Abstract
F‐box and WD repeat domain‐containing 7 (FBW7) is a SCF‐type E3 ubiquitin ligase targeting a multitude of oncoproteins for degradation. Acting as one of the most important tumor suppressors, it is frequently inactivated in various tumors. In this study we aimed to evaluate the relationship of FBW7 with glioma pathology and prognosis, and examine its effect in glioma malignancies and temozolomide (TMZ)‐based therapy. Clinical tissues and TCGA database analysis revealed that FBW7 expression was correlated inversely with glioma histology and positively with patient survival time. Lentivirus transfection‐induced FBW7 overexpression significantly suppressed proliferation, invasion and migration of U251 and U373 cells, whereas knockdown of FBW7 by targeted shRNA promoted proliferation, invasion and migration of glioma cells. Most importantly, the expression level of FBW7 was found to affect the 50% inhibitory concentration (IC50) of U251 and the TMZ‐resistant variant. Combining TMZ with FBW7 overexpression notably increased the cytotoxicity compared to TMZ treatment alone, which was conversely attenuated by FBW7 knockdown. Moreover, flow cytometry (FC) analysis showed overexpression of FBW7, TMZ or the combination‐increased proportion of G2/M arrest and the apoptotic rate, whereas FBW7 inhibition reduced G2/M arrest and apoptosis in U251 cells. Finally, mechanistic study found that FBW7 overexpression downregulated Aurora B, Mcl1 and Notch1 levels in a time‐dependent pattern and this expressional suppression was independent of TMZ. These findings collectively demonstrate the critical role of FBW7 as a prognostic factor and a potential target to overcome chemoresistance of glioblastoma.
Keywords: Aurora B, FBW7, glioma, Mcl1, temozolomide
1. INTRODUCTION
Glioma, especially the most common subtype, glioblastoma multiforme (GBM), is among the most malignant human tumors. Currently, despite the standard treatment regimen including the maximum extent of surgical resection plus postoperative chemoradiotherapy, the improvement of the median survival period of GBM is still unsatisfactory, which is largely attributed to the therapeutic resistance to temozolomide (TMZ).1 Previous research has already implicated a complicated molecular network of signaling pathways in mediating the malignance of glioma. Monotherapy targeting 1 gene or signaling pathway usually leads to other compensative oncogenic activation, thus causing therapeutic resistance.2 Hence, exploring such therapeutic targets that are capable of regulating multiple molecules or signaling pathways in gliomagenesis and TMZ resistance will be beneficial for improvement of glioma treatment.
Since being identified in 1996, the ubiquitin proteasome system (UPS) has been regarded as an important therapeutic target in tumor therapy for its roles in regulating many aspects of the cancer biology by controlling protein turnover.3, 4 The function of UPS depends on a cascade activation of ubiquitin‐related enzymes, which includes ubiquitin‐activating enzyme E1, ubiquitin‐conjugating enzyme E2, and ubiquitin ligase E3. By direct binding to substrates, E3 enzyme determines the specificity of UPS regulation, which includes 2 main subtypes: the anaphase promoting complex (APC) type and the SKP1‐CUL1‐F‐box complex (SCF) type.5
FBW7 is the substrate binding unit of SCF‐type E3 ligase which recognizes substrates with conserved phospho‐degron motifs (CPD) through its WD40 repeats domain.5, 6 It is an important tumor suppressor with dominant‐negative mutations in a multitude of tumors, including gastric, colorectal, breast, pancreatic, liver, prostate and endometrial cancers, as well as leukemia and osteosarcoma.5, 7 To date, multiple substrates of FBW7 have been identified, which include a series of important oncoproteins like Cyclin E, c‐Myc, Jun, Mcl1 and Sox9 that are involved in the regulation of cell division, apoptosis, differentiation and some classical oncogenic signaling pathways, such as Notch and mTOR.8, 9, 10, 11, 12, 13, 14 Among them, the distinct regulation of several substrates needs to be examined in glioma cells. FBW7 gene encodes 3 distinctive protein isoforms (α, β, γ) each from its own promoter. Although it has been shown that FBW7β is mostly suppressed in human glioma cell lines, others have controversially reported that FBW7α and γ, instead of isoform β, inhibited growth of glioma cells,15, 16 which implied that the isoform‐specific cellular functions of FBW7 in glioma could be comprehensive. Moreover, the impact of FBW7 expression on gliomagenesis and therapeutic resistance as well as the mechanism by which FBW7 affects the glioma malignancy and related substrates remain far from fully elucidated.
In this study, we examined the expression pattern of FBW7 and its prognostic valve in glioma specimens, tested the inhibitory effect of FBW7 overexpression on cell malignancies of glioblastoma with special regard to TMZ chemoresistance, and, finally, explored the underlying mechanisms of FBW7 regulation in cell cycle, apoptosis and the related substrates. Our research aimed to demonstrate that targeting FBW7 is promising for the treatment of glioma.
2. MATERIALS AND METHODS
2.1. Cell culture and cell lines
The U251 and U373 cells used in this study were purchased from Cell Bank of Chinese Academy of Science (Shanghai, China). All cell lines, including the parental glioblastoma cell line and the modified lines described below, were grown in DMEM (Hyclone, USA) supplemented with 10% FBS (Gibco, USA), 100 U/mL penicillin G, 100 mg/mL streptomycin and 1.5 mmol/L L‐glutamine. Cells were maintained in monolayer culture at 37°C, in humidified air with 5% CO2.
For resistant cell isolation, parental U251 cells were consecutively re‐exposed to an incremental TMZ (Tasly, Tianjin, China) pulse commencing at 1 μg/mL to reach the desired concentration for a period of 6 months. Finally, a variant glioma cell line with a 4‐fold 50% inhibitory concentration (IC50) was isolated according to our previous experimental procedures and termed U251/TMZ.
2.2. Analyzation of TCGA data
The expression data of FBW7 expression and the clinical information of patients from the Affymetrix microarray were downloaded from the official website of TCGA (https://tcga-data.nci.nih.gov/tcga/dataacce2|LINETAL.ssMatrix.htm?modecApplyFilter&showMatrix=true&diseaseType=GBM&tumorNormal=TN&tumorNormal=T&tumorNormal=NT). The chip data contained 455 GBM samples and 27 low‐grade glioma (LGG) samples with complete survival information. The average value of relative mRNA expressions from different nucleic acid probes was used to compare the difference in FBW7 expression between GBM and LGG samples by student's t test, and to acquire the prognostic significance of FBW7 in GBM patients by Kaplan‐Meier survival analysis.
2.3. Immunohistochemistry assay
For the present study, 55 tissue samples of different grades of glioma (grade II: 15; grade III: 10; grade IV: 30) and 3 tissue samples of normal brain were collected from the Department of Neurosurgery at Shanghai Changzheng Hospital. All the samples were made into paraffin slices and then stained with specific anti‐FBW7 antibody (Abcam, USA) and anti‐Ki‐67 antibody (Abcam, USA), respectively, according to our previous procedures.17 The score of the staining rate was graded by the percentage of positively stained cells: 0% = 0, 1%‐25% = 1, 26%‐75% = 2 and 76%‐100% = 3. The score of staining intensity was also divided into 4 levels: negative = 0, weak = 1, medium = 2 and strong = 3. The final score was the arithmetic product of percentage score and intensity score, with <1 as low expression, 2‐4 as medium expression, 5‐7 as high expression, and ≥8 as extremely high expression.
2.4. Constructs and transfection
Human FBW7 complementary DNA reversely‐transcribed from the longest transcript NM_013233 containing all 3 isoform‐encoding sequences (GAGGATCCCCGGGTACCGGTCGCCACCATGAATC) was tagged with enhanced green fluorescent protein (eGFP) and then cloned into the lentiviral vector GV358 (Genechem, Shanghai, China) to create the complete functional overexpression plasmid named as Lv‐FBW7‐OE. An shRNA sequence directed against a shared exon of FBW7 with a loop structure (GATCCGCAACAACGACGCC GAATT ACTTCAAGAGACTTTGCCAAAGGTTCCCTTGCTTTTTTG) was constructed and cloned into GV358 vector, and named as shRNA. Another control lentiviral vector tagged with eGFP was also constructed as Lv‐GFP. Conditioned medium containing lentiviruses was harvested 48 hours from transfected 293T cells and prepared for further transfection. An immunoblotting assay was used for verification of FBW7 overexpression and downexpression. In this way, modified U251 cell lines were isolated either expressing GFP, overexpressing FBW7, or expressing a FBW7‐specific shRNA, which led to stable downexpression of FBW7. Similarly, different U373 cell lines transfected with Lv‐FBW7‐OE, shRNA or Lv‐GFP were isolated as described above.
2.5. Proliferation, invasion and migration assay
Cell proliferation was determined by MTT colorimetric assay. U251 cells and U373 cells in the logarithmic phase were reseeded in 96‐well plates at a density of 4 × 103cells/well. At daily intervals (24, 48, 72, 96 and 120 hours after plating), the culture medium was replaced by MTT for 4 hours, avoiding light and at 37°C, and then shaken in dimethyl sulfoxide (DMSO) for 15 minutes to fully dissolve the formazan crystals. Absorbance of cells was measured at 590 nm with a microplate reader (Tecan, Switzerland). For the invasion assay, U251 cells and U373 cells were seeded at 2.5 × 105cells per Matrigel‐coated transwell chamber. After 24 hours, the cells remaining in Matrigel were fixed with 4% paraformaldehyde for 20 minutes and then stained with crystal violet for 15 minutes. Pictures were taken using a 100× optical microscope and the cell number was counted. A wound healing assay was used for cell migration. Cells were seeded into a 24‐well plate until confluent growth in a monolayer. We gently scratched the monolayer with a 1‐mL pipette tip across the center of each well. After additional growth for 18 and 36 hours, cells were washed, fixed and stained with crystal violet in ethanol for 30 minutes. The final gap distance was measured under a microscope and quantified using ImageJ (http://rsb.info.nih.gov/ij/download.htm). All experiments were repeated 3 times.
2.6. Western blot assay
The whole cell lysate was extracted in RIPA buffer (Beyotime Biotechnology, China) on ice box and then boiled with 5× loading buffer for 5 minutes. Next, 20 μg of total cell proteins were added into the 12% precast gels and separated by SDS PAGE by 120 V for 75 minutes. After transferring the proteins to the PVDF member, the band of specific protein was detected by monoclonal or polyclonal antibodies. All the antibodies were purchased from Abcam as follows: FBW7 (ab109617), Ki‐67 (ab15580), Aurora B (ab45145), Mcl‐1 (ab32087), Notch‐1 (ab52627), Caspase‐3 (ab13586) and beta‐actin (ab6276).
2.7. Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) assay
Total RNA was extracted from U251 cells and U251/TMZ cells using an RNeasy Mini Kit (Qiagen, Venlo, the Netherlands). First‐strand complementary DNA was synthesized by reverse transcription with the PrimeScript RT Master Mix Kit (TaKaRa, Japan), then amplified by SYBR Premix Ex Taq (Tli RNaseH Plus) kit (TaKaRa, Japan). The forward primer of FBW7 was: 5′‐GTCCCGAGAAGCGGTTT GATA‐3′ and the reverse primer of FBW7 was: 5′‐TGCTCAGGCACGTCAGAAAAG‐3′; the forward primer of β‐actin was: 5′‐GTCTGCCTTGGTAGTGGATAATG‐3′ and the reverse primer of β‐actin was: 5′‐TCGAGGACGCCCTATCATGG‐3′.
2.8. Temozolomide sensitivity assay
The U251 cells and U251/TMZ cells were trypsinized and diluted to 5 × 105 cells/mL in DMEM with 10% FBS. We added 100 μL of single‐cell suspension into each well of the 96‐well plates. We mixed the 4‐mL fresh DMEM medium with a 4‐μL stock solution of TMZ to a final concentration of 128 μg/mL. Then the TMZ working solution was diluted into 64, 32, 16, 8, 4, 2 and 1 μg/mL using the double‐dilution method; 1 μL DMSO served as control. We added the different working solutions into the cell suspension and cultured for 72 hours continuously. IC50 was calculated according to the OD450 nm by MTT assay as previously described.18 For in vitro cytoxicity testing, U251 cells were cultured in 6‐cm dishes until they had reached nearly half confluence and were then treated with Lv‐GFP (control), TMZ (8 μg/mL) + LV‐GFP, Lv‐FBW7‐OE, TMZ (8 μg/mL) + Lv‐FBW7‐OE, shRNA or shRNA + TMZ (8 μg/mL), respectively. After 36 and 72 hours, all the groups of cells were observed for qualitative analysis under a phase contrast microscope. For each time point, cells were harvested and replated onto 96‐well plates for subsequent MTT assay. The formula of the in vitro cell inhibition rate = (1‐OD450 nm of treatment group/OD450 nm of control group) × 100%.
2.9. Flow cytometric analysis
For analysis of the cell cycle, cells were treated with Lv‐GFP alone, Lv‐FBW7‐OE alone, Lv‐GFP + TMZ (8 μg/mL), Lv‐FBW7‐OE + TMZ (8 μg/mL), shRNA alone or shRNA + TMZ (8 μg/mL) for 48 hours before being trypsinized and harvested, washed in PBS and then stained with 5 μL propidium iodide (PI) for 30 minutes in a dark at room temperature. Then the distribution of the cell cycle was determined by Cytomics FC 500 Flow Cytometer (Beckman, USA). For analysis of apoptosis, cells with the same grouping as above were trypsinized and stained with 5 μL propidium iodide (PI) and 5 μL annexin V‐APC simultaneously for 15 minutes in the dark at room temperature. The cells were then analyzed by flow cytometer.
2.10. Statistics
Data analysis was performed using GraphPad Prism software (Version 7, CA, USA). The alpha level for the type I error was set at 0.05 for rejecting the null hypothesis. Data were expressed as means ± standard deviation (SD). Overall and progress‐free survival time of glioma patients from TCGA were analyzed by Kaplan‐Meier and log‐rank test. The immunoactivity of FBW7 and Ki67 in between HGG and LGG was compared using the Kruskal‐Wallis test. The correlation between FBW7 and Ki‐67 immunoactivity was examined by Spearman's correlation analysis. IC50 and FBW7 expression were compared with Student's t test. Biological behavior, chemosensitivity and FACS results were compared using ANOVA followed by Bonferroni's multiple comparison.
3. RESULTS
3.1. Expression of FBW7 was downregulated in glioma
The distribution analysis of TCGA data revealed that relative FBW7 mRNA expressions of all the samples including high grade glioma (HGG) and LGG were concentrated within the lower region (medium = 0.5993, skewness = 1.057, Kurtosis = 2.155; Figure 1A). The relative expression of FBW7 mRNA in low grade glioma (1.494 ± 0.1934) was significantly higher than that in high grade glioma (HGG) according to Student's t test (P‐value <.001, Figure 1B). Study of the TCGA data showed that the lower expression of FBW7 was associated with the shorter overall survival (OS) and progress‐free survival (PFS) (P‐value <.05, log‐rank HR = 1.220 and 1.366, respectively; Figure 1C,D). In the immunohistochemistry assay, the morphological and statistical results confirmed that the expression level of FBW7 was significantly associated with tumor grade (P‐value <.001, Figure 1D,E). In addition, the FBW7 staining scores of LGG were obviously higher than those of HGG with a statistical difference (P‐value <.001), and were negatively correlated with the Ki‐67 staining scores (P‐value <.01, Figure 1F,G). Moreover, we observed that the localization of FBW7 is scattered from cytoplasm, nucleus to nucleolus, which was due to the used antibody binding to the conserved FBW7 structure determined by the shared exons.
Figure 1.
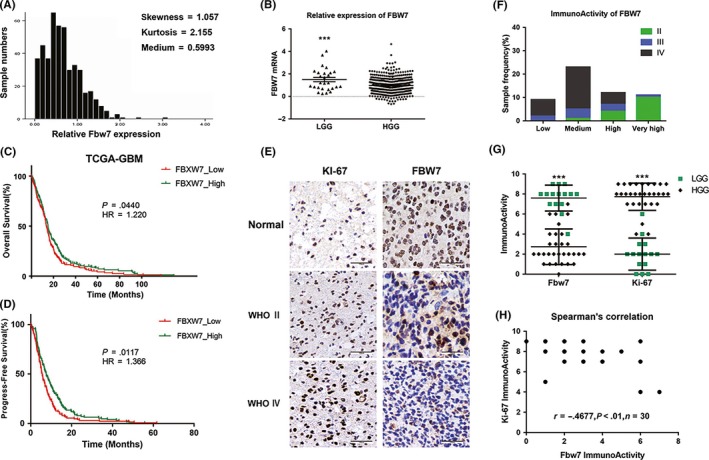
FBW7 subexpression in glioma samples. A, Measures of distribution of FBW7 expression according to data obtained from TCGA. B, Relative expression of FBW7 mRNA in low‐grade glioma (LGG) and high‐grade glioma (HGG) tissues (***P < .001). C,D, Overall and progress‐free survival curve by data of FBW7 expression. Statistical difference existed between the FBW7_high group and the FBW7_low group (*P < .05). E, Representative pictures of FBW7 and Ki‐67 immunohistochemical staining in glioma tissues at different histological grades. Scale bars = 100 μm. F, Sample frequency of FBW7 immunoactivity in glioma tissues of different grades. G, Respective data of FBW7 and Ki‐67 immunoactivity in both LGG group and HGG groups (***P < .001). H, Relevance of FBW7 and Ki‐67 immunoactivity (**P < .01, r = .4677)
3.2. Maneuvered expression of FBW7 suppressed the aggressive biological behaviors of glioma cells
The transfection efficiency of Lv‐FBW7‐OE was verified by western blot assay in U251 and U373 cells, in which the expressional abundance of FBW7 is different, by qRT‐PCR (data not shown). The expression level of FBW7 protein was significantly increased in both cell lines by stable transfection with Lv‐FBW7‐OE compared with the untransfected parental cells, whereas the expression level was almost unchanged in the Lv‐GFP transfected cell line. Conversely, transfection of FBW7‐targeted shRNA resulted in a notably reduced FBW7 protein level in both U251 and U373 cell lines (Figure 2A). Malignant proliferation was among the most distinct features of glioma (ie. GBM). The MTT assay results revealed that proliferation activity of U251 cells transfected with Lv‐FBW7‐OE cells was notably reduced at 48 hours compared with parental cells and cells transfected with Lv‐GFP (P‐value <.05). Later there was a more proliferation reduction at 72, 96 and 120 hours (P‐value <.001) in cells overexpressing FBW7. There was no significant difference between untreated U251 cell line and Lv‐GFP transfected cell line of U251. The trend was comparable in cell proliferation to that of U373 line after FBW7 overexpression, with remarkable growth reduction at 48, 72, 96 or 120 hours (Figure 2B). By contrast, knockdown of FBW7 by interfering shRNA resulted in a time‐dependent increase of the proliferation rate, which was expected, from 72 to 120 hours in U251 line and from 96 to 120 hours in U373 line. Invasion and migration capability were another 2 aspects of glioma cells to exhibit high malignance or intracranial metastasis and cause treatment failure, and it is reported by Balamurugan et al. that the mTOR/AKT/HIF1α axis regulated by FBW7 could promote invasion and metastasis in breast tumor cells.19 To clarify the role of FBW7 overexpression in the invasiveness and migration capability of glioma cells, wound healing and transwell assays were used. Upregulating the expression level of FBW7 in U251 and U373 significantly reduced the number of cells transferred to the lower side of the chamber (P‐value <.001 compared with control). Although no significant increase of invasion was observed in the FBW7‐specific shRNA‐transfected U373 line, that of the U251 cell line was considerably increased after FBW7 downexpression (P < .01, Figure 2C,D). As for the migration assay, the migration distance in U251‐Lv‐FBW7‐OE and U373‐Lv‐FBW7‐OE at both 18 and 36 hours after lentivirus transfection was significantly less than that of the other 2 control cell lines (P‐value <.001, Figure 2E,G). As expected, there was a similarly increased number of migrated cells at 36 hours by FBW7 downexpression in these 2 cell lines (P < .05, Figure 2E,G).
Figure 2.
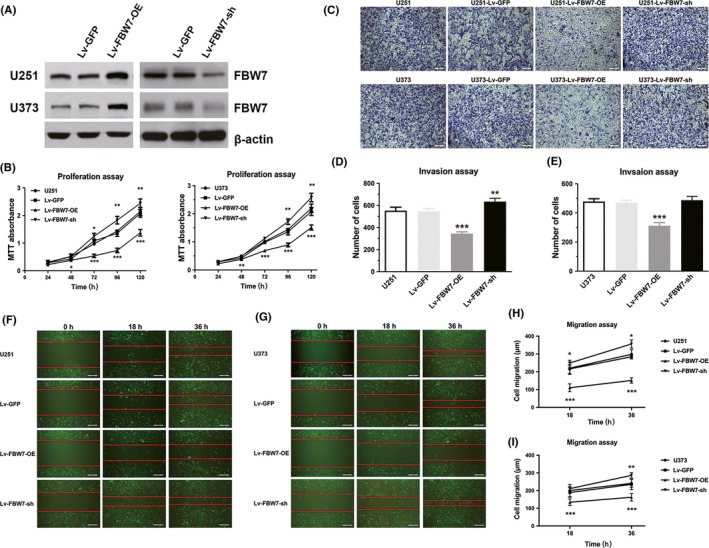
FBW7 overexpression inhibited the malignant biological behaviors of glioblastoma multiforme (GBM). A, The efficiency of FBW7 overexpression (OE) and shRNA lentivirus vector in U251 and U373 cell lines were detected by western blot assay. B, The cell viability of U251 and U373 was assessed by MTT assay in non‐transfected cells, cells transfected with eGFP lentivirus, cells stably overexpressing FBW7 and cells stably transfected with FBW7‐specific shRNA. C‐E, Representative photographs of invasion cells in both U251 and U373 cell lines by transwell assay. Scale bars = 100 μm. Data of invasive cell numbers from 3 independent experiments are presented in the form of histograms. F‐I, Representative fluorescent pictures with red strip to show the result of wound healing assay in E and F. Scale bars = 100 μm. Data of migrated cell numbers of different groups were analyzed in the form of line charts. (*P < .05, **P < .01, ***P < .001 compared to blank control)
3.3. Overexpression of FBW7 increased the sensitivity of glioma cells to temozolomide
Temozolomide has been the first‐line drug for chemotherapy of glioma due to its high penetrability to cross the blood‐brain barrier and strong DNA toxicity. The cytotoxic effect of TMZ can be affected by complex molecular pathways.20 Herein, we observed that the expression of FBW7 was markedly inhibited at both protein and mRNA levels in U251/TMZ, a cell line which was isolated in our previous experiment with 4‐fold 50% inhibition rate (IC50; data not shown), compared to the parental U251 cell line (P‐value <.01 for quantitative RT‐PCR, Figure 3A,B), suggesting the potential role of FBW7 in mediating TMZ response. Furthermore, this finding was convinced by the results that re‐expression of FBW7 in the variant U251/TMZ line partly reversed its resistance to TMZ (P‐value <.01, Figure 3C), while reducing FBW7 expression in U251 cells promoted its resistance to TMZ (P‐value <.01, Figure 3D).
Figure 3.
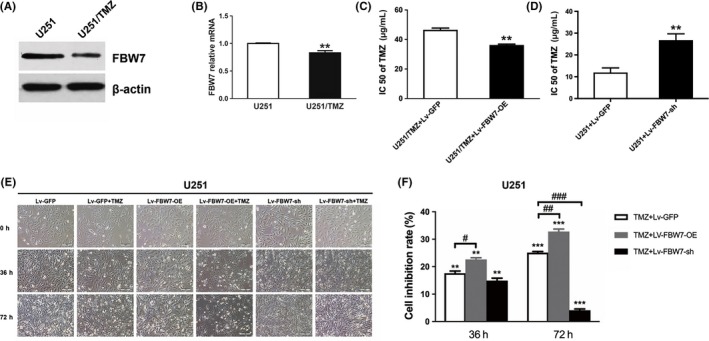
Overexpression of FBW7 enhanced the sensitivity of U251 cells to temozolomide (TMZ). A,B, Expression levels of FBW7 protein and mRNA in U251 cells and TMZ‐resistant U251/TMZ cells were assessed by western bolt assay (**P < .01 between groups). C,D, Respective effect of FBW7 overexpression and knockdown on IC50 of TMZ‐resistant U251/TMZ and U251 cell line (**P < .01 between groups). E, Representative photos of phase contrast microscopy displayed the time‐dependent cell status in cells with different treatment modalities at 0, 36 and 72 h, respectively. Scale bars = 100 μm. F, Inhibition rates of different treatment groups were assessed by MTT assay at 36 and 72 h in 3 independent experiments (**P < .01, ***P < .001 compared to Lv‐GFP; ##P < .01 compared to TMZ)
To further evaluate the synergistic effect of TMZ and FBW7 overexpression, we performed a cytotoxicity test in U251 cells to mimic the in vivo experiment. From both the morphological and colorimetric observations, TMZ significantly reduced viable U251 cells at both 36 and 72 hours post‐treatment, which was consistent with the theory that TMZ induced cell death usually after 1 cycle of cell division for at least 24 hours. Interestingly, when TMZ was given with FBW7 transfection, cell viability was dramatically inhibited at 36 and 72 hours compared to the control, with a much greater inhibition rate than TMZ alone at 72 hours (Figure 3D,E). As expected, specific shRNA‐induced FBW7 knockdown attenuated the cytotoxity of TMZ, as shown in Figure 3D,E, with a reduced inhibition rate in the TMZ + Lv‐FBW7‐OE‐sh group relative to the TMZ alone group at 72 hours.
3.4. Overexpression of FBW7 increased G2/M arrest and apoptotic death
Previous studies have proved that FBW7 can regulate several cell cycle‐associated proteins, such as kinase Aurora B and cyclin E.8, 21 In light of determining the regulating effect of FBW7 on cell division of glioma, FC analysis was applied. It was shown that U251 cells stably transfected with Lv‐FBW7‐OE exerted higher proportion of G2/M phase than Lv‐GFP transfected or parental control (P‐value <.001). It is commonly accepted that G2/M arrest is critical for TMZ toxicity.22 Consistent with this, TMZ alone caused a much higher percentage of G2/M phase than other groups, which was further increased when co‐treated with Lv‐FBW7‐OE transfection (Figure 4B). In contrast, FBW7 downexpression in U251 cells and TMZ treated cells dramatically decreased the G2 phase proportion relative to control or cells with TMZ alone.
Figure 4.
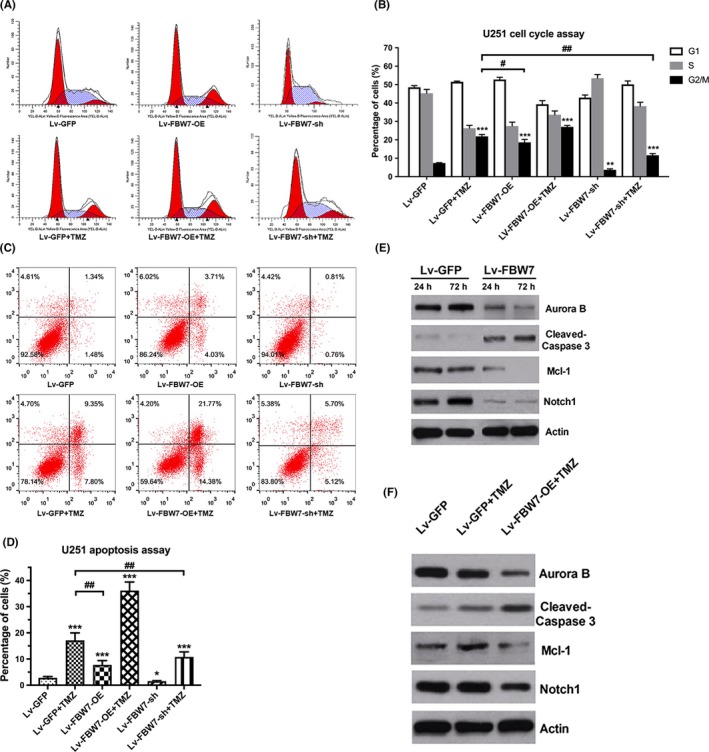
Overexpression of FBW7 arrested cell cycle in G2/M phase and induced apoptosis. A,B, Representative flow cytometry pictures of cell cycle distribution of U251 in different treatment groups. Quantitative analysis of cell cycle is shown in the form of histograms. C,D, Representative flow cytometry pictures of cell apoptosis in different groups, which are quantitatively presented in histograms. E, Protein changes of Aurora B, cleaved‐caspase3, Mcl‐1 and Notch1 in U251 cells were detected by western blot at 24 and 72 h, respectively. F, The protein levels of above FBW7 substrates with temozolomide (TMZ) and TMZ plus Lv‐FBW7‐OE treatment by western blot assay. (*P < .05, **P < .01, ***P < .001 compared to Lv‐GFP; ##P < .01 compared to TMZ)
Apoptosis has been reported to be tightly linked with TMZ‐induced G2/M arrest other than autophagy.22, 23 We next tested the possible effect of FBW7 on cell apoptosis with TMZ treatment. As a result, we found that U251 cells with TMZ treatment or FBW7 transfection alone had obviously greater apoptosis rates compared to Lv‐GFP transfected cells (P‐value <.001). Interestingly, combined treatment more significantly increased apoptotic death compared to either treatment alone (Figure 4C‐F). As shown in the histogram figure, downregulating FBW7 by shRNA transfection suppressed cell apoptosis. In addition, combining shRNA transfection with TMZ markedly reduced the apoptotic rate induced by TMZ alone. Taken together, these results strongly demonstrated the synergistic effect of FBW7 expression with TMZ on cell cycle and apoptosis of glioma cells.
3.5. Suppression of several cycle and apoptosis associated proteins with FBW7 overexpression was time‐dependent and independent of temozolomide
FBW7 has been reported to target a group of phosphorylated proteins for ubiquitination and degradation.5 Theoretically, their abnormal regulation due to FBW7 loss of function may underscore the malignant phenotypes of glioma; however, only a tiny portion of them have been observed in glioma cells. To this end, given the critical role of FBW7 in controlling the cell cycle (ie. G2/M) and apoptosis in glioma cells, we were prompted to examine the molecular mechanisms by western blot. First, we revealed that FBW7 suppressed the expression of pro‐G2/M transition kinase Aurora B and anti‐apoptotic Mcl1, whereas it upregulated the apoptotic effector Caspase‐3 at 24 and 72 hours in a time‐dependent manner. These findings were in accordance with the viability‐time curve in Figure 2 and may partially explain the proliferation inhibition of glioma cells. Notably, it was found that Notch1 expression was regulated at an earlier time after FBW7 transfection, suggesting a distinct role of Notch1 in the inhibition of cell migration in less than 24 hours with FBW7 overexpression (Figure 4G).
Next, we added TMZ to examine the effect of FBW7 and TMZ treatment on those substrates. As revealed by western blot, TMZ treatment alone upregulated the expression of Mcl1 and slightly increased the cleaved Caspase‐3 level without altering the expression level of Aurora B and Notch1, indicative of the apoptotic alteration in TMZ therapy. When FBW7 was overexpressed before TMZ treatment by lentivirus transfection in U251 cells, obvious reduction of Mcl1 and Aurora B expression with cleaved Caspase‐3 upregulation was observed and the Notch1 expression was decreased slightly (Figure 4H). These observations together suggest that the FBW7‐mediated inhibition of Aurora B, Mcl1 and Notch1 was independent of TMZ.
4. DISCUSSION
Although FBW7 has been intensively studied in a series of human malignancies, the impact of FBW7 on the malignant behavior of glioma remains elusive. Consistent with the function in other cancer types, our study also indicated a tumor suppressor role of FBW7 in glioma by exploration of its expression pattern, prognostic value, biological functions and potential mechanisms. In particular, we found that FBW7 expression was tightly associated with the resistance of glioma cells to TMZ chemotherapy. FBW7 overexpression could significantly increase the TMZ chemosensitivity of glioma in vitro. Our results demonstrated a promising pharmaceutical potential of FBW7 in improving the chemotherapeutic outcome in glioma treatment.
Prior research has demonstrated that FBW7 is frequently inactivated for its mutation in a multitude of tumors, including gastric, colorectal, breast, pancreatic, liver cancers and leukemia. With a mutation rate of 6% to 35%, it is considered to be the most common tumor suppressor right behind p53 and PTEN.7 In addition, many upstream molecules, such as transcriptional factors, kinases and microRNA, can also regulate FBW7 either transcriptionally or post‐translationally.24 For example, 1 study has reported that a p53‐FBW7‐Aurora B axis, which identified FBW7 as a transcriptional target of p53.21 Another study found that FBW7 could be phosphorylated and destabilized by ERK kinase.25 More intriguingly, just before this work was finished, Yang's team discovered a novel circ‐FBW7 in glioma cells, which is able to antagonize the inhibitory effect of USP28 on FBW7α so as to degrade c‐Myc.26 In the present study, we found that FBW7 was both downregulated at mRNA level by interrogating the TCGA database and at protein level by immunohistochemistry in glioma tissues. This might be the comprehensive result of mutation and upstream deregulation of FBW7 and requires further investigation. Our data also revealed a diffuse cellular distribution of FBW7 including the cytoplasm, nucleus and nucleolus, which was consistent with the diverse subcellular location of the 3 FBW7 isoforms reported in previous studies.16
The lethality of glioma is largely attributed to its malignant phenotypes, such as proliferation, migration and invasion, which are controlled by multiple molecules or signaling pathways. Cell proliferation is tightly associated with the cell cycle, which is rigidly regulated by a series of kinases, among which Cyclin E and Aurora B are 2 important FBW7 substrates involved in the promotion of G1/S and G2/M transition, respectively.5, 8, 21 Previous studies have implicated the role of FBW7 in the regulation of cell proliferation in colon, breast and kidney cancers.7 In our study, we found that forced FBW7 expression inhibited the proliferation of glioma cells. It increased the ratio of G2/M and led to time‐dependent Aurora B downregulation, whereas FBW7 knockdown promoted cell proliferation with significantly reduced G2/M proportion, which suggests that the effect of FBW7 on glioma tumorigenesis could be partially explained by FBW7‐mediated Aurora B expression. We also noticed that there were similar changes in G1 proportion with Lv‐FBW7‐OE or shRNA transfection, suggesting that cyclin E is perhaps involved in FBW7‐induced cycle distribution; however, the trend was much less significant than G2/M. Cell migration and invasion are controlled by many biological processes. One of the most common mechanisms is the induction of epithelial‐mesenchymal transition (EMT). In several malignancies, such as gastric and colorectal tumors, FBW7 has also been suggested to regulate cell migration and invasion by inhibition of EMT.19, 27 Notch signaling pathway has been widely recognized for its vital roles in cell differentiation, cancer stemness, as well as EMT. Among these the cleaved intracellular domain of Notch1, NICD, is the most important intermediate for Notch signaling transduction, which is another substrate of FBW7.13 From the western blot assay, the mechanism of inhibited migration and invasion upon FBW7 elevation in our study can be partially explained by the early downregulation of Notch1. In addition, an antiapoptotic member of the Bcl‐2 family, Mcl1, was reduced in a similar time‐dependent pattern with Aurora B in U251 cell line, and apoptosis was found to be enhanced with FBW7 overexpression. Considering the critical role of apoptosis deregulation in tumor growth and metastasis,28 our findings also implicated FBW7 in the regulation of Mcl1, and apoptosis possibly contributes to the inhibition of cell proliferation and migration.
Since the establishment of standard TMZ chemotherapy in glioma treatment, acquired chemotherapeutic resistance has emerged as one of the most important obstacles leading to tumor reoccurrence and refractoriness, which can be regulated by a series of mechanisms such as aberrantly enhanced DNA repair, cell stemness conversion, alteration of metabolism, elevated multidrug resistant phenotype, and imbalance of apoptosis or autophagy.17, 18, 29 Therefore, enhancing the sensitivity to TMZ was a pivotal measure to improve the treatment outcome of glioma. Here, for the first time, we demonstrate that FBW7 is involved in the spontaneous defensive strategy of glioma cells undergoing TMZ treatment and intervening its expression can bidirectionally regulate glioma TMZ resistance. Previously, our protein microarray analysis found that Aurora B expression was markedly increased in TMZ‐resistant cells compared with parental cells, which indicates that Aurora B also mediates TMZ resistance (data not shown). Mcl1 is another substrate of FBW7, and the FBW7‐Mcl1 axis has been widely reported to be positively correlated with chemoresistance in ovarian cancer and T lymphocytic leukemia.11 In our study, we not only found significant G2/M arrest and apoptosis was increased but also confirmed that Aurora B and Mcl1 were significantly downregulated in U251 cell line by combining FBW7 overexpression and TMZ. Considering that TMZ cytotoxicity is largely dependent on the cell apoptosis caused by DNA damage‐triggered mismatch repair within the G2/M phase,22, 23 we speculate that the mechanisms for FBW7‐mediated enhanced chemosensitivity can be both ascribed to the arrest of the G2/M transition and attenuation of apoptosis by suppression of Aurora B and Mcl‐1 expression, respectively (Figure 5). It should be noted that Mcl1, rather than Aurora B and Notch1, was obviously upregulated by 72‐hours TMZ treatment. On the one hand, this indicated that the latter 2 substrates of FBW7 were not as important as Mcl1 in the autonomous survival pathways of U251 cells. On the other hand, it could be possible that a period of 72 hours is insufficient for these 2 substrates to express in response to TMZ. Although the exact relationship between FBW7 and chemoresistance is not known, the current work theoretically demonstrates the clinical value of FBW7 in reinforcing the traditional TMZ‐based chemotherapy.
Figure 5.
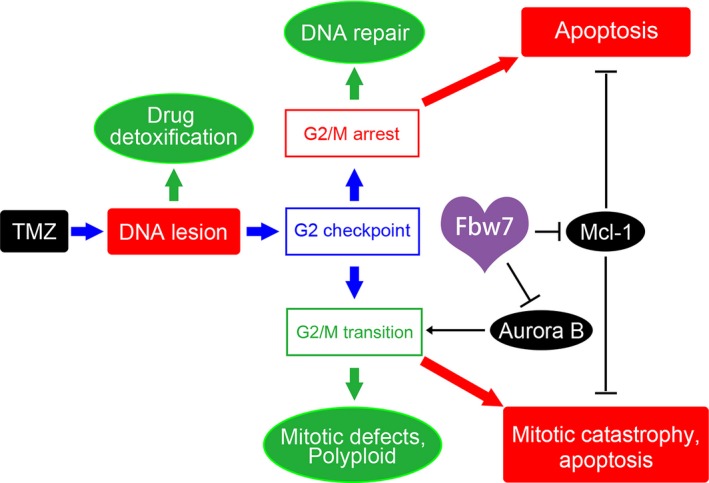
Schematic illustration of FBW7 regulating chemosensitivity of glioma cells to temozolomide (TMZ). On one hand, by regulating Aurora B, FBW7 inhibits G2/M transition and results in G2/M arrest and subsequent apoptosis. On the other hand, by regulating Mcl1, FBW7 enhances apoptotic cell death. (Green indicates tumor survival strategy and red indicates TMZ cytotoxic effect)
In a future study, the tumor suppressor function and the effect of FBW7 on chemosensitivity will be further validated in more glioma cell lines as well as in a patient‐derived xenograft model in vivo. More substrates in the upstream and downstream pathways of FBW7 will be identified by genomic and proteomic analysis of high‐throughput microarrays to better understand its regulatory mechanisms in glioma.
CONFLICT OF INTEREST
The authors have no conflict of interest to declare.
Lin J, Ji A, Qiu G, et al. FBW7 is associated with prognosis, inhibits malignancies and enhances temozolomide sensitivity in glioblastoma cells. Cancer Sci. 2018;109:1001–1011. https://doi.org/10.1111/cas.13528
Funding information
Project of National Natural Science Foundation (grant number 30930094 awarded to Yicheng Lu, grant number 81302187 awarded to Hua He and grant number 81702944 awarded to Guanzhong Qiu); the Project of the Ministry of Health of the People's Liberation Army (grant number CWS14C063), and the State Key Laboratory of Drug Research (grant number SIMM1705KF‐10) awarded to Hua He, Medical Project of Sichuan Province Medical Association (grant number S15005 awarded to Chaoli Song); Special Presidential Foundation of General Hospital of Jinan Military Command (grant number 2016BS04 to Guanzhong Qiu).
Lin, Ji and Qiu contributed equally to this work.
Contributor Information
Chaoli Song, Email: songcl1976@126.com.
Hua He, Email: hehuapanda@163.com.
Yicheng Lu, Email: lycczyy@163.com.
REFERENCES
- 1. Van Meir EG, Hadjipanayis CG, Norden AD, et al. Exciting new advances in neuro‐oncology: the avenue to a cure for malignant glioma. CA Cancer J Clin. 2010;60:166‐193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bartek J Jr, Ng K, Bartek J, et al. Key concepts in glioblastoma therapy. J Neurol Neurosurg Psychiatry. 2012;83:753‐760. [DOI] [PubMed] [Google Scholar]
- 3. Nakayama KI, Nakayama K. Ubiquitin ligases: cell‐cycle control and cancer. Nat Rev Cancer. 2006;6:369‐381. [DOI] [PubMed] [Google Scholar]
- 4. Qiu GZ, Sun W, Jin MZ, et al. The bad seed gardener: deubiquitinases in the cancer stem‐cell signaling network and therapeutic resistance. Pharmacol Ther. 2017;172:127‐138. [DOI] [PubMed] [Google Scholar]
- 5. Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8:83‐93. [DOI] [PubMed] [Google Scholar]
- 6. Hao B, Oehlmann S, Sowa ME, et al. Structure of a Fbw7‐Skp1‐cyclin E complex: multisite‐phosphorylated substrate recognition by SCF ubiquitin ligases. Mol Cell. 2007;26:131‐143. [DOI] [PubMed] [Google Scholar]
- 7. Cao J, Ge MH, Ling ZQ. Fbxw7 tumor suppressor: a vital regulator contributes to human tumorigenesis. Medicine (Baltimore). 2016;95:e2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ekholm‐Reed S, Spruck CH, Sangfelt O, et al. Mutation of hCDC4 leads to cell cycle deregulation of cyclin E in cancer. Cancer Res. 2004;64:795‐800. [DOI] [PubMed] [Google Scholar]
- 9. Yada M, Hatakeyama S, Kamura T, et al. Phosphorylation‐dependent degradation of c‐Myc is mediated by the F‐box protein Fbw7. EMBO J. 2004;23:2116‐2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Musti AM, Treier M, Bohmann D. Reduced ubiquitin‐dependent degradation of c‐Jun after phosphorylation by MAP kinases. Science. 1997;275:400‐402. [DOI] [PubMed] [Google Scholar]
- 11. Inuzuka H, Shaik S, Onoyama I, et al. SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature. 2011;471:104‐109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Suryo Rahmanto A, Savov V, Brunner A, et al. FBW7 suppression leads to SOX9 stabilization and increased malignancy in medulloblastoma. EMBO J. 2016;35:2192‐2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Oberg C, Li J, Pauley A, et al. The Notch intracellular domain is ubiquitinated and negatively regulated by the mammalian Sel‐10 homolog. J Biol Chem. 2001;276:35847‐35853. [DOI] [PubMed] [Google Scholar]
- 14. Fu L, Kim YA, Wang X, et al. Perifosine inhibits mammalian target of rapamycin signaling through facilitating degradation of major components in the mTOR axis and induces autophagy. Cancer Res. 2009;69:8967‐8976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hagedorn M, Delugin M, Abraldes I, et al. FBXW7/hCDC4 controls glioma cell proliferation in vitro and is a prognostic marker for survival in glioblastoma patients. Cell Div. 2007;2:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gu Z, Inomata K, Ishizawa K, et al. The FBXW7 beta‐form is suppressed in human glioma cells. Biochem Biophys Res Commun. 2007;354:992‐998. [DOI] [PubMed] [Google Scholar]
- 17. Cui Y, Wang Q, Wang J, et al. Knockdown of AKT2 expression by RNA interference inhibits proliferation, enhances apoptosis, and increases chemosensitivity to the anticancer drug VM‐26 in U87 glioma cells. Brain Res. 2012;1469:1‐9. [DOI] [PubMed] [Google Scholar]
- 18. Cui Y, Lin J, Zuo J, et al. AKT2‐knockdown suppressed viability with enhanced apoptosis, and attenuated chemoresistance to temozolomide of human glioblastoma cells in vitro and in vivo. Onco Targets Ther. 2015;8:1681‐1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Balamurugan K, Wang JM, Tsai HH, et al. The tumour suppressor C/EBPdelta inhibits FBXW7 expression and promotes mammary tumour metastasis. EMBO J. 2010;29:4106‐4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kanzawa T, Germano IM, Komata T, et al. Role of autophagy in temozolomide‐induced cytotoxicity for malignant glioma cells. Cell Death Differ. 2004;11:448‐457. [DOI] [PubMed] [Google Scholar]
- 21. Teng CL, Hsieh YC, Phan L, et al. FBXW7 is involved in Aurora B degradation. Cell Cycle. 2012;11:4059‐4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hirose Y, Berger MS, Pieper RO. p53 effects both the duration of G2/M arrest and the fate of temozolomide‐treated human glioblastoma cells. Cancer Res. 2001;61:1957‐1963. [PubMed] [Google Scholar]
- 23. Hayashi T, Adachi K, Ohba S, et al. The Cdk inhibitor flavopiridol enhances temozolomide‐induced cytotoxicity in human glioma cells. J Neurooncol. 2013;115:169‐178. [DOI] [PubMed] [Google Scholar]
- 24. Wang L, Ye X, Liu Y, et al. Aberrant regulation of FBW7 in cancer. Oncotarget. 2014;5:2000‐2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ji S, Qin Y, Shi S, et al. ERK kinase phosphorylates and destabilizes the tumor suppressor FBW7 in pancreatic cancer. Cell Res. 2015;25:561‐573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang Y, Gao X, Zhang M, et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst. 2018;1:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang X, Zhang J, Zhou L, et al. Fbxw7 regulates hepatocellular carcinoma migration and invasion via Notch1 signaling pathway. Int J Oncol. 2015;47:231‐243. [DOI] [PubMed] [Google Scholar]
- 28. Martin SS, Ridgeway AG, Pinkas J, et al. A cytoskeleton‐based functional genetic screen identifies Bcl‐xL as an enhancer of metastasis, but not primary tumor growth. Oncogene. 2004;23:4641‐4645. [DOI] [PubMed] [Google Scholar]
- 29. Happold C, Roth P, Wick W, et al. Distinct molecular mechanisms of acquired resistance to temozolomide in glioblastoma cells. J Neurochem. 2012;122:444‐455. [DOI] [PubMed] [Google Scholar]


