mediated suppressions depicted in the left panel, leading to the increased resistance to CRT. In conclusion, SIM2 is frequently suppressed in ESCC, and is involved in CRT sensitivity through differentiation of PDPN‐positive tumor basal cells and repression of DNA repair and antioxidant enzymes by cooperation with ARNT. Therefore, the downstream signal pathways repressed by the SIM2‐ARNT complex and the upstream signal pathways of SIM2 may provide therapeutic targets in ESCC.
CONFLICTS OF INTEREST
Authors declare no conflicts of interest for this article.
Supporting information
ACKNOWLEDGMENTS
The authors thank Mr Richard De Lapp for editorial comments. This work was supported by the National Institute of Biomedical Innovation, Health and Nutrition (for the Advanced Research for Medical Products Mining Programme), Japan Agency for Medical Research and Development (Practical Research for Innovative Cancer Control), and National Cancer Center Research and Development Fund.
Tamaoki M, Komatsuzaki R, Komatsu M, et al. Multiple roles of single‐minded 2 in esophageal squamous cell carcinoma and its clinical implications. Cancer Sci. 2018;109:1121–1134. https://doi.org/10.1111/cas.13531
Funding information
Japan Agency for Medical Research and Development (Grant/Award Number: ‘Practical Research for Innovative Cancer Control’), National Cancer Center (Grant/Award Number: ‘National Cancer Center Research and Development Fu’), National Institute of Biomedical Innovation, Health and Nutrition (Grant/Award Number: ‘Advanced Research for Medical Products Mining Prog’)
REFERENCES
- 1. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69‐90. [DOI] [PubMed] [Google Scholar]
- 2. Ando N, Kato H, Igaki H, et al. A randomized trial comparing postoperative adjuvant chemotherapy with cisplatin and 5‐fluorouracil versus preoperative chemotherapy for localized advanced squamous cell carcinoma of the thoracic esophagus (JCOG9907). Ann Surg Oncol. 2012;19:68‐74. [DOI] [PubMed] [Google Scholar]
- 3. Kato K, Muro K, Minashi K, et al. Phase II study of chemoradiotherapy with 5‐fluorouracil and cisplatin for Stage II‐III esophageal squamous cell carcinoma: JCOG trial (JCOG 9906). Int J Radiat Oncol Biol Phys. 2011;81:684‐690. [DOI] [PubMed] [Google Scholar]
- 4. Broders AC. Epithelioma of the genito‐urinary organs. Ann Surg. 1922;75:574‐604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Arthur JF, Fenner ML. The influence of histological grading on prognosis in carcinoma of the tongue (a computer analysis of 299 cases). Clin Radiol. 1966;17:384‐396. [DOI] [PubMed] [Google Scholar]
- 6. Rowe DE, Carroll RJ, Day CL Jr. Prognostic factors for local recurrence, metastasis, and survival rates in squamous cell carcinoma of the skin, ear, and lip. Implications for treatment modality selection. J Am Acad Dermatol. 1992;26:976‐990. [DOI] [PubMed] [Google Scholar]
- 7. Tanaka Y, Aoyagi K, Minashi K, et al. Discovery of a good responder subtype of esophageal squamous cell carcinoma with cytotoxic T‐lymphocyte signatures activated by chemoradiotherapy. PLoS One. 2015;10:e0143804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chrast R, Scott HS, Chen H, et al. Cloning of two human homologs of the Drosophila single‐minded gene SIM1 on chromosome 6q and SIM2 on 21q within the Down syndrome chromosomal region. Genome Res. 1997;7:615‐624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bersten DC, Sullivan AE, Peet DJ, Whitelaw ML. bHLH–PAS proteins in cancer. Nat Rev Cancer. 2013;13:827‐841. [DOI] [PubMed] [Google Scholar]
- 10. Swanson HI, Chan WK, Bradfield CA. DNA binding specificities and pairing rules of the Ah receptor, ARNT, and SIM proteins. J Biol Chem. 1995;270:26292‐26302. [DOI] [PubMed] [Google Scholar]
- 11. Moffett P, Pelletier J. Different transcriptional properties of mSim‐1 and mSim‐2. FEBS Lett. 2000;466:80‐86. [DOI] [PubMed] [Google Scholar]
- 12. Farrall AL, Whitelaw ML. The HIF1alpha‐inducible pro‐cell death gene BNIP3 is a novel target of SIM2s repression through cross‐talk on the hypoxia response element. Oncogene. 2009;28:3671‐3680. [DOI] [PubMed] [Google Scholar]
- 13. Metz RP, Kwak HI, Gustafson T, Laffin B, Porter WW. Differential transcriptional regulation by mouse single‐minded 2s. J Biol Chem. 2006;281:10839‐10848. [DOI] [PubMed] [Google Scholar]
- 14. DeYoung MP, Tress M, Narayanan R. Identification of Down's syndrome critical locus gene SIM2‐s as a drug therapy target for solid tumors. Proc Natl Acad Sci USA. 2003;100:4760‐4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kwak HI, Gustafson T, Metz RP, Laffin B, Schedin P, Porter WW. Inhibition of breast cancer growth and invasion by single‐minded 2s. Carcinogenesis. 2007;28:259‐266. [DOI] [PubMed] [Google Scholar]
- 16. DeYoung MP, Tress M, Narayanan R. Down's syndrome‐associated Single Minded 2 gene as a pancreatic cancer drug therapy target. Cancer Lett. 2003;200:25‐31. [DOI] [PubMed] [Google Scholar]
- 17. Halvorsen OJ, Rostad K, Øyan AM, et al. Increased expression of SIM2‐s protein is a novel marker of aggressive prostate cancer. Clin Cancer Res. 2007;13:892‐897. [DOI] [PubMed] [Google Scholar]
- 18. Laffin B, Wellberg E, Kwak HI, et al. Loss of singleminded‐2s in the mouse mammary gland induces an epithelial‐mesenchymal transition associated with up‐regulation of slug and matrix metalloprotease 2. Mol Cell Biol. 2008;28:1936‐1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Scribner KC, Behbod F, Porter WW. Regulation of DCIS to invasive breast cancer progression by Singleminded‐2s (SIM2s). Oncogene. 2013;32:2631‐2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wellberg E, Metz RP, Parker C, Porter WW. The bHLH/PAS transcription factor singleminded 2s promotes mammary gland lactogenic differentiation. Development. 2010;137:945‐952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aleman MJ, DeYoung MP, Tress M, Keating P, Perry GW, Narayanan R. Inhibition of Single Minded 2 gene expression mediates tumor‐selective apoptosis and differentiation in human colon cancer cells. Proc Natl Acad Sci USA. 2005;102:12765‐12770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Arredouani MS, Lu B, Bhasin M, et al. Identification of the transcription factor single‐minded homologue 2 as a potential biomarker and immunotherapy target in prostate cancer. Clin Cancer Res. 2009;15:5794‐5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Long Q, Johnson BA, Osunkoya AO, et al. Protein‐coding and microRNA biomarkers of recurrence of prostate cancer following radical prostatectomy. Am J Pathol. 2011;179:46‐54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yamaki A, Tochigi J, Kudoh J, Minoshima S, Shimizu N, Shimizu Y. Molecular mechanisms of human single‐minded 2 (SIM2) gene expression: identification of a promoter site in the SIM2 genomic sequence. Gene. 2001;270:265‐275. [DOI] [PubMed] [Google Scholar]
- 25. Isohata N, Aoyagi K, Mabuchi T, et al. Hedgehog and epithelial‐mesenchymal transition signaling in normal and malignant epithelial cell of the esophagus. Int J Cancer. 2009;125:1212‐1221. [DOI] [PubMed] [Google Scholar]
- 26. Tong L, Yuan S, Feng F, Zhang H. Role of podoplanin expression in esophageal squamous cell carcinoma: a retrospective study. Dis Esophagus. 2012;25:72‐80. [DOI] [PubMed] [Google Scholar]
- 27. Chuang WY, Yeh CJ, Wu YC, et al. Tumor cell expression of podoplanin correlates with nodal metastasis in esophageal squamous cell carcinoma. Histol Histopathol. 2009;24:1021‐1027. [DOI] [PubMed] [Google Scholar]
- 28. Atsumi N, Ishii G, Kojima M, Sanada M, Fujii S, Ochiai A. Podoplanin, a novel marker of tumor‐initiating cells in human squamous cell carcinoma A431. Biochem Biophys Res Commun. 2008;373:36‐41. [DOI] [PubMed] [Google Scholar]
- 29. Yoshikawa M, Tsuchihashi K, Ishimoto T, et al. xCT inhibition depletes CD44v‐expressing tumor cells that are resistant to EGFR‐targeted therapy in head and neck squamous cell carcinoma. Cancer Res. 2013;73:1855‐1866. [DOI] [PubMed] [Google Scholar]
- 30. Yamaki A, Kudoh J, Shimizu N, Shimizu Y. A novel nuclear localization signal in the human single‐minded proteins SIM1 and SIM2. Biochem Biophys Res Commun. 2004;313:482‐488. [DOI] [PubMed] [Google Scholar]
- 31. Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415‐428. [DOI] [PubMed] [Google Scholar]
- 32. Rahadiani N, Ikeda J, Makino T, et al. Tumorigenic role of podoplanin in esophageal squamous‐cell carcinoma. Ann Surg Oncol. 2010;17:1311‐1323. [DOI] [PubMed] [Google Scholar]
- 33. Huen MS, Sy SM, Chen J. BRCA1 and its toolbox for the maintenance of genome integrity. Nat Rev Mol Cell Biol. 2010;11:138‐148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim H, D'Andrea AD. Regulation of DNA cross‐link repair by the Fanconi anemia/BRCA pathway. Genes Dev. 2012;26:1393‐1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Thacker J, Zdzienicka MZ. The XRCC genes: expanding roles in DNA double‐strand break repair. DNA Repair (Amst). 2004;3:1081‐1090. [DOI] [PubMed] [Google Scholar]
- 36. Zelko IN, Mariani TJ, Folz RJ. Superoxide dismutase multigene family: a comparison of the CuZn‐SOD (SOD1), Mn‐SOD (SOD2), and EC‐SOD (SOD3) gene structures, evolution, and expression. Free Radic Biol Med. 2002;33:337‐349. [DOI] [PubMed] [Google Scholar]
- 37. Kamijo T, Yokose T, Hasebe T, et al. Potential role of microvessel density in predicting radiosensitivity of T1 and T2 stage laryngeal squamous cell carcinoma treated with radiotherapy. Clin Cancer Res. 2000;6:3159‐3165. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


