Abstract
Engineered nanomaterials (ENM) are now used in a wide variety of fields, and, thus, their safety should urgently be assessed and secured. It has been suggested that inflammatory responses via the phagocytosis of ENM by macrophages is a key mechanism for their genotoxicity. The present study was conducted to establish a mechanism‐based assay to evaluate the genotoxicity of ENM under conditions simulating an in vivo situation, featuring a co‐culture system of murine lung resident cells (GDL1) and immune cells (RAW264.7). GDL1 were cultured with or without RAW264.7, exposed to a multi‐walled carbon nanotube (MWCNT), and then analyzed for mutagenicity and underlying mechanisms. Mutation frequencies induced in GDL1 by the MWCNT were significantly greater with the co‐existence of RAW264.7 than in its absence. Mutation spectra observed in GDL1 co‐cultured with RAW264.7 were different from those seen in GDL1 cultured alone, but similar to those observed in the lungs of mice exposed to the MWCNT in vivo. Inflammatory cytokines, such as IL‐1β and TNF‐α, were produced from RAW264.7 cells treated with the MWCNT. The generation of reactive oxygen species and the formation of 8‐oxodeoxyguanosine in GDL1 exposed to the MWCNT were greater in the co‐culture conditions than in the single culture conditions. Based on these findings, it is indicated that inflammatory responses are involved in the genotoxicity of MWCNT, and that the presently established, novel in vitro assay featuring a co‐culture system of tissue resident cells with immune cells is suitable to evaluate the genotoxicity of ENM.
Keywords: co‐culture, GDL1 cells, genotoxicity, multi‐walled carbon nanotube, nanomaterials
1. INTRODUCTION
Engineered nanomaterials (ENM) are used in various fields of the human environment, and assessment of their safety is, therefore, urgently required. So far, we have examined the genotoxicity of various ENM by using in vitro and in vivo assays, which indicated that inflammatory response, induced by macrophages and other immune cells, is considered a key mechanism for the genotoxicity and other toxic properties of ENM.1, 2, 3, 4, 5 In general, a single culture of tissue resident cells has been used for in vitro assays to evaluate genotoxicity of ENM. Based on the aforementioned genotoxic mechanisms of ENM, however, the interactions between tissue resident cells and immune cells seems to be very important. Thus, it was thought that a single culture was not necessarily sufficient to evaluate the genotoxicity of ENM. In fact, we recently reported that DNA damage was induced in A549 cells, derived from human lung cancer cells, by exposure to a nanomaterial, kaolin, and the magnitude of the DNA damage formation was markedly greater with the co‐existence of kaolin‐exposed RAW264, derived from murine macrophage‐like immune cells, than in its absence.6 Therefore, it is suggested that a co‐culture of tissue resident cells and immune cells could be a suitable in vitro assay to evaluate the genotoxicity of ENM because such a condition well simulates the in vivo situation. In the present study, we conducted a co‐culture of gpt delta L1 (GDL1) cells and a murine macrophage, RAW264.7. A GDL1 cell line was established by Takeiri et al7 from gpt delta transgenic mouse lung fibroblasts, and this cell line could be a useful tool to evaluate mutagenicity induced by various chemicals.
It has been well documented that a multi‐walled carbon nanotube (MWCNT) including MWNT‐7, originally supplied by Mitsui, Ibaragi, Japan, induces pulmonary carcinogenicity.8, 9, 10, 11, 12 Based on the available data that carcinogenic properties vary among MWCNT products and depend on their physicochemical characteristics,11, 12 the International Agency for Research on Cancer has categorized the consistently carcinogenic MWNT‐7 into group 2B, “possibly carcinogenic to humans,” while the other MWCNT products have been placed in group 3, “not classifiable as to their carcinogenicity to humans.”13 However, we have revealed the genotoxicity of MWNT‐7 as micronucleus induction, chromosome aberration and DNA damage, using in vitro and in vivo assays.4
Taken together, the present study was conducted to establish a mechanism‐based assay to evaluate the genotoxicity of ENM under conditions simulating the in vivo situation, featuring a co‐culture system of tissue resident cells, GDL1, and immune cells, RAW264.7. GDL1 cells were cultured with or without RAW264.7, exposed to a test ENM, MWNT‐7, and then analyzed for mutagenicity and underlying mechanisms.
2. MATERIALS AND METHODS
2.1. Materials and cells
MWNT‐7 was kindly provided by Mitsui (Ibaragi, Japan). The size distribution of MWNT‐7 used in the present study was analyzed by dynamic light scattering (DLS) using FPAR‐1000 (Otsuka Electronics, Osaka, Japan). MWNT‐7 was shown to have ranges of 914.85‐1323.77 nm, and the most abundant size was at 1104.9 ± 86.2 nm. MWNT‐7 was suspended in saline containing 0.05% of Tween 80 (Nacalai Tesque, Kyoto, Japan), at a concentration of 2.0 mg/mL.
GDL1 cells, established from gpt delta mice lung fibroblasts,7 were kindly gifted from Dr Akira Takeiri, Chugai Pharmaceutical. RAW264 was purchased from RIKEN Cell Bank.
2.2. Cytotoxicity of multi‐walled carbon nanotube
Cell viability was determined using the MTT assay (Sigma‐Aldrich, Tokyo, Japan) according to the manufacturer's instructions. Briefly, cells (2.0‐4.0 × 104 cells/well) were incubated with various concentrations of MWNT‐7 for 24 hours at 37°C. Thiazolyl blue tetrazolium bromide (MTT) solution (5 mg/mL) was added to each well at a final concentration of 0.5 mg/mL and incubated for 3 hours. After aspiration of the culture medium, insoluble formazan was dissolved with DMSO, then cell viability was measured using a plate reader at 570 nm (Viento XS, DS Pharma Biomedical, Suita, Osaka, Japan). In addition to single‐culture conditions, cell viability of GDL1 cells co‐cultured with RAW264 was also performed using the procedure as described below.
2.3. Incorporation of multi‐walled carbon nanotube into the cells
The incorporation rates of MWNT‐7 into the cells were measured using flow cytometry (FCM) analysis as described in previous research.6, 14 Briefly, GDL1 and RAW264.7 treated with MWNT‐7 for 3 hours were trypsinized and suspended in fresh culture medium. Propidium iodide (0.5 μg/mL) was added to each cell suspension to determine cell survival rates. The numbers of cells incorporating MWNT‐7 were analyzed with FACSCaliber (Becton Dickinson, Mountain View, CA, USA). In the FCM analysis, forward‐scattered light indicating cell size and side‐scattered (SS) light indicating intracellular complexity were observed. The cells increasing SS were considered as incorporating cells, and the change in SS was an index of incorporation rates as described previously.6, 14
2.4. Co‐culture system of lung resident fibroblasts and macrophage‐like immune cells
To establish the biomimetic system of lungs, GDL1 and RAW264.7 were co‐cultured in the same culture medium as described previously.6 Briefly, in the case of the single culture condition (SC), GDL1 cells were cultured in MEM (Nacalai Tesque) supplemented with 10% FBS, penicillin (100 U/mL) and streptomycin (0.1 mg/mL) for 24 hours, empty cell culture inserts (pore size; 0.4 μm, high density, Greiner Bio‐One, St. Gallen, Switzerland) were set, and 24 hours thereafter MWNT‐7 at appropriate concentrations was administered into the medium outside of the inserts to treat only GDL1 cells (Figure 1A) for 24 hours. In the case of the co‐culture conditions (CC), GDL cells were cultured for 24 hours, and RAW264.7 cells were subsequently seeded into the inserts. Another 24 hours thereafter, MWNT‐7 with appropriate concentrations was administered into the medium in the inserts to treat only RAW264.7 cells (Figure 1B), or into both in and outside the inserts to treat both cells (Figure 1C) for 24 hours. After the end of the treatment, MWNT‐7 was removed and GDL1 cells were harvested for assays as described below.
Figure 1.
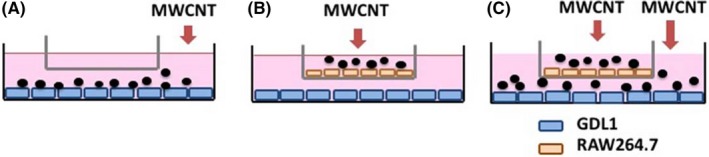
Co‐culture model. GDL1 cells were cultured in MEM supplemented with 10% FBS, penicillin and streptomycin for 24 h, setting empty cell culture inserts, and MWNT‐7 at appropriate concentrations was administered into the medium outside the inserts to treat only GDL1 cells for 24 h (A). GDL cells were cultured for 24 h, RAW264.7 cells were subsequently seeded into the inserts, and 24 h thereafter MWNT‐7 at appropriate concentrations was administered into the medium in the inserts to treat only RAW264.7 cells (B), or both in and outside the inserts to treat both cells (C) for 24 h. After the end of the treatment, MWNT‐7 was removed, and GDL1 cells were harvested for assays as described in the Materials and Methods
2.5. Mutagenicity of multi‐walled carbon nanotube, a gpt mutation assay
For the mutation analysis, appropriate concentrations of MWNT‐7 were exposed to only GDL1 cells (n = 3, SC, Figure 1A) or RAW264.7 cells (n = 3, CC‐R, Figure 1B), and both GDL1 and RAW264.7 cells (n = 3, CC‐RG, Figure 1C). The control samples (n = 3) were prepared with the solvent only treatment under the SC and CC conditions. After 24 hours exposure, MWNT‐7 was removed and GDL1 cells were trypsinized and reseeded, then cultured for a further 7‐9 days, including subcultures. The cells were then re‐harvested and stored at −80°C, until the DNA was isolated for the mutation assay. High‐molecular‐weight genomic DNA was extracted from the cultured cells using a RecoverEase DNA Isolation Kit (Stratagene, La Jolla, CA, USA) according to the supplier's instructions. Lambda EG10 phages were rescued using a Transpack Packaging Extract (Stratagene). The gpt mutagenesis assay was performed according to the previously described method.3, 4, 5, 6, 7 The mutation spectra of the gpt coding sequence were assessed by PCR and direct sequencing. Briefly, a 739‐bp DNA fragment containing gpt was amplified by PCR as described previously,3, 6, 15 and the sequencing analysis was done at Takara Bio (Mie, Japan).
2.6. Generation and release from RAW264.7 by multi‐walled carbon nanotube of inflammatory cytokines
Inflammatory cytokines in the culture supernatant of RAW264.7 were measured. RAW264.7 cells were exposed to 20 or 80 μg/mL of MWNT‐7 for 24 hours, and subsequently interleukin (IL)‐1β and tumor necrosis factor (TNF)‐α were quantified using immunoassay kits, Mouse IL‐1β (BioSource International, Camarillo, CA, USA) and Quantikine Mouse TNF‐α (R&D Systems, Minneapolis, MN, USA), respectively, according to the manufacturer's protocols.
2.7. Generation by multi‐walled carbon nanotube of reactive oxygen species
The generation of reactive oxygen species (ROS) in the cells was measured using FCM with 2′,7′‐dichlorodihydrofluorescin diacetate (DCFH‐DA) as described previously.6, 16 DCFH‐DA without fluorescence permeates cell membrane, is subsequently hydrolyzed by unspecific esterases and is converted into DCF with fluorescence in the presence of ROS. RAW264 cells were treated with MWNT‐7 at a dose of 80 μg/mL for 3 hours. GDL1 cells were also treated with MWNT‐7 under both SC and CC‐RG conditions for 3 hours. After washing out the MWNT‐7 with PBS, both cells were trypsinized and suspended, then DCFH‐DA was added into the cell suspensions and incubated at 37°C for 30 minutes. The cell suspensions were applied to FCM analysis, and the fluorescence was detected at an excitation wavelength of 485 nm and emission wavelength of 530 nm. The numbers of cells increasing fluorescence were measured as ROS generating cells.
2.8. Formation by multi‐walled carbon nanotube of oxidative stress‐related DNA adduct, 8‐oxodeoxyguanosine
The formation of 8‐oxodeoxyguanosine (8‐oxo‐dG) was measured by the same procedure described previously.4 Briefly, MWNT‐7 was exposed to GDL1 cells under both SC and CC‐RG conditions for 24 hours. Control samples were obtained from the vehicle‐exposed GDL1 cells. DNA was extracted and purified using a Gentra Puregene tissue kit (QIAGEN, Valencia, CA) according to the manufacturer's instructions, except that desferrioxamine (final concentration: 0.1 mmol/L) was added to all solutions to avoid the formation of oxidative adducts during the purification step. The extracted DNA was stored at −80°C until analysis for DNA adducts. DNA (40 μg) was extracted from vehicle (n = 3) and MWNT‐7‐treated (n = 3) cells were enzymatically digested, and 8‐oxo‐dG was analyzed and quantified using LC‐MS/MS.
2.9. Statistical analysis
For the statistical comparison of the experimental and control group data from the gpt mutation and DNA adduct assays, expressed as means ± SD, the F‐test was initially performed. If the F‐test revealed that the variances were unequal, the P‐value was determined using Welch's t test. If not, Student's t test was used instead. In the case of the mutation spectrum analysis, P‐values were determined using Fisher's exact test according to Carr and Gorelick.17 In any case, P‐values lower than .05 were considered to indicate statistical significance.
3. RESULTS
3.1. Cytotoxicity of multi‐walled carbon nanotube
A 24‐hour treatment of more than 40 μg/mL of MWNT‐7 caused significant but rather small growth inhibition against both cells compared with the vehicle control (Figure S1). To examine whether the MWNT‐7 cytotoxicity against GDL1 was affected by co‐existing RAW264.7, we also analyzed MTT assay under the co‐culture conditions (Figure 2). When low (20 μg/mL) or high (80 μg/mL) doses of MWNT‐7 were administered only to RAW264.7, no obvious cytotoxicity was observed in GDL1 cells. However, when MWNT‐7 was exposed to only GDL1 cells, the cells were killed. When MWNT‐7 was simultaneously exposed to both cells, the cell viability of GDL1 cells was slightly but significantly decreased. Furthermore, in this case, the cytotoxicity of MWNT‐7 on GDL1 cells was enhanced by the co‐existence of RAW264.7. To determine appropriate levels of MWNT‐7, therefore, doses of 20 and 80 μg/mL of MWNT‐7, which do not markedly decrease cell viability, were chosen for subsequent experiments.
Figure 2.
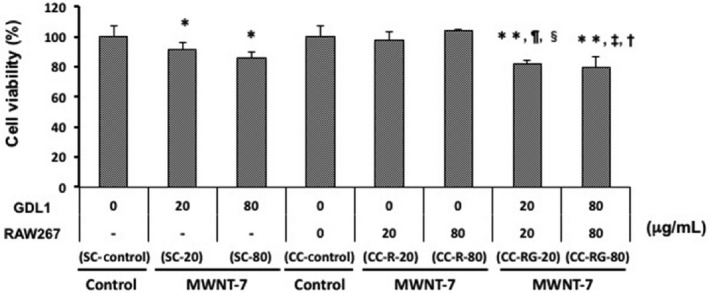
Cytotoxicity of multi‐walled carbon nanotube (MWCNT) in GDL1 with/without RAW264.7. Data are expressed as the means ± SD. *P < .05 vs SC‐control, **P < .05 vs CC‐control, ¶ P < .05 vs SC‐CNT‐20, ‡ P < .05 vs SC‐CNT‐80, § P < .05 vs CC‐CNT‐20, † P < .05 vs CC‐R‐CNT‐80
3.2. Incorporation of multi‐walled carbon nanotube into the cells
After 3 hours exposure of MWNT‐7 to RAW264 or GDL1 cells, incorporation rates into the cells were analyzed by FCM analysis according to the procedure described in the Materials and Methods. MWNT‐7 was incorporated into both RAW264.7 and GDL1 cells in a dose‐dependent manner, with comparable rates (Figure 3).
Figure 3.
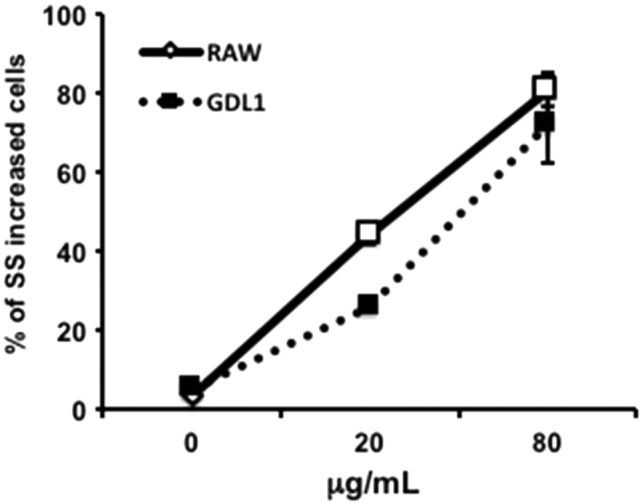
Incorporation rates of multi‐walled carbon nanotube into the cells. Data are expressed as the means ± SD (n = 3). SS, side‐scattered
3.3. In vitro mutagenicity of multi‐walled carbon nanotube assessed in the co‐culture system
gpt mutant frequencies (MF) in the single cultured GDL1 cells exposed to both 20 and 80 μg/mL of MWNT‐7 (SC‐CNT‐20 and SC‐CNT‐80, respectively) significantly increased from that of the control group (SC‐control) (Figure 4 and Table S1). MF in GDL1 co‐cultured with RAW264.7 and not exposed to MWNT‐7 (CC‐control) was comparable to that of the SC‐control (Figure 4 and Table S1). When 20 and 80 μg/mL of MWNT‐7 was exposed to only RAW264.7 (CC‐R‐CNT‐20 and CC‐R‐CNT‐80), MF in GDL1 were slightly increased but not statistically significantly. In contrast, when MWNT‐7 was exposed to both GDL1 and co‐existing RAW264.7 (CC‐RG‐CNT‐20 and CC‐RG‐CNT‐80, respectively), MF in GDL1 increased in a dose‐dependent manner (Figure 4 and Table S1). While MF in GDL1 of CC‐RG‐CNT‐20 was in the same range as that of SC‐CNT‐20, MF in GDL1 of CC‐RG‐CNT‐80 was significantly greater than that of SC‐CNT‐80 (Figure 4 and Table S1).
Figure 4.
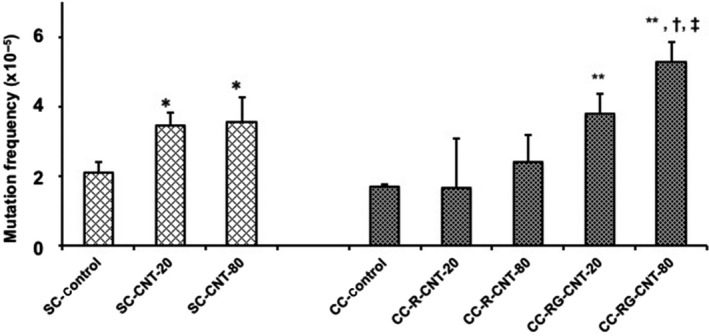
gpt mutant frequencies by exposure to multi‐walled carbon nanotube in GDL1 with or without co‐existing RAW264.7. The data represent the means ± SD. Control represents treatment with vehicle (saline containing 0.05% of Tween 80). *P < .05 vs SC‐control, **P < .05 vs CC‐control, †, P < .05 vs CC‐R‐CNT‐80, ‡, P < .05 vs SC‐CNT‐80
The classes of the mutations found in the gpt gene of GDL1 cells are summarized in Figure 5 and Table S2. Base substitutions predominated in both MWNT‐7‐induced and spontaneous cases. Under single culture conditions, the numbers of G:C to C:G, A:T to T:A and deletions tended to increase by MWNT‐7 treatment, even without statistical significance. Under the co‐culture conditions, G:C to A:T and G:C to C:G were significantly increased by MWNT‐7 treatment. Moreover, the mutation spectra observed in MWNT‐7‐treated GDL1 under co‐cultured conditions were different from those in MWNT‐7‐treated GDL1 under single culture conditions; both G:C to A:T and G:C to C:G mutations were statistically greater in CC‐RG‐CNT‐80 than in SC‐CNT‐80. Interestingly, the mutation spectrum of CC‐RG‐CNT‐80, but not SC‐CNT‐80, was similar to that observed in the lungs of mice exposed to MWNT‐7 in vivo in Kato et al.4 However, an Spi− mutation assay was not performed in the present study because we have previously reported that Spi− mutation frequency was not increased in the lungs of gpt delta mice intratracheally instilled with MWNT‐7.4
Figure 5.

gpt mutation spectra by exposure to multi‐walled carbon nanotube of GDL1 with or without co‐existing RAW264.7. Numbers in parentheses indicate the total numbers of analyzed clones. The column at the right end demonstrates our published data of the mutation spectrum observed in the lung of mice exposed to MWNT‐7 in vivo4
3.4. Production and release of inflammatory cytokines from RAW264.7 by multi‐walled carbon nanotube
When exposed to 20 and 80 μg/mL of MWNT‐7 for 24 hours (CNT‐20 and CNT‐80, respectively), RAW264.7 cells significantly generated and released both IL‐1β and TNF‐α (Figure 6).
Figure 6.

Generation and release from RAW264.7 of inflammatory cytokines, TNF‐α (A) and IL‐1β (B), by multi‐walled carbon nanotube. The values represent the means of 3 independent studies ± SD. *P < .05 vs control
3.5. Generation of reactive oxygen species in multi‐walled carbon nanotube‐exposed cells
The generation of ROS in single cultured RAW264.7 was enhanced almost 2‐fold by exposure to 80 μg/mL of MWNT‐7 (Figure 7A). In the case of GDL1 cells, whereas ROS were not apparently generated even after exposure to MWNT‐7 under single culture conditions, they were significantly generated when co‐cultured with RAW264.7 (Figure 7B).
Figure 7.
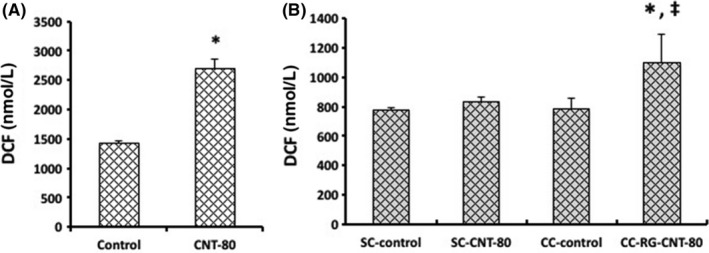
Generation of reactive oxygen species in multi‐walled carbon nanotube‐exposed cells, single cultured RAW264.7 (A) or GDL1 cultured with or without co‐existence of RAW264.7 (B). The values represent the means ± SD. *P < .05 vs control (A) or CC‐control (B), ‡ P < .05 vs SC‐CNT‐80
3.6. Formation of oxidative DNA adducts in GDL1 cells exposed to multi‐walled carbon nanotube
Formation of 8‐oxo‐dG was increased by MWNT‐7 treatment in GDL1 either with or without co‐existence of RAW264.7, and was greater under the co‐culture conditions than under the single culture conditions (Figure 8).
Figure 8.
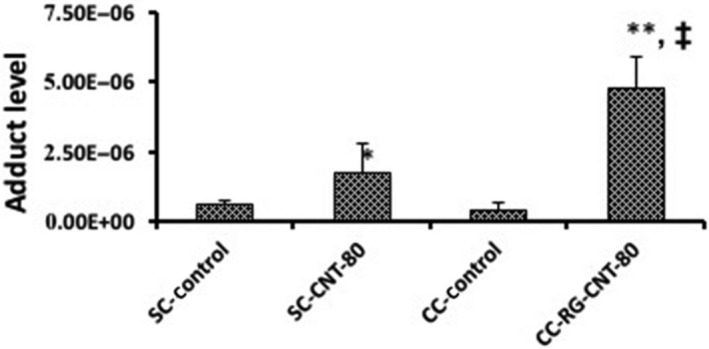
Formation of oxidative stress‐related DNA adducts by multi‐walled carbon nanotube in GDL1 cells with/without the co‐existence of RAW264.7 cells. The values represent the means of 3 independent studies ± SD. *P < .05 vs SC‐control, **P < .05 vs CC‐control, ‡ P < .05 vs SC‐CNT‐80
4. DISCUSSION
Engineered nanomaterials (ENM) have been introduced into the human environment as innovative and promising manufactured materials. Because development had been given priority, assessment of the safety of ENM tended to be put off. However, recently, large and global efforts have been made to assess and manage the risks of ENM. It has been pointed out that ENM may come to possess their biological functions, including toxicity, by the different modes of action from overscaled counterparts, and that inflammatory response by virtue of immune cells, such as macrophages, and oxidative stress and signaling agents, such as cytokines, play major roles in the mechanism underlying effects of ENM on tissue resident cells.4, 5, 6 In Kato et al. (2017) we revealed that the interaction between tissue resident cells and immune cells is important for the genotoxic mechanisms of nanomaterials. This was done by applying a fine particulate matter, kaolin, to a novel assay to evaluate (geno)toxicity featuring a co‐culture system of human lung epithelial cells, A549, and murine macrophage‐like immune cells, RAW264.7.6 GDL1 cells were established from lung resident fibroblasts of gpt delta transgenic mice, and are, thus, capable of separately detecting point mutations and deletions using gpt and Spi− assays, respectively.7 Based on this, the present study was conducted to establish a useful assay to evaluate genotoxicity of ENM using GDL1 cells under co‐culture conditions with immune cells. This system is expected to simulate the in vivo conditions of ENM influence better than the single culture conditions. MWNT‐7 was used as a test material because of its “positive control” property for the toxicity of MWCNT, including genotoxicity4 and carcinogenicity.8, 9, 10, 11, 12
The present study clearly demonstrates that: (i) MWNT‐7 was incorporated into both RAW264.7 and GDL1; (ii) RAW264.7 produced IL‐1β and TNF‐α by exposure to MWNT‐7; (iii) RAW264.7 and GDL1 only when co‐cultured with RAW264.7 generated ROS by MWNT‐7 exposure; and (iv) 8‐oxo‐dG and the gpt mutation were induced in MWNT‐7‐exposed GDL1 cells more with the co‐existence of MWNT‐7‐exposed RAW264.7 cells than in its absence. Asbestos and silica nanoparticles activate Nlrp3 inflammasomes in macrophages, and the macrophages subsequently release IL‐1β.18, 19 Li et al20 have demonstrated that ROS are induced in A549 cells by treatment with IL‐1β. It has also been noted that IL‐1β induces superoxide through the activation of NADPH oxidase on the endosomal membrane when macrophages incorporate exogenous materials.21, 22, 23, 24 Moreover, proinflammatory cytokines, such as IL‐1β and TNF‐α, could induce activation of nuclear factor (NF)‐kB, a key transcription factor for regulating inflammatory genes, then induce the expression of inducible nitric oxide synthase, and finally production of reactive nitric oxide species (RNOS).25 Taken together, it is suggested that the phagocytosis of MWNT‐7 by RAW264.7 may enhance the mutagenicity of the ENM towards GDL1 by virtue of the inflammatory reaction and ROS‐originated oxidative stress.
It has recently been shown that MWCNT is incorporated into A549 cells via endocytosis, and leads to inflammatory reactions and induces RNOS.26 However, Hiraku et al27 demonstrate that MWCNT induces RNOS and 8‐nitro‐dG, but not in A549 cells, and indicate that the cell culture conditions may alter the balance of the generation of hydroxyl radicals and nitric oxide. It should be considered, therefore, that ROS‐originated and RNOS‐originated oxidative stresses are both involved in the (geno)toxicity of MWCNT and ENM in general. When GDL1 was singly cultured and exposed to MWNT‐7, the number of G:C to C:G transversions, but not that of G:C to T:A transversions, was increased. The major mutation resulting from 8‐oxo‐dG is G:C to T:A; however the proportion of these mutations tends to decrease in MWNT‐7 exposed cells compared with the control (Figure 5). Borghini et al. demonstrate that repair activity of oxidative damaged DNA was increased by MWNT‐7 exposure.28 Although the 8‐oxo‐dG levels were markedly increased in the cells after exposure to MWNT‐7 for 24 hours in the present study, it is likely that this adduct would be repaired before fixation of mutations, and G:C to T:A mutations would ultimately decrease. However, increasing G:C to C:G transversions by MWNT‐7 exposure is suggested to be due to oxidative stress because ROS affects guanine to form not only 8‐oxo‐dG but also a variety of other products, such as imidazolone (Iz), oxazolone (Oz), spiroiminodihydantoin (Sp) and guanidinohydantoin (Gh), and these products can cause G to C transversions through translesion synthesis systems.29, 30, 31, 32, 33, 34, 35, 36, 37 Moreover, these oxidative DNA adducts have also been detected in model organisms;38, 39 therefore, it is suggested that G:C to C:G transversions induced by MWNT‐7 might involve the formation of oxidized DNA adducts other than 8‐oxo‐dG. In addition to the base change mutations, deletions were also increased. Copper nanoparticles have been shown to induce inflammation via mitochondrial stress, and to promote DNA fragmentation in lung fibroblast cell lines.40 We have previously reported that MWNT‐7 consistently induces micronuclei in cultured mammalian cells.4 It is, thus, suggested that DNA fragmentation may partly be involved in the deletion mutation in GDL1 cells by MWNT‐7.
In contrast, a different mutation spectrum was observed in the co‐culture system of GDL1 and RAW264.7, where the mutations occurring at G:C base pairs were markedly increased, especially for G:C to A:T transition and the G:C to C:G transversion, which were dominant. The mechanisms for induction of G:C to C:G mutation is suggested to be production of oxidized DNA adducts, such as Iz, Oz, Sp and Gh, as described above. The proportion of G:C to A:T transition is also dominant in controls; however, the specific MF for G:C to A:T is much higher in CC‐RG‐CNT‐80 than that of controls (11.0 × 10−6 for CC‐RG‐CNT‐80 vs 5.1 × 10−6 for CC control; Table S2). Interestingly, this mutation spectrum was similar to that observed in the lungs of mice exposed to MWNT‐7 in vivo.4 The G to A transition is commonly observed in spontaneously‐occurring and chemically‐induced mutants, and the deamination of 5‐methylcytosine or the alkylation of guanine are its potential origin.41, 42 Nitric oxide induces DNA damage by producing dinitrogen trioxide and then diazonium ions lead to the DNA deamination and 8‐nitro‐dG formation, and 8‐nitro‐dG preferentially causes the G:C to A:T transition through production of an apurinic site.27, 37, 43 However, nitric oxide is produced by activated macrophages in inflamed organs in vivo. In the in vivo toxicity tests of ENM, macrophage phagocytosing test substances are frequently observed.5, 44 Similarly, MWNT‐7 was phagocytosed by RAW264.7 in the present study. It is, thus, suggested that the gpt mutation with the predominance of the G:C to A:T transition seen in GDL1 co‐cultured with RAW264.7, as well as the situation of the lungs of mice in vivo, may be due to oxidative stress originated by RNOS that macrophage‐like immune cells phagocytosing MWNT‐7 produce and transmit to tissue resident cells.
In conclusion, it is indicated that inflammatory responses derived by macrophage‐like immune cells are involved in the genotoxicity of MWNT‐7 to lung resident cells, and that the presently established, novel in vitro assay featuring a co‐culture system of tissue resident cells with immune cells is mechanism‐based, extrapolative to the in vivo situation, and, thus, suitable to evaluate the genotoxicity of ENM. Further studies using a variety of other ENM are required to verify the assay.
CONFLICT OF INTEREST
This study was funded by AMED, the Ministry of Health, Labour and Welfare of Japan, JSPS and JCIA.
Supporting information
ACKNOWLEDGMENTS
We received technical support from Mr Shuntaro Akimoto.
Fukai E, Sato H, Watanabe M, Nakae D, Totsuka Y. Establishment of an in vivo simulating co‐culture assay platform for genotoxicity of multi‐walled carbon nanotubes. Cancer Sci. 2018;109:1024–1031. https://doi.org/10.1111/cas.13534
Funding information
Research on Global Health Issues (U.S.‐Japan Cooperative Medical Sciences Program) and Research on Regulatory Science of Pharmaceuticals and Medical Devices from the Japan Agency for Medical Research and Development; Research on Risk of Chemical Substances from the Ministry of Health, Labour and Welfare of Japan; Grant‐in‐Aid for Scientific Research (B) or (C) from the Japan Society for the Promotion of Science (JSPS); Japan Chemical Industry Association Long‐range Research Initiative grant
REFERENCES
- 1. Watanabe M, Yoneda M, Morohashi A, et al. Effects of Fe3O4 magnetic nanoparticles on A549 cells. Int J Mol Sci. 2013;14:15546‐15560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kawanishi M, Ogo S, Ikemoto M, et al. Genotoxicity and reactive oxygen species production induced by magnetite nanoparticles in mammalian cells. J Toxicol Sci. 2013;38:503‐511. [DOI] [PubMed] [Google Scholar]
- 3. Totsuka Y, Kato T, Masuda S, et al. In vitro and in vivo genotoxicity induced by fullerene (C60) and kaolin. Genes Environ. 2011;1:14‐20. [Google Scholar]
- 4. Kato T, Totsuka Y, Ishino K, et al. Genotoxicity of multi‐walled carbon nanotubes in both in vitro and in vivo assay systems. Nanotoxicology. 2013;7:452‐461. [DOI] [PubMed] [Google Scholar]
- 5. Totsuka Y, Ishino K, Kato T, et al. Magnetite nanoparticles induce genotoxicity in the lung of mice via inflammatory response. Nanomaterials. 2014;4:175‐188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kato T, Toyooka T, Ibuki Y, Masuda S, Watanabe M, Totsuka Y. Effect of physicochemical character differences on the genotoxic potency of kaolin. Genes Environ. 2017;39:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Takeiri A, Mishima M, Tanaka K, et al. A newly established GDL1 cell line from gpt delta mice well reflects the in vivo mutation spectra induced by mitomycin C. Mutat Res. 2006;609:102‐115. [DOI] [PubMed] [Google Scholar]
- 8. Sakamoto Y, Nakae D, Fukumori N, et al. Induction of mesothelioma by a single intrascrotal administration of multi‐wall carbon nanotube in intact male Fischer 344 rats. J Toxicol Sci. 2009;34:65‐76. [DOI] [PubMed] [Google Scholar]
- 9. Suzui M, Futakuchi M, Fukamachi K, et al. Multiwalled carbon nanotubes intratracheally instilled into the rat lung induce development of pleural malignant mesothelioma and lung tumors. Cancer Sci. 2016;107:924‐935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kasai T, Umeda Y, Ohnishi M, et al. Lung carcinogenicity of inhaled multi‐walled carbon nanotube in rats. Part Fibre Toxicol. 2016;13:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nagai H, Okazaki Y, Chew SH, et al. Diameter and rigidity of multiwalled carbon nanotubes are critical factors in mesothelial injury and carcinogenesis. Proc Natl Acad Sci U S A. 2011;108:E1330‐E1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nagai H, Okazaki Y, Chew SH, et al. Intraperitoneal administration of tangled multiwalled carbon nanotubes of 15 nm in diameter does not induce mesothelial carcinogenesis in rats. Pathol Int. 2013;63:457‐462. [DOI] [PubMed] [Google Scholar]
- 13. Grosse Y, Loomis D, Guyton KZ, et al. Carcinogenicity of fluoro‐edenite, silicon carbide fibres and whiskers, and carbon nanotubes. Lancet Oncol. 2014;15:1427‐1428. [DOI] [PubMed] [Google Scholar]
- 14. Suzuki H, Toyooka T, Ibuki Y. Simple and easy method to evaluate uptake potential of nanoparticles in mammalian cells using a flow cytometric light scatter analysis. Environ Sci Technol. 2007;41:3018‐3024. [DOI] [PubMed] [Google Scholar]
- 15. Nohmi T, Suzuki T, Masumura K. Recent advances in the protocols of transgenic mouse mutation assays. Mutat Res. 2000;455:191‐215. [DOI] [PubMed] [Google Scholar]
- 16. Könczöl M, Ebeling S, Goldenberg E, et al. Cytotoxicity and genotoxicity of size‐fractionated iron oxide (magnetite) in A549 human lung epithelial cells: role of ROS, JNK, and NF‐κB. Chem Res Toxicol. 2011;24:1460‐1475. [DOI] [PubMed] [Google Scholar]
- 17. Carr GJ, Gorelick NJ. Mutational spectra in transgenic animal research: data analysis and study design based upon the mutant or mutation frequency. Environ Mol Mutagen. 1996;28:405‐413. [DOI] [PubMed] [Google Scholar]
- 18. Dostert C, Pétrilli V, Van Bruggen R, Steele C, Mossman BT, Tschopp J. Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science. 2008;320:674‐677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yazdi AS, Guarda G, Riteau N, et al. Nanoparticles activate the NLR pyrin domain containing 3 (Nlrp3) inflammasome and cause pulmonary inflammation through release of IL‐1α and IL‐1β. Proc Natl Acad Sci USA. 2010;107:19449‐19454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li WJ, Wang TK. Calcitonin gene‐related peptide inhibits interleukin‐1beta‐induced interleukin‐8 secretion in human type II alveolar epithelial cells. Acta Pharmacol Sin. 2006;27:1340‐1345. [DOI] [PubMed] [Google Scholar]
- 21. Brigelius‐Flohé R, Banning A, Kny M, Böl GF. Redox events in interleukin‐1 signaling. Arch Biochem Biophys. 2004;423:66‐73. [DOI] [PubMed] [Google Scholar]
- 22. Kermanizadeh A, Chauché C, Brown DM, Loft S, Møller P. The role of reactive oxygen and nitrogen species in the response of airway epithelium to particulates. Environ Health Perspect. 1997;105(suppl 5):1301‐1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Forman HJ, Torres M. Reactive oxygen species and cell signaling: respiratory burst in macrophage signaling. Am J Respir Crit Care Med. 2002;166:S4‐S8. [DOI] [PubMed] [Google Scholar]
- 24. Forman HJ, Torres M. Signaling by the respiratory burst in macrophages. IUBMB Life. 2001;51:365‐371. [DOI] [PubMed] [Google Scholar]
- 25. Singer II, Kawka DW, Scott S, et al. Expression of inducible nitric oxide synthase and nitrotyrosine in colonic epithelium in inflammatory bowel disease. Gastroenterology. 1996;111:871‐885. [DOI] [PubMed] [Google Scholar]
- 26. Guo F, Ma N, Horibe Y, Kawanishi S, Murata M, Hiraku Y. Nitrative DNA damage induced by multi‐walled carbon nanotube via endocytosis in human lung epithelial cells. Toxicol Appl Pharmacol. 2012;260:183‐192. [DOI] [PubMed] [Google Scholar]
- 27. Hiraku Y, Guo F, Ma N, et al. Multi‐walled carbon nanotube induces nitrative DNA damage in human lung epithelial cells via HMGB1‐RAGE interaction and Toll‐like receptor 9 activation. Part Fibre Toxicol. 2016;13:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Borghini A, Roursgaard M, Andreassi MG, Kermanizadeh A, Møller P. Repair activity of oxidatively damaged DNA and telomere length in human lung epithelial cells after exposure to multi‐walled carbon nanotubes. Mutagenesis. 2017;32:173‐180. [DOI] [PubMed] [Google Scholar]
- 29. Kornyushyna O, Berges AM, Muller JG, Burrows CJ. In vitro nucleotide misinsertion opposite the oxidized guanosine lesions spiroiminodihydantoin and guanidinohydantoin and DNA synthesis past the lesions using Escherichia coli DNA polymerase I (Klenow fragment). Biochemistry. 2002;41:15304‐15314. [DOI] [PubMed] [Google Scholar]
- 30. Cadet J, Berger M, Buchko GW, Joshi PC, Raoul S, Ravanat JL. 2,2‐Diamino‐4‐[(3,5‐di‐O‐acetyl‐2‐deoxy‐β‐D‐erythro‐pentofuranosyl)amino]‐5‐(2H)‐oxazolone: a novel and predominant radical oxidation product of 3′,5′‐di‐O‐acetyl‐2′‐deoxyguanosine. J Am Chem Soc. 1994;116:7403‐7404. [Google Scholar]
- 31. Goyal RN, Jain N, Garg DK. Electrochemical and enzymic oxidation of guanosine and 8‐hydroxyguanosine and the effects of oxidation products in mice. Bioelectrochem Bioenerg. 1997;43:105‐114. [Google Scholar]
- 32. Burrows CJ, Muller JG, Kornyushyna O, et al. Structure and potential mutagenicity of new hydantoin products from guanosine and 8‐oxo‐7,8‐dihydroguanine oxidation by transition metals. Environ Health Perspect. 2002;110(suppl 5):713‐717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kino K, Sugiyama H. UVR‐induced G‐C to C‐G transversions from oxidative DNA damage. Mutat Res. 2005;571:33‐42. [DOI] [PubMed] [Google Scholar]
- 34. Kino K, Sugiyama H. Possible cause of G‐C–>C‐G transversion mutation by guanine oxidation product, imidazolone. Chem Biol. 2001;8:369‐378. [DOI] [PubMed] [Google Scholar]
- 35. Kino K, Ito N, Sugasawa K, Sugiyama H, Hanaoka F. Translesion synthesis by human DNA polymerase eta across oxidative products of guanine. Nucleic Acids Symp Ser. 2004;48:171‐172. [DOI] [PubMed] [Google Scholar]
- 36. Ye Y, Muller JG, Luo W, et al. Formation of 13C‐, 15N‐, and 18O‐labeled guanidinohydantoin from guanosine oxidation with singlet oxygen. Implications for structure and mechanism. J Am Chem Soc. 2003;125:13926‐13927. [DOI] [PubMed] [Google Scholar]
- 37. Loeb LA, Preston BD. Mutagenesis by apurinic/apyrimidinic sites. Annu Rev Genet. 1986;20:201‐230. [DOI] [PubMed] [Google Scholar]
- 38. Hailer MK, Slade PG, Martin BD, Sugden KD. Nei deficient Escherichia coli are sensitive to chromate and accumulate the oxidized guanine lesion spiroiminodihydantoin. Chem Res Toxicol. 2005;18:1378‐1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Matter B, Malejka‐Giganti D, Csallany AS, Tretyakova N. Quantitative analysis of the oxidative DNA lesion, 2,2‐diamino‐4‐(2‐deoxy‐beta‐D‐erythro‐pentofuranosyl)amino]‐5(2H)‐oxazolone (oxazolone), in vitro and in vivo by isotope dilution‐capillary HPLC‐ESI‐MS/MS. Nucleic Acids Res. 2006;34:5449‐5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lerner CA, Rutagarama P, Ahmad T, Sundar IK, Elder A, Rahman I. Electronic cigarette aerosols and copper nanoparticles induce mitochondrial stress and promote DNA fragmentation in lung fibroblasts. Biochem Biophys Res Commun. 2016;477:620‐625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shen JC, Rideout WM 3rd, Jones PA. The rate of hydrolytic deamination of 5‐methylcytosine in double‐stranded DNA. Nucleic Acids Res. 1994;22:972‐976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Swann PF. Why do O 6‐alkylguanine and O 4‐alkylthymine miscode? The relationship between the structure of DNA containing O 6‐alkylguanine and O 4‐alkylthymine and the mutagenic properties of these bases. Mutat Res. 1990;233:81‐94. [DOI] [PubMed] [Google Scholar]
- 43. Burney S, Caulfield JL, Niles JC, Wishnok JS, Tannenbaum SR. The chemistry of DNA damage from nitric oxide and peroxynitrite. Mutat Res. 1999;424:37‐49. [DOI] [PubMed] [Google Scholar]
- 44. Totsuka Y, Higuchi T, Imai T, et al. Genotoxicity of nano/microparticles in in vitro micronuclei, in vivo comet and mutation assay systems. Part Fibre Toxicol. 2009;6:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


