Abstract
Helicobacter pylori is a bacterium that has infected more than half of the human population worldwide. This bacterium is closely associated with serious human diseases, such as gastric cancer, and identifying and understanding factors that predict bacterial virulence is a priority. In addition, this pathogen shows high genetic diversity and co-evolution with human hosts. H. pylori population genetics, therefore, has emerged as a tool to track human demographic history. As the number of genome sequences available is increasing, studies on the evolution and virulence of H. pylori are gaining momentum. This review article summarizes the most recent findings on H. pylori virulence factors and population genetics.
Keywords: genetic population, Helicobacter pylori, virulence factor
1. Introduction
Helicobacter pylori (H. pylori) is a Gram-negative spiral-shaped bacterium found in the gastric epithelium of humans (Suerbaum and Michetti, 2002). Since its first description by Marshall and Warren (1984), this species has gained considerable research attention because of its clinical and evolutionary importance (Marshall and Warren, 1984; Suerbaum and Achtman, 1999; Yamaoka, 2010).
The bacterium is primarily transmitted within families and acquired during childhood. In the absence of adequate treatment, life-long gastric colonization can result in several diseases such as chronic gastritis, peptic ulcer, and gastric cancer (Yamaoka, 2010). The association between gastric cancer—one of the most common malignancies in the world—and H. pylori infection has attracted great interest worldwide, with the International Agency for Research on Cancer (IARC), a subordinate organization of the World Health Organization (WHO), classifying H. pylori as a “group 1 (definite carcinogen)” in 1994 (1994). Therefore, many studies have explored bacterial factors to explain the link between gastric cancer and this bacterium. Several bacterial virulence factors have been identified that predict severe clinical outcomes and explain the global geographic distribution of gastric cancer (Yamaoka, 2010). Thus far, the biological function and structure of these virulence factors, as well as the discovery of new candidate virulence factors, has continued to occupy most H. pylori -related research.
H. pylori and humans have co-evolved for at least 100,000 years, long before human ancestors left Africa (Moodley et al., 2012). During this long history in its hostile gastric niche in humans, H. pylori has developed a wide spectrum of strategies to persist in and adapt to changing conditions in and around its host (Suerbaum and Achtman, 1999). Thus, H. pylori is one of the most diverse bacterial species and arguably the most successful human pathogen known to date (Falush et al., 2003b; Suerbaum and Achtman, 1999). However, despite of their high genetic diversity, H. pylori strains appear genetically structured, exhibiting phylogeographic patterns that consistently correlate with that of their human hosts. Therefore, the population genetics and phylogenetic relationships among isolates have enabled accurate mapping of the local and global demographic histories of human evolution (Falush et al., 2003b; Linz et al., 2007; Moodley et al., 2012; Moodley et al., 2009). H. pylori genetics is promising to shed light on yet unknown dynamics of human evolution. Therefore, an increasing amount of resources are being devoted to detailed analyses of H. pylori populations with the aim of describing human history.
In addition, the increasing availability of H. pylori whole-genome sequences is enabling more genomic analyses than ever before. Such analyses are empower efforts to detect new virulence factors as well as detailed studies of population genetics. The present literature review addresses the most recent and important findings on bacterial virulence factors and genetic populations of H. pylori. Scientific data, mostly that reported in the last three years, are summarized, with the aim of highlighting expected future developments in H. pylori-related molecular epidemiological research.
2. New insights on H. pylori virulence factors
The pathogenesis of H. pylori is driven by several virulence factors that facilitate colonization, induce inflammation, and damage host cells. These virulence factors have been linked to the risk of developing severe gastric diseases and include the cag pathogenicity island (PAI), vacuolating cytotoxin (VacA), outer membrane proteins (OMPs), serine protease HtrA, and many others.
2.1. VacA
VacA is an exotoxin that was named for its capacity to induce host cell vacuolation (Cover and Blaser, 1992). At the time of its discovery, no bacterial toxin with similar activity had yet been described, and since this discovery, many studies have been conducted to clarify its function and structure.
VacA have been described as a multi-receptor protein that has pleiotropic effects, including membrane depolarization, mitochondrial dysfunction, autophagy, activation of mitogen-activated protein kinases, inhibition of T cell function, and induction of apoptosis (Foegeding et al., 2016). These functions contribute to the persistent colonization of H. pylori and its pathogenesis in several upper digestive tract diseases. Recently, additional VacA-related pathways and functions have been reported. Amilon et al. (2015) described a putative stem-loop structure in the 5′ untranslated region that affects transcription of vacA and leads to higher expression and toxicity of VacA (Amilon et al., 2015). An extra-digestive location of functional VacA in lungs has led to the suggestion that VacA plays a role in the pathogenesis of respiratory diseases by inducing IL-8 and IL-6 (Nakashima et al., 2015). In addition, new host factors that interact with or regulate the VacA-induced apoptosis have been reported. Yahiro et al. (2015) described a new signaling pathway for VacA-induced apoptosis that is mediated by cytoplasmic accumulation of connexin 43 (Cx43), a ubiquitous connexin family member that plays a role in gap junction and cell-cell channel formation (Yahiro et al., 2015). In addition, Chang et al. (2016) described the role of cortactin, an actin-binding protein, in the regulation of apoptosis induced by VacA (Chang et al., 2016).
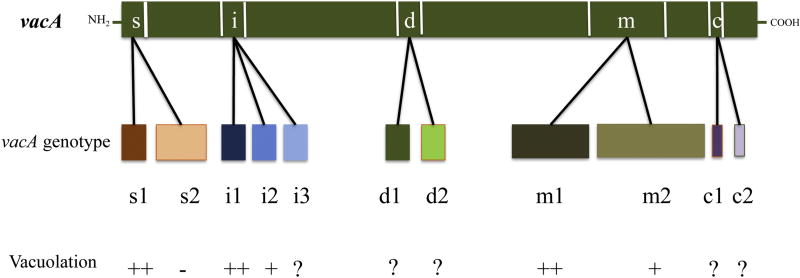
VacA includes a 33-kDa N-terminal domain associated with cytotoxicity and a 55-kDa C-terminal domain involved in binding of the bacterium to cell surface receptors (Yahiro et al., 2015). Almost all H. pylori strains harbor the vacA gene, and allelic polymorphisms of the protein show clinical significance and toxic activity that are associated with specific combination of its three regions: the signal peptide (s1 and s2 variants), the intermediate region (i1, i2, and i3 variants), and the middle region (m1 and m2 variants). Molecular epidemiological studies have revealed two novel polymorphic sites, the deletion (d1 and d2 variants) and c-region (c1 and c2 variants) located in the 3′-end region of VacA (Fig. 2) (Thi Huyen Trang et al., 2016). Similar to sites described previously, some variants of these two novel regions have been associated with high risk of gastric cancer (Bakhti et al., 2016; Ogiwara et al., 2009). However, the contributions of these regions to different VacA functions such as vacuole formation have not yet been identified.
Figure 2. Current allelic diversity inferred from the VacA sequence.
The VacA structure comprises five regions of sequence diversity referred to as the signal (s), intermediate (i) middle (m), deletion (d), and c (c) regions. The vacuolating activity of VacA varies with different alleles. In vitro, vacA s1/m1/i1 alleles show higher levels of vacuole formation than s2/m2/i2. The functions of novel polymorphic regions c and d, as well as the i3 subtype, have not yet been studied.
2.2. cag PAI
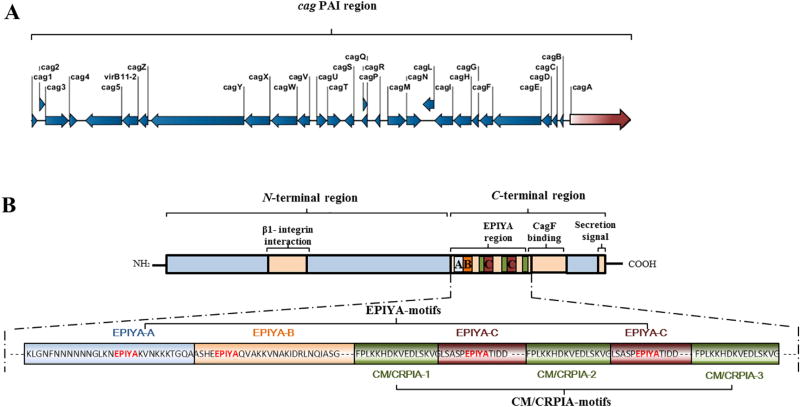
cag PAI is a chromosomic region of approximately 37 kb that encodes the cag type IV secretion system (cag-T4SS), including cytotoxin-associated gene A (cagA) (Fig. 1A). CagA is a 120–140-kDa cellular effector that is translocated into host cells through the cag-T4SS and interacts with a large repertoire of cellular signaling pathways, including those leading to carcinogenesis (Tegtmeyer et al., 2017a). CagA was discovered prior to the cag PAI and was named for its presumed link with the vacuolating cytotoxin activity of the VacA protein that had been discovered two years before (Tummuru et al., 1993). Since then, CagA and cag PAI have shown no effect on vacuolating cytotoxin production, suggesting the possible functional independence of vacA and cagA, two genes located approximately 300 kb apart on the H. pylori chromosome (Tummuru et al., 1994).
Figure 1. Overall structure of the cag PAI (A) and CagA protein (B) in H. pylori strain P12.
A. Structure of the cag PAI region (~37 kb). The region comprises 28 genes that encode components of the cag-T4SS, including the CagA protein effector.
B. Structure of the CagA protein (1,214 amino acid residues). The N-terminal part of CagA harbors a putative β-integrin-binding region. The C-terminal region comprises the EPIYA region, which contains EPIYA ABCC motifs and three MKI/CM/CRPIA motifs, regions that bind to the secretion chaperone CagF and contain the C-terminal secretion signal.
The cag PAI is currently the most extensively studied H. pylori virulence factor. Its epidemiological role has been discussed previously (Suzuki et al., 2012; Yamaoka, 2010), and numerous studies have shown that cag PAI-positive H. pylori strains are associated with an increased risk of gastric cancer compared to strains that lack cag PAI (Yamaoka, 2010). The risk of gastric cancer is determined by several cag PAI-related features, including cag PAI-positivity, sequence variation within CagA (such as the number and type of EPIYA motifs), and the presence or absence of a functional cag type IV secretion system (which translocates CagA into host cells) (Suzuki et al., 2012; Tegtmeyer et al., 2017a; Yamaoka, 2010). Host c-Src and c-Abl kinases control hierarchic phosphorylation and CagA function in Western and East Asian H. pylori strains (Mueller et al., 2012). Recently, the function and structure of the CagA and cag-T4SS have been elucidated further.
First, the crystal structure of the N-terminal segment of the CagA molecule, which harbors a unique structure with no sequence homology to any known proteins, was recently obtained (Hayashi et al., 2012). The structured N-terminal part of CagA consists of several domains and harbors the putative integrin-binding region (Hayashi et al., 2012; Kaplan-Turkoz et al., 2012). The unstructured C-terminal region displays recognized repeated sections, EPIYA (Glu-Pro-Ile-Tyr-Ala), and CM (CagA multimerization) or CRPIA (conserved repeat responsible for phosphorylation-independent activity) motifs, as well as a region that binds to the secretion chaperone CagF, and a C-terminal secretion signal (Fig. 1B) (Schindele et al., 2016).
Second, further insights in the molecular mechanisms regulating cagA function through the cag PAI have been released. In fact, the cagA promoter region, which had been described previously (Loh et al., 2011), was further characterized by Ferreira et al. (Ferreira et al., 2016). This study identified specific sequence motifs located in the promoter region (+59 AATAAGATA and −10 TATAATGA sequence motifs) that are linked to CagA expression levels and interleukin-8 (IL-8) secretion in an infected gastric cell line, as well as to severe clinical outcomes (Ferreira et al., 2016). Because these sequence variations can be used to discriminate between two different levels of gastric cancer risk associated with Colombian strains and those with European and African origins, the discussion should be extended in future studies to strains from other geographical origins. Another important cagA-related feature identified recently is the number of copies of this gene found in different strains. Jang et al. (2017) showed that H. pylori isolates can carry multiple tandem copies of cagA that affect CagA expression and activity and may impact the development of gastric disease (Jang et al., 2017). Consistent with the findings of Jang et al. (2017), Draper et al. (2017) showed, using close strains named PMSS1 and SS1, that the number of cagA changes dynamically and modulates CagA activity (Draper et al., 2017). Thus, future epidemiological studies should address not only the sequence variation within CagA (EPIYA and CM/CRPIA motifs) but also the functionality of the whole cag PAI/T4SS in determining the biological effects of CagA, as well cagA promoter variants and the number of cagA copies as useful markers for disease risk. A β-lactamase-dependent reporter system that allows a precise and quantitative determination of CagA translocation into host cells has just been developed (Schindele et al., 2016). This phosphorylation-independent assay has opened the door to further insights into the in vivo function and epidemiological role played by H. pylori cag-T4SS.
Finally, an integrative model of the activity of translocated CagA was recently developed (Tegtmeyer et al., 2017a). This model summarizes the data available on highly complex signaling pathways altered by translocated CagA through multiple receptor kinases (c-Met and EGFR) and non-receptor kinases (Src, Abl, Csk, aPKC, Par1, PI3K, Akt, FAK, GSK-3, JAK, PAK1, PAK2, and MAP kinases) in the human gastric epithelium that manipulate processes ranging from cell adhesion and polarity to apoptosis, inflammation, and cell cycle progression (Tegtmeyer et al., 2017a).
2.3. OMPs
Several OMPs have been predicted, based on the structure of the H. pylori genome, to play a role in bacterial adherence to the gastric mucosa (Alm et al., 2000). Though only a few OMPs have been established as adhesins (e.g., BabA/B, SabA, AlpA/B, OipA, and HopQ) with known host receptors (e.g., BabA/B, SabA, AlpA/B, and HopQ), their role in the virulence of H. pylori is increasingly evident (Aspholm et al., 2006; Bjornham et al., 2005; Javaheri et al., 2016; Koniger et al., 2016; Senkovich et al., 2011; Yamaoka et al., 2000). Based on epidemiological data, highly virulent H. pylori strains are known to express OMPs along with proteins from the cag PAI. This is consistent with observations showing that high pathogenic strains, especially those encoding the cag PAI, are also highly adherent strains harboring numerous OMPs with the ability to enhance the expression of OMP ligands on gastric epithelial cells (Marcos et al., 2008). Thus, adhesins likely promote intimate contact between H. pylori and gastric epithelial cells, while the cag-T4SS, which forms an extracellular pilus-like structure, facilitates translocation of the effector protein CagA to induce pathogenicity leading to severe gastroduodenal diseases such as gastric ulcers and gastric cancer (Koniger et al., 2016). In addition to their adherence function, new roles for OMPs in bacterial virulence have been discovered recently through studies analyzing their interactions with cag-T4SS or their direct effects on host cells.
Blood group antigen-binding adhesin (BabA) is the most studied H. pylori OMP. It mediates bacterial adherence to ABO/Lewis b (Leb) blood group antigens in the gastric pit region of the human stomach mucosa (Ansari and Yamaoka, 2017). Evidence for a virulence role for BabA rely on epidemiological data associating the babA gene to other virulence genes and to severe gastroduodenal diseases, as well as on the fact that BabA-mediated adherence of H. pylori can be a potentiator of cag-T4SS activity and inducer of proinflammatory cytokines (e.g., CCL5 and IL-8) and precancer-related factors (e.g., CDX2 and MUC2) (Ansari and Yamaoka, 2017; Ishijima et al., 2011). Recently, additional evidence for this virulence role was provided by an epidemiological study that showed a significant association between the combination of OipA, BabA, and SabA and a diagnosis of H. pylori-associated gastric cancer (Su et al., 2016). However, recent investigations have reiterated that H. pylori BabA sequences, expression, and corresponding binding phenotypes are highly diverse and dynamic (Bugaytsova et al., 2017; Hansen et al., 2017; Kable et al., 2017; Moonens and Remaut, 2017; Subedi et al., 2014; Sweeney and Guillemin, 2016). Thus, caution is needed when suggesting an association between babA and clinical outcomes based on epidemiological studies. Sweeney and Guillemin (2016) rightly suggested that the discussion sections in some reports should be extended to babA sequence and expression variation, host glycans, and disease incidence in different host and H. pylori populations (Sweeney and Guillemin, 2016).
Helicobacter outer membrane protein Q (HopQ) is an OMP that was first called HP1177 or omp27 after H. pylori was completely sequenced but before its presence on the surfaces of bacteria cells, influencing the adherence to human epithelial cells, was demonstrated (Loh et al., 2008). Since two alleles of hopQ were described as having an epidemiological association with the hopQ I allele and cagA gene (Cao et al., 2005; Ohno et al., 2009), this OMP is attracting increasingly interest among researchers. By screening a large number of H. pylori mutants, Belogolova et al. (2013) identified HopQ as a non-cag PAI-encoded co-factor of T4SS function that is essential for CagA translocation and CagA-mediated host cell responses such as the formation of a hummingbird phenotype and cell scattering. Their work also showed that deletion of hopQ reduces the T4SS-dependent activation of NF-κB, induction of MAPK signaling, and secretion of IL-8 in host cells (Belogolova et al., 2013). Moreover, Jiménez-Soto LF et al. (2013) identified HopQ along with other H. pylori OMPs such as HopI and AlpAB as factors restricting and controlling subsequent CagA translocation into host cells, independently of β1 integrin receptor availability (Jiménez-Soto et al., 2013). Efforts to clarify the molecular mechanisms underlying OMP interactions and CagA translocation have led to HopQ being implicated in cag PAI-related virulence-associated interactions with human receptors from the carcinoembryonic antigen-related cell adhesion molecule (CEACAMs) family (Javaheri et al., 2016; Koniger et al., 2016).
The H. pylori outer inflammatory protein A (OipA) adhesion function and its ability to induce a pro-inflammatory response in a gastric epithelial cell line or animal models are controversial. However, based on epidemiological studies, functional OipA correlates with highly virulent strains expressing the cag PAI and vacA-s1/m-1 (Matsuo et al., 2017). The OipA receptor has not yet been identified, but OipA-related host cell signaling has been reported. This OMP is believed to trigger pathways related to inflammation induction, actin remodeling, and cell apoptosis through epidermal growth factor receptor (EGFR)/focal adhesion kinase (FAK), phosphoinositide-3 kinase (PI3K)-dependent Akt activation, and Forkhead transcription factors of class O (FoxO) (Tabassam et al., 2008, 2012). Recently, new arguments in factor of a virulence role for H. pylori OipA have been presented based on the treatment of gastric cell lines with various concentrations of OipA (Teymournejad et al., 2017). While confirming the binding property of OipA, this group showed toxic effects, as well as an apoptosis-triggered cascade via signaling that affected the Bax/Bcl-2 protein ratio and cleaved-caspase 3 level, leading to a mitochondrial apoptotic cascade (Teymournejad et al., 2017).
2.4. H. pylori prophages
Bacteriophages (phages) are viruses that infect bacteria (Canchaya et al., 2003). Phage-bacteria interplay includes a step in which the phage is inserted into the host genome, leading to either bacterial lysis or prophage domestication. Thus, the host genome might be shaped in terms of the evolution of diversity or virulence in conjunction with phage infection (Rodriguez-Valera et al., 2009). Bacteriophages were initially reported in H. pylori by Schmid et al. (1990), who hypothesized phage-mediated virulence of this species (Schmid et al., 1990). Subsequent observations of phages in Helicobacter spp. have been relatively rare (Arnold et al., 2011; Eppinger et al., 2006; Heintschel von Heinegg et al., 1993; Vale et al., 2008). The putative pathogenic role of H. pylori prophages has been revived since the first isolation of an integrated prophage, which was similar to phages of the Siphoviridae family, from a patient with gastric mucosa-associated lymphoid tissue lymphoma (Lehours et al., 2011). Recently, Kyrillos et al. (2016), by screening phage-orthologous H. pylori sequences from 335 strains, found a correlation between the presence of phage-related sequences, likely acquired by horizontal gene transfer, and that of the two major virulence factors CagA and VacA (Kyrillos et al., 2016). Another H. pylori prophage has been reported in the genome of a cag PAI-negative strain, which was isolated from a patient suffering from gastric cancer (Mucito-Varela et al., 2016). Because non-pyloric Helicobacter prophages have been revealed to encode antibiotic resistance genes and virulence factors (Qumar et al., 2017), we can predict that the genetic content and putative pathogenic role of H. pylori prophages will attract increasing attention from researchers.
2.5. H. pylori HtrA
High temperature requirement A (HtrA) is a serine protease released in the extracellular environment by H. pylori during infection. Extracellular H. pylori HtrA had been shown to cleave the cell adhesion protein through proteolysis, and its possible crosstalk with CagA and direct effects on the infection process have been intensively studied (Hoy et al., 2012; Hoy et al., 2010). Identification of tumor suppressor E-cadherin as an HtrA substrate came emphasized the significant role of HtrA activity in H. pylori-induced carcinogenesis and disruption of adherens junctions, which allows bacterial transmigration across the epithelium (Hoy et al., 2010). Recently, Schmidt et al. (2016a, 2016b) further elucidated the activity of HtrA on E-cadherin (Schmidt et al., 2016a; Schmidt et al., 2016b). Tegtmeyer et al (2016). showed that the htrA gene locus was conserved among 992 H. pylori clinical isolates and that the proteolytic activity of HtrA was essential for bacterial survival (Tegtmeyer et al., 2016). Finally, Harrer et al., by successfully introducing a second functional htrA gene in H. pylori strains P12 and 26695, showed that the overexpression of HtrA enhances cleavage of E-cadherin, bacterial transmigration, and delivery of the cag-T4SS effector protein CagA into polarized epithelial cells (Harrer et al., 2017). Taken together, these findings suggests a novel model that relies on H. pylori access to the basolateral compartment for deployment of cag-T4SS and injection of the oncoprotein CagA into host cells (Harrer et al., 2017; Tegtmeyer et al., 2017b). Therefore, H. pylori HtrA-triggered E-cadherin is, without doubt, a bacterial virulence factor. Further studies will elucidate the epidemiological role of such an important virulent factor.
3. Human migration tracked through H. pylori genetic diversity
The isolation of genes (e.g., cagA and vacA), as well as a multilocus sequence typing (MLST) of a set of seven concatenated housekeeping genes (atpA, efp, mutY, ppa, trpC, ureI, and yphC) or cag PAI genes, have been used as tools to track human migration throughout history. For details, see our previous review paper (Suzuki et al., 2012). Recently, further interesting insights have been gained by genome-based analyses to identify genetic populations of H. pylori genetic. For examples, new potential tools for tracking human migration have been found in H. pylori prophage sequences.
3.1. Genome-based inference of H. pylori population structure
H. pylori strains from diverse geographical regions have shown a quasi-panmictic structure, with the lack of gene allele clonality resulting from frequent genetic recombination (Suerbaum et al., 1998). However, this species could be split into distinct bacterial populations that exhibit close relationships with the ethno-geographical distribution of its human host (Falush et al., 2003b; Linz et al., 2007). Thus, the identification of genetic populations of H. pylori has arisen as a valuable tool for analyzing H. pylori population differentiation and selection, as well as for inferring human demographic history (Moodley and Linz, 2009). The software most often used to identify H. pylori population structure using genetic data is STRUCTURE, developed by Falush et al. (Falush et al., 2003a; Falush et al., 2003b). Additionally, phylogenetic tree construction has been also used (Moodley et al., 2012). Based on these methods, bacterial populations, including three from Africa (hpNEAfrica, hpAfrica1, and hpAfrica2), one from Europe (hpEurope), and three from Asia (hpEAsia, hpAsia2, and hpSahul) had been distinguished (Falush et al., 2003b; Linz et al., 2007; Moodley and Linz, 2009; Moodley et al., 2012). Different subpopulations have been shown in hpEastAsia (hspAmerind, hspEAsia, and hsp Maori), hpNEAfrica (hspEastNEAfrica and hspCentralNEAfrica), and hpAfrica1 (hspSAfrica, hspWAfrica, and hspCAfrica) (Moodley et al., 2012; Nell et al., 2013). However, the phylogenetic approach is limited to making a rough assessment of populations, and it is not designed to infer the number of populations directly (Yahara et al., 2013). STRUCTURE, even with a linkage model option, is limited by the need for satisfactory convergence that requires prior specification of the K number of populations (Lawson and Falush, 2012; Lawson et al., 2013). This may lead to a violation of the methodological assumptions and to biased results (Lawson et al., 2013).
To circumvent the limitations of these approaches and to improve the accuracy and reliability population and subpopulation identification, new approaches have been developed for use with genome-wide sequence data instead of sequences of isolated genes. A new tool called fineSTRUCTURE, introduced by Lawson et al. and based on the chromosome painting method (Lawson and Falush, 2012), has been used successfully by Yahara et al. (2013) to describe H. pylori populations (Yahara et al., 2013). Expectedly, the fineSTRUCTURE approach conducted under linkage and admixture models provided a population structure consistent with those reached using STRUCTURE and phylogenetic approaches. However, it revealed the structure of H. pylori populations at a finer scale (Yahara et al., 2013). Thus, novel subgroups, even singletons, could be found within subpopulations defined by previous methods. Moreover, the new approach is able to elucidate the genetic flux between populations, while providing faster and more efficient results based on both small and large sets of genome-wide sequences (Yahara et al., 2013). Data supporting consistency between the different approaches, specifically identification of main genetic populations at a genomic level, have been shown by Montano et al. (Montano et al., 2015). More recently, by applying the fineSTRUCTURE approach, Thorell et al. (2017) uncovered more dynamic evolution of H. pylori in four new subpopulations of H. pylori: hspAfrica1NAmerica, hspAfrica1Nicaragua, hspEuropeColombia, and hspMiscAmerica (Thorell et al., 2017). Thus, these new subpopulations seen as the result of genetic drift and admixture events occurring over a relatively short period (~500 years), are suggestive of rapid evolution of this bacterium in the Americas (Thorell et al., 2017). More evidence for this rapid evolution was provided by analysis of MLST and phylogenetic data on cag PAI and seven housekeeping genes in isolates from Latin America and Portuguese-speakers worldwide (Munoz-Ramirez et al., 2017; Oleastro et al., 2017).
These new data have strengthened the view that H. pylori genetic diversity can be used as a marker for historical human migration events (summarized in Table 1 and Table 2). Future studies will be conducted to infer a global demographic history by extending the analysis to isolates from underrepresented regions such as Central Africa, Northeast Africa, Central Asia, and the Siberian region. Moreover, the fine-scale structure of H. pylori populations revealed by fineSTRUCTURE would likely be needed to elucidate further details in human evolutionary events, as well as the regional distribution of gastric cancer risk.
Table 1.
Geographic range of modern populations and subpopulations of H. pylori
| Population | Subpopulation | Geographic range/location | Main references |
|---|---|---|---|
| hpAfrica1 | hspWAfrica | Africa: West to North, then Eastward to South | (Nell et al., 2013) |
| hspCAfrica | Africa: West to center towards Southwest | (Nell et al., 2013) | |
| hspSAfrica | Africa: South-East (+ Madagascar) to South | (Nell et al., 2013) | |
| hpAfrica1/hpEurope hybrid | hspEuropeColombia | South America | (Thorell et al., 2017) |
| hspAfrica1NAmerica | America: North, Center, and South | (Thorell et al., 2017) | |
| hspAfrica1Nicaragua | America: Center | (Thorell et al., 2017) | |
| hspMiscAmerica | America: Center and South | (Thorell et al., 2017) | |
| hpAfrica2 | hspNorthSan | Africa: South-West | (Linz et al., 2007; Moodley et al., 2012) |
| hspSouthSan | Africa: South Africa | (Moodley et al., 2012) | |
| hpNEAfrica | hspENEAfrica | Africa: Horn of Africa, westward to the Nile Valley, and to the Northwest | (Nell et al., 2013) |
| hspCNEAfrica | Africa: Nile Valley, toward the West and Northwest | (Nell et al, 2013) | |
| hpSahu1 | hspAustralia | Australia | (Moodley et al., 2009) |
| hspNGuinea | Papua New Guinea | (Moodley et al., 2009) | |
| hpAsia2 | hspLadakh | Asia: Himalayan region of Northern India | (Breurec et al., 2011; Tay et al., 2009) |
| hspIndia | India, Bangladesh, Malaysia, Thailand, the Philippines, and even Northern Europe | (Breurec et al., 2011; Tay et al., 2009) | |
| hpEurope | hspEuropeS, hspEuropeN | Europe, as far East as South-East Asia | (Thorell et al., 2017) |
| ~ | Australia, America, South Africa, Australia, and the South Pacific Islands | (Falush et al., 2003b) | |
| hpEastAsia | hspEAsia | Asia: East Asia, toward Japan | (Falush et al., 2003b; Moodley et al., 2009) |
| hspAmerind | Asia and America: East Asia, toward North and South America | (Falush et al., 2003b) | |
| hspMaori | Asia: Taiwan and the Pacific Islands | (Linz et al., 2007; Moodley et al., 2009) |
Table 2.
Main demographic events inferred from modern populations and subpopulations of H.pylori
| Population | Subpopulation | Main human demographic events so far inferred | Main references |
|---|---|---|---|
| hpAfrica1 | hspWAfrica | Prehistoric expansion of Bantu speakers (Eastern route), ~3,000 to 5,000 BP | (Nell et al., 2013) |
| hspCAfrica | Prehistoric expansion of bantu speakers (Western route) | (Nell et al., 2013) | |
| hspSAfrica | Specific prehistoric Bantu migration of Nguni speakers from central West Africa along East coast | (Nell et al., 2013) | |
| hpAfrica1/hpEurope hybrid | hspEuropeColombia | Post-Colombian migration of Europeans and Africans toward the New World | (Nell et al., 2013) |
| hspAfrica1NAmerica,, | Migration of Africans (along with Europeans) toward the New World during the colonial-era slave trade | (Thorell et al., 2017) | |
| hspAfrica1Nicaragua | Migration of Africans (along with Europeans) toward the New World during the colonial-era slave trade | (Thorell et al., 2017) | |
| hspMiscAmerica | Migration of Africans (along with Europeans) toward the New World during the colonial-era slave trade | (Thorell et al., 2017) | |
| hpAfrica2 | hspNorthSan | Colonization of specific groups of Khoisan peoples (Xun and Khwe) | (Thorell et al., 2017) |
| hspSouthSan | Colonization of specific groups of Khoisan peoples (Khomani) | (Linz et al., 2007; Moodley et al., 2012) | |
| hpNEAfrica | hspENEAfrica | Prehistoric expansion of Nilo-Saharan speakers | (Moodley et al., 2012) |
| hspCNEAfrica | Prehistoric expansion of Nilo-Saharan speakers during the Holocene humid period | (Nell et al., 2013) | |
| hpSahu1 | hspAustralia | Peopling of Sahul and isolation in Australia after the Last Glacial Maximum | (Nell et al., 2013) |
| hspNGuinea | Peopling of Sahul and isolation in New Guinea after the Last Glacial Maximum | (Moodley et al., 2009) | |
| hpAsia2 | hspLadakh | Past peopling of the eastern Eurasia and isolation in Himalayas | (Breurec et al., 2011; Tay et al., 2009) |
| hspIndia | Past peopling of the eastern Eurasia as well as spread of Uralic speakers out of Siberia | (Breurec et al., 2011; Tay et al., 2009) | |
| hpEurope | hspEuropeS, hspEuropeN | Prehistoric mixing of populations (from Africa and Asia) during colonization of Europe | (Thorell et al., 2017) |
| Post-Colombian expansion of Europeans during the colonial era, ~500 BP | (Falush et al., 2003b) | ||
| hpEastAsia | hspEAsia | Expansion of Chinese speakers (sino-Tibetan), ~3000 years ago during the Zhou Dynasty | (Falush et al., 2003b; Moodley et al., 2009) |
| hspAmerind | Prehistoric migration across the Bering strait and colonization of the Americas, >12,000 BP | (Falush et al., 2003b) | |
| hspMaori | Expansion of Austronesian Speakers into the Pacific | (Linz et al., 2007; Moodley et al.,2009) |
3.2. H. pylori prophages and the geographical origin of strains
An increasing number of H. pylori prophage sequences are being identified and analyzed (Secka et al., 2017; Uchiyama et al., 2016; Vale et al., 2017). Vale et al. (2015) are the first to show that the diversity of two concatenated H. pylori prophage genes (integrase and holin) allow strains to be differentiated according to their geographic origins, in agreement with MLST-based classification using seven H. pylori housekeeping genes (Vale et al., 2015). More interestingly, analysis of H. pylori prophage genes has revealed two European subpopulations of H. pylori, hpNEurope and hpSWEurope, whereas the traditional MLST-based method identified only hpEurope (Vale et al., 2015). However, whether these new prophage-based subpopulations of H. pylori coincide with those found by genome-based analyses still needs to be analyzed. To gain more insight into the phylogenetic relationships between prophages sequences and bacterial geographic distribution, Vale et al. (2017) just analyzed a complete prophage genome (Vale et al., 2017). This analysis shows similarities with prophage-isolated genes and a refinement of phylogeographic clustering assigned by STRUCTURE (Vale et al., 2017). Additional observations have been made for H. pylori strains from Gambia (Secka et al., 2017). Moreover, the insertion sites of prophages suggest ancient acquisition and vertical transfer, with a single ancestral prophage likely responsible for sequences inserted at similar loci in different bacterial genomes (Vale et al., 2017). Therefore, H. pylori prophages and their host bacteria appear to share a complex evolutionary history that is shaped by human populations, possibly providing a more detailed approach to population genetic analysis than MLST.
4. Conclusion
Fascinating developments in H. pylori-related research, especially regarding bacterial virulence factors and evolution, should be expected. New virulence factors, as well as interconnections between known ones, will be brought to light. Detailed population genetic studies will indicate previously unknown dynamics of local and more recent demographic processes in human evolution. This research will increasingly be impacted by access to genome-wide data.
Highlights.
Virulence factors are ever key points for H. pylori related pathogenesis
The genetics of H. pylori prophage sequences bring interesting features
New H. pylori populations will enounce the usefulness for tracing human migrations.
Acknowledgments
This report is based on work supported in part by grants from the grants-in-aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan (26640114, 15H02657 and 16H05191).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Potential competing interests: The authors declare that they have no competing interests.
References
- Schistosomes, liver flukes and Helicobacter pylori. IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Lyon, 7–14 June 1994. IARC monographs on the evaluation of carcinogenic risks to humans. 1994;61:1–241. [PMC free article] [PubMed] [Google Scholar]
- Alm RA, Bina J, Andrews BM, Doig P, Hancock RE, Trust TJ. Comparative genomics of Helicobacter pylori: analysis of the outer membrane protein families. Infection and immunity. 2000;68:4155–4168. doi: 10.1128/iai.68.7.4155-4168.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amilon KR, Letley DP, Winter JA, Robinson K, Atherton JC. Expression of the Helicobacter pylori virulence factor vacuolating cytotoxin A (vacA) is influenced by a potential stem-loop structure in the 5' untranslated region of the transcript. Molecular microbiology. 2015;98:831–846. doi: 10.1111/mmi.13160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari S, Yamaoka Y. Helicobacter pylori BabA in adaptation for gastric colonization. World journal of gastroenterology : WJG. 2017;23:4158–4169. doi: 10.3748/wjg.v23.i23.4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspholm M, Olfat FO, Norden J, Sonden B, Lundberg C, Sjostrom R, Altraja S, Odenbreit S, Haas R, Wadstrom T, Engstrand L, Semino-Mora C, Liu H, Dubois A, Teneberg S, Arnqvist A, Boren T. SabA is the H. pylori hemagglutinin and is polymorphic in binding to sialylated glycans. PLoS pathogens. 2006;2:e110. doi: 10.1371/journal.ppat.0020110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhti SZ, Latifi-Navid S, Mohammadi S, Zahri S, Bakhti FS, Feizi F, Yazdanbod A, Siavoshi F. Relevance of Helicobacter pylori vacA 3'-end Region Polymorphism to Gastric Cancer. Helicobacter. 2016;21:305–316. doi: 10.1111/hel.12284. [DOI] [PubMed] [Google Scholar]
- Belogolova E, Bauer B, Pompaiah M, Asakura H, Brinkman V, Ertl C, Bartfeld S, Nechitaylo TY, Haas R, Machuy N, Salama N, Churin Y, Meyer TF. Helicobacter pylori outer membrane protein HopQ identified as a novel T4SS-associated virulence factor. Cellular microbiology. 2013;15:1896–1912. doi: 10.1111/cmi.12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornham O, Fallman E, Axner O, Ohlsson J, Nilsson UJ, Boren T, Schedin S. Measurements of the binding force between the Helicobacter pylori adhesin BabA and the Lewis b blood group antigen using optical tweezers. Journal of biomedical optics. 2005;10:44024. doi: 10.1117/1.1989227. [DOI] [PubMed] [Google Scholar]
- Bugaytsova JA, Bjornham O, Chernov YA, Gideonsson P, Henriksson S, Mendez M, Sjostrom R, Mahdavi J, Shevtsova A, Ilver D, Moonens K, Quintana-Hayashi MP, Moskalenko R, Aisenbrey C, Bylund G, Schmidt A, Aberg A, Brannstrom K, Koniger V, Vikstrom S, Rakhimova L, Hofer A, Ogren J, Liu H, Goldman MD, Whitmire JM, Aden J, Younson J, Kelly CG, Gilman RH, Chowdhury A, Mukhopadhyay AK, Nair GB, Papadakos KS, Martinez-Gonzalez B, Sgouras DN, Engstrand L, Unemo M, Danielsson D, Suerbaum S, Oscarson S, Morozova-Roche LA, Olofsson A, Grobner G, Holgersson J, Esberg A, Stromberg N, Landstrom M, Eldridge AM, Chromy BA, Hansen LM, Solnick JV, Linden SK, Haas R, Dubois A, Merrell DS, Schedin S, Remaut H, Arnqvist A, Berg DE, Boren T. Helicobacter pylori Adapts to Chronic Infection and Gastric Disease via pH-Responsive BabA-Mediated Adherence. Cell host & microbe. 2017;21:376–389. doi: 10.1016/j.chom.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canchaya C, Proux C, Fournous G, Bruttin A, Brussow H. Prophage genomics. Microbiology and molecular biology reviews : MMBR. 2003;67:238–276. doi: 10.1128/MMBR.67.2.238-276.2003. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao P, Lee KJ, Blaser MJ, Cover TL. Analysis of hopQ alleles in East Asian and Western strains of Helicobacter pylori. FEMS microbiology letters. 2005;251:37–43. doi: 10.1016/j.femsle.2005.07.023. [DOI] [PubMed] [Google Scholar]
- Chang H, Chen D, Ni B, Zuo Q, Wang C, Han R, Lan C. Cortactin Mediates Apoptosis of Gastric Epithelial Cells Induced by VacA Protein of Helicobacter pylori. Digestive diseases and sciences. 2016;61:80–90. doi: 10.1007/s10620-015-3836-0. [DOI] [PubMed] [Google Scholar]
- Cover TL, Blaser MJ. Purification and characterization of the vacuolating toxin from Helicobacter pylori. The Journal of biological chemistry. 1992;267:10570–10575. [PubMed] [Google Scholar]
- Draper JL, Hansen LM, Bernick DL, Abedrabbo S, Underwood JG, Kong N, Huang BC, Weis AM, Weimer BC, van Vliet AH, Pourmand N, Solnick JV, Karplus K, Ottemann KM. Fallacy of the Unique Genome: Sequence Diversity within Single Helicobacter pylori Strains. mBio. 2017;8 doi: 10.1128/mBio.02321-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003a;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Wirth T, Linz B, Pritchard JK, Stephens M, Kidd M, Blaser MJ, Graham DY, Vacher S, Perez-Perez GI, Yamaoka Y, Megraud F, Otto K, Reichard U, Katzowitsch E, Wang X, Achtman M, Suerbaum S. Traces of human migrations in Helicobacter pylori populations. Science. 2003b;299:1582–1585. doi: 10.1126/science.1080857. [DOI] [PubMed] [Google Scholar]
- Ferreira RM, Pinto-Ribeiro I, Wen X, Marcos-Pinto R, Dinis-Ribeiro M, Carneiro F, Figueiredo C. Helicobacter pylori cagA Promoter Region Sequences Influence CagA Expression and Interleukin 8 Secretion. The Journal of infectious diseases. 2016;213:669–673. doi: 10.1093/infdis/jiv467. [DOI] [PubMed] [Google Scholar]
- Foegeding NJ, Caston RR, McClain MS, Ohi MD, Cover TL. An Overview of Helicobacter pylori VacA Toxin Biology. Toxins. 2016;8 doi: 10.3390/toxins8060173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen LM, Gideonsson P, Canfield DR, Boren T, Solnick JV. Dynamic Expression of the BabA Adhesin and Its BabB Paralog during Helicobacter pylori Infection in Rhesus Macaques. Infection and immunity. 2017;85 doi: 10.1128/IAI.00094-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrer A, Boehm M, Backert S, Tegtmeyer N. Overexpression of serine protease HtrA enhances disruption of adherens junctions, paracellular transmigration and type IV secretion of CagA by Helicobacter pylori. Gut pathogens. 2017;9:40. doi: 10.1186/s13099-017-0189-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Senda M, Morohashi H, Higashi H, Horio M, Kashiba Y, Nagase L, Sasaya D, Shimizu T, Venugopalan N, Kumeta H, Noda NN, Inagaki F, Senda T, Hatakeyama M. Tertiary structure-function analysis reveals the pathogenic signaling potentiation mechanism of Helicobacter pylori oncogenic effector CagA. Cell host & microbe. 2012;12:20–33. doi: 10.1016/j.chom.2012.05.010. [DOI] [PubMed] [Google Scholar]
- Hoy B, Geppert T, Boehm M, Reisen F, Plattner P, Gadermaier G, Sewald N, Ferreira F, Briza P, Schneider G, Backert S, Wessler S. Distinct roles of secreted HtrA proteases from gram-negative pathogens in cleaving the junctional protein and tumor suppressor E-cadherin. The Journal of biological chemistry. 2012;287:10115–10120. doi: 10.1074/jbc.C111.333419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoy B, Lower M, Weydig C, Carra G, Tegtmeyer N, Geppert T, Schroder P, Sewald N, Backert S, Schneider G, Wessler S. Helicobacter pylori HtrA is a new secreted virulence factor that cleaves E-cadherin to disrupt intercellular adhesion. EMBO reports. 2010;11:798–804. doi: 10.1038/embor.2010.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishijima N, Suzuki M, Ashida H, Ichikawa Y, Kanegae Y, Saito I, Boren T, Haas R, Sasakawa C, Mimuro H. BabA-mediated adherence is a potentiator of the Helicobacter pylori type IV secretion system activity. The Journal of biological chemistry. 2011;286:25256–25264. doi: 10.1074/jbc.M111.233601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S, Su H, Blum FC, Bae S, Choi YH, Kim A, Hong YA, Kim J, Kim JH, Gunawardhana N, Jeon YE, Yoo YJ, Merrell DS, Ge L, Cha JH. Dynamic Expansion and Contraction of cagA Copy Number in Helicobacter pylori Impact Development of Gastric Disease. mBio. 2017;8 doi: 10.1128/mBio.01779-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javaheri A, Kruse T, Moonens K, Mejias-Luque R, Debraekeleer A, Asche CI, Tegtmeyer N, Kalali B, Bach NC, Sieber SA, Hill DJ, Koniger V, Hauck CR, Moskalenko R, Haas R, Busch DH, Klaile E, Slevogt H, Schmidt A, Backert S, Remaut H, Singer BB, Gerhard M. Helicobacter pylori adhesin HopQ engages in a virulence-enhancing interaction with human CEACAMs. Nature microbiology. 2016;2:16189. doi: 10.1038/nmicrobiol.2016.189. [DOI] [PubMed] [Google Scholar]
- Jiménez-Soto LF, Clausen S, Sprenger A, Ertl C, Haas R. Dynamics of the Cag-type IV secretion system of Helicobacter pylori as studied by bacterial co-infections. Cellular microbiology. 2013;15:1924–1937. doi: 10.1111/cmi.12166. [DOI] [PubMed] [Google Scholar]
- Kable ME, Hansen LM, Styer CM, Deck SL, Rakhimova O, Shevtsova A, Eaton KA, Martin ME, Gideonsson P, Boren T, Solnick JV. Host Determinants of Expression of the Helicobacter pylori BabA Adhesin. Scientific reports. 2017;7:46499. doi: 10.1038/srep46499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan-Turkoz B, Jimenez-Soto LF, Dian C, Ertl C, Remaut H, Louche A, Tosi T, Haas R, Terradot L. Structural insights into Helicobacter pylori oncoprotein CagA interaction with beta1 integrin. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:14640–14645. doi: 10.1073/pnas.1206098109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koniger V, Holsten L, Harrison U, Busch B, Loell E, Zhao Q, Bonsor DA, Roth A, Kengmo-Tchoupa A, Smith SI, Mueller S, Sundberg EJ, Zimmermann W, Fischer W, Hauck CR, Haas R. Helicobacter pylori exploits human CEACAMs via HopQ for adherence and translocation of CagA. Nature microbiology. 2016;2:16188. doi: 10.1038/nmicrobiol.2016.188. [DOI] [PubMed] [Google Scholar]
- Kyrillos A, Arora G, Murray B, Rosenwald AG. The Presence of Phage Orthologous Genes in Helicobacter pylori Correlates with the Presence of the Virulence Factors CagA and VacA. Helicobacter. 2016;21:226–233. doi: 10.1111/hel.12282. [DOI] [PubMed] [Google Scholar]
- Lawson DJ, Falush D. Population identification using genetic data. Annual review of genomics and human genetics. 2012;13 doi: 10.1146/annurev-genom-082410-101510. [DOI] [PubMed] [Google Scholar]
- Lawson LP, Vernesi C, Ricci S, Rovero F. Evolutionary history of the grey-faced Sengi, Rhynchocyon udzungwensis, from Tanzania: a molecular and species distribution modelling approach. PloS one. 2013;8:e72506. doi: 10.1371/journal.pone.0072506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehours P, Vale FF, Bjursell MK, Melefors O, Advani R, Glavas S, Guegueniat J, Gontier E, Lacomme S, Alves Matos A, Menard A, Megraud F, Engstrand L, Andersson AF. Genome sequencing reveals a phage in Helicobacter pylori. mBio. 2011;2 doi: 10.1128/mBio.00239-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linz B, Balloux F, Moodley Y, Manica A, Liu H, Roumagnac P, Falush D, Stamer C, Prugnolle F, van der Merwe SW, Yamaoka Y, Graham DY, Perez-Trallero E, Wadstrom T, Suerbaum S, Achtman M. An African origin for the intimate association between humans and Helicobacter pylori. Nature. 2007;445:915–918. doi: 10.1038/nature05562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh JT, Shaffer CL, Piazuelo MB, Bravo LE, McClain MS, Correa P, Cover TL. Analysis of cagA in Helicobacter pylori strains from Colombian populations with contrasting gastric cancer risk reveals a biomarker for disease severity. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2011;20:2237–2249. doi: 10.1158/1055-9965.EPI-11-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh JT, Torres VJ, Algood HM, McClain MS, Cover TL. Helicobacter pylori HopQ outer membrane protein attenuates bacterial adherence to gastric epithelial cells. FEMS microbiology letters. 2008;289:53–58. doi: 10.1111/j.1574-6968.2008.01368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcos NT, Magalhaes A, Ferreira B, Oliveira MJ, Carvalho AS, Mendes N, Gilmartin T, Head SR, Figueiredo C, David L, Santos-Silva F, Reis CA. Helicobacter pylori induces beta3GnT5 in human gastric cell lines, modulating expression of the SabA ligand sialyl-Lewis x. The Journal of clinical investigation. 2008;118:2325–2336. [Google Scholar]
- Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;1:1311–1315. doi: 10.1016/s0140-6736(84)91816-6. [DOI] [PubMed] [Google Scholar]
- Matsuo Y, Kido Y, Yamaoka Y. Helicobacter pylori Outer Membrane Protein-Related Pathogenesis. Toxins. 2017;9 doi: 10.3390/toxins9030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montano V, Didelot X, Foll M, Linz B, Reinhardt R, Suerbaum S, Moodley Y, Jensen JD. Worldwide Population Structure, Long-Term Demography, and Local Adaptation of Helicobacter pylori. Genetics. 2015;200:947–963. doi: 10.1534/genetics.115.176404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moodley Y, Linz B. Helicobacter pylori Sequences Reflect Past Human Migrations. Genome dynamics. 2009;6:62–74. doi: 10.1159/000235763. [DOI] [PubMed] [Google Scholar]
- Moodley Y, Linz B, Bond RP, Nieuwoudt M, Soodyall H, Schlebusch CM, Bernhoft S, Hale J, Suerbaum S, Mugisha L, van der Merwe SW, Achtman M. Age of the association between Helicobacter pylori and man. PLoS pathogens. 2012;8:e1002693. doi: 10.1371/journal.ppat.1002693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moodley Y, Linz B, Yamaoka Y, Windsor HM, Breurec S, Wu JY, Maady A, Bernhoft S, Thiberge JM, Phuanukoonnon S, Jobb G, Siba P, Graham DY, Marshall BJ, Achtman M. The peopling of the Pacific from a bacterial perspective. Science. 2009;323:527–530. doi: 10.1126/science.1166083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moonens K, Remaut H. Evolution and structural dynamics of bacterial glycan binding adhesins. Current opinion in structural biology. 2017;44:48–58. doi: 10.1016/j.sbi.2016.12.003. [DOI] [PubMed] [Google Scholar]
- Mucito-Varela E, Castillo-Rojas G, Cevallos MA, Lozano L, Merino E, Lopez-Leal G, Lopez-Vidal Y. Complete Genome Sequence of Helicobacter pylori Strain 29CaP Isolated from a Mexican Patient with Gastric Cancer. Genome announcements. 2016;4 doi: 10.1128/genomeA.01512-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller D, Tegtmeyer N, Brandt S, Yamaoka Y, De Poire E, Sgouras D, Wessler S, Torres J, Smolka A, Backert S. c-Src and c-Abl kinases control hierarchic phosphorylation and function of the CagA effector protein in Western and East Asian Helicobacter pylori strains. The Journal of clinical investigation. 2012;122:1553–1566. doi: 10.1172/JCI61143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Ramirez ZY, Mendez-Tenorio A, Kato I, Bravo MM, Rizzato C, Thorell K, Torres R, Aviles-Jimenez F, Camorlinga M, Canzian F, Torres J. Whole Genome Sequence and Phylogenetic Analysis Show Helicobacter pylori Strains from Latin America Have Followed a Unique Evolution Pathway. Frontiers in cellular and infection microbiology. 2017;7:50. doi: 10.3389/fcimb.2017.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima S, Kakugawa T, Yura H, Tomonaga M, Harada T, Hara A, Hara S, Nakano M, Yamasaki E, Sakamoto N, Ishimatsu Y, Isomoto H, Gochuico BR, Suffredini AF, Mukae H, Kurazono H, Hirayama T, Moss J, Kohno S. Identification of Helicobacter pylori VacA in human lung and its effects on lung cells. Biochemical and biophysical research communications. 2015;460:721–726. doi: 10.1016/j.bbrc.2015.03.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nell S, Eibach D, Montano V, Maady A, Nkwescheu A, Siri J, Elamin WF, Falush D, Linz B, Achtman M, Moodley Y, Suerbaum S. Recent acquisition of Helicobacter pylori by Baka pygmies. PLoS genetics. 2013;9:e1003775. doi: 10.1371/journal.pgen.1003775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiwara H, Sugimoto M, Ohno T, Vilaichone RK, Mahachai V, Graham DY, Yamaoka Y. Role of deletion located between the intermediate and middle regions of the Helicobacter pylori vacA gene in cases of gastroduodenal diseases. Journal of clinical microbiology. 2009;47:3493–3500. doi: 10.1128/JCM.00887-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno T, Sugimoto M, Nagashima A, Ogiwara H, Vilaichone RK, Mahachai V, Graham DY, Yamaoka Y. Relationship between Helicobacter pylori hopQ genotype and clinical outcome in Asian and Western populations. Journal of gastroenterology and hepatology. 2009;24:462–468. doi: 10.1111/j.1440-1746.2008.05762.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleastro M, Rocha R, Vale FF. Population genetic structure of Helicobacter pylori strains from Portuguese - speaking countries. Helicobacter. 2017 doi: 10.1111/hel.12382. [DOI] [PubMed] [Google Scholar]
- Qumar S, Majid M, Kumar N, Tiwari SK, Semmler T, Devi S, Baddam R, Hussain A, Shaik S, Ahmed N. Genome Dynamics and Molecular Infection Epidemiology of Multidrug-Resistant Helicobacter pullorum Isolates Obtained from Broiler and Free-Range Chickens in India. Applied and environmental microbiology. 2017;83 doi: 10.1128/AEM.02305-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Valera F, Martin-Cuadrado AB, Rodriguez-Brito B, Pasic L, Thingstad TF, Rohwer F, Mira A. Explaining microbial population genomics through phage predation. Nature reviews. Microbiology. 2009;7:828–836. doi: 10.1038/nrmicro2235. [DOI] [PubMed] [Google Scholar]
- Schindele F, Weiss E, Haas R, Fischer W. Quantitative analysis of CagA type IV secretion by Helicobacter pylori reveals substrate recognition and translocation requirements. Molecular microbiology. 2016;100:188–203. doi: 10.1111/mmi.13309. [DOI] [PubMed] [Google Scholar]
- Schmid EN, von Recklinghausen G, Ansorg R. Bacteriophages in Helicobacter (Campylobacter) pylori. Journal of medical microbiology. 1990;32:101–104. doi: 10.1099/00222615-32-2-101. [DOI] [PubMed] [Google Scholar]
- Schmidt TP, Goetz C, Huemer M, Schneider G, Wessler S. Calcium binding protects E-cadherin from cleavage by Helicobacter pylori HtrA. Gut pathogens. 2016a;8:29. doi: 10.1186/s13099-016-0112-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt TP, Perna AM, Fugmann T, Bohm M, Jan H, Haller S, Gotz C, Tegtmeyer N, Hoy B, Rau TT, Neri D, Backert S, Schneider G, Wessler S. Identification of E-cadherin signature motifs functioning as cleavage sites for Helicobacter pylori HtrA. Scientific reports. 2016b;6:23264. doi: 10.1038/srep23264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secka O, Vale FF, Buissonniere A, Thomas JE, Megraud F, Lehours P. Phylogeographic agreement between prophage and bacterial housekeeping genes in Helicobacter pylori strains from The Gambia. Helicobacter. 2017;22 doi: 10.1111/hel.12394. [DOI] [PubMed] [Google Scholar]
- Senkovich OA, Yin J, Ekshyyan V, Conant C, Traylor J, Adegboyega P, McGee DJ, Rhoads RE, Slepenkov S, Testerman TL. Helicobacter pylori AlpA and AlpB bind host laminin and influence gastric inflammation in gerbils. Infection and immunity. 2011;79:3106–3116. doi: 10.1128/IAI.01275-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YL, Huang HL, Huang BS, Chen PC, Chen CS, Wang HL, Lin PH, Chieh MS, Wu JJ, Yang JC, Chow LP. Combination of OipA, BabA, and SabA as candidate biomarkers for predicting Helicobacter pylori-related gastric cancer. Scientific reports. 2016;6:36442. doi: 10.1038/srep36442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subedi S, Moonens K, Romao E, Lo A, Vandenbussche G, Bugaytsova J, Muyldermans S, Boren T, Remaut H. Expression, purification and X-ray crystallographic analysis of the Helicobacter pylori blood group antigen-binding adhesin BabA. Acta crystallographica. Section F, Structural biology communications. 2014;70:1631–1635. doi: 10.1107/S2053230X14023188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suerbaum S, Achtman M. Evolution of Helicobacter pylori: the role of recombination. Trends in microbiology. 1999;7:182–184. doi: 10.1016/s0966-842x(99)01505-x. [DOI] [PubMed] [Google Scholar]
- Suerbaum S, Smith JM, Bapumia K, Morelli G, Smith NH, Kunstmann E, Dyrek I, Achtman M. Free recombination within Helicobacter pylori. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:12619–12624. doi: 10.1073/pnas.95.21.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki R, Shiota S, Yamaoka Y. Molecular epidemiology, population genetics, and pathogenic role of Helicobacter pylori. Infection, genetics and evolution : journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2012;12:203–213. doi: 10.1016/j.meegid.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney EG, Guillemin K. H. pylori's BabA Embraces Change. Cell host & microbe. 2016;19:5–7. doi: 10.1016/j.chom.2015.12.013. [DOI] [PubMed] [Google Scholar]
- Tabassam FH, Graham DY, Yamaoka Y. OipA plays a role in Helicobacter pylori-induced focal adhesion kinase activation and cytoskeletal re-organization. Cellular microbiology. 2008;10:1008–1020. doi: 10.1111/j.1462-5822.2007.01104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabassam FH, Graham DY, Yamaoka Y. Helicobacter pylori-associated regulation of forkhead transcription factors FoxO1/3a in human gastric cells. Helicobacter. 2012;17:193–202. doi: 10.1111/j.1523-5378.2012.00939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegtmeyer N, Moodley Y, Yamaoka Y, Pernitzsch SR, Schmidt V, Traverso FR, Schmidt TP, Rad R, Yeoh KG, Bow H, Torres J, Gerhard M, Schneider G, Wessler S, Backert S. Characterisation of worldwide Helicobacter pylori strains reveals genetic conservation and essentiality of serine protease HtrA. Molecular microbiology. 2016;99:925–944. doi: 10.1111/mmi.13276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegtmeyer N, Neddermann M, Asche CI, Backert S. Subversion of host kinases: A key network in cellular signaling hijacked by Helicobacter pylori CagA. Molecular microbiology. 2017a doi: 10.1111/mmi.13707. [DOI] [PubMed] [Google Scholar]
- Tegtmeyer N, Wessler S, Necchi V, Rohde M, Harrer A, Rau TT, Asche CI, Boehm M, Loessner H, Figueiredo C, Naumann M, Palmisano R, Solcia E, Ricci V, Backert S. Helicobacter pylori Employs a Unique Basolateral Type IV Secretion Mechanism for CagA Delivery. Cell host & microbe. 2017b;22:552–560. e555. doi: 10.1016/j.chom.2017.09.005. [DOI] [PubMed] [Google Scholar]
- Teymournejad O, Mobarez AM, Hassan ZM, Talebi Bezmin Abadi A. Binding of the Helicobacter pylori OipA causes apoptosis of host cells via modulation of Bax/Bcl-2 levels. Scientific reports. 2017;7:8036. doi: 10.1038/s41598-017-08176-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thi Huyen Trang T, Thanh Binh T, Yamaoka Y. Relationship between vacA Types and Development of Gastroduodenal Diseases. Toxins. 2016;8:182. doi: 10.3390/toxins8060182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorell K, Yahara K, Berthenet E, Lawson DJ, Mikhail J, Kato I, Mendez A, Rizzato C, Bravo MM, Suzuki R, Yamaoka Y, Torres J, Sheppard SK, Falush D. Rapid evolution of distinct Helicobacter pylori subpopulations in the Americas. PLoS genetics. 2017;13:e1006546. doi: 10.1371/journal.pgen.1006546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tummuru MK, Cover TL, Blaser MJ. Cloning and expression of a high-molecular-mass major antigen of Helicobacter pylori: evidence of linkage to cytotoxin production. Infection and immunity. 1993;61:1799–1809. doi: 10.1128/iai.61.5.1799-1809.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tummuru MK, Cover TL, Blaser MJ. Mutation of the cytotoxin-associated cagA gene does not affect the vacuolating cytotoxin activity of Helicobacter pylori. Infection and immunity. 1994;62:2609–2613. doi: 10.1128/iai.62.6.2609-2613.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama J, Takemura-Uchiyama I, Kato S, Takeuchi H, Sakaguchi Y, Ujihara T, Daibata M, Shimakura H, Okamoto N, Sakaguchi M, Matsuzaki S. Screening of KHP30-like prophages among Japanese Helicobacter pylori strains, and genetic analysis of a defective KHP30-like prophage sequence integrated in the genome of the H. pylori strain NY40. FEMS microbiology letters. 2016;363 doi: 10.1093/femsle/fnw157. [DOI] [PubMed] [Google Scholar]
- Vale FF, Nunes A, Oleastro M, Gomes JP, Sampaio DA, Rocha R, Vitor JM, Engstrand L, Pascoe B, Berthenet E, Sheppard SK, Hitchings MD, Megraud F, Vadivelu J, Lehours P. Genomic structure and insertion sites of Helicobacter pylori prophages from various geographical origins. Scientific reports. 2017;7:42471. doi: 10.1038/srep42471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale FF, Vadivelu J, Oleastro M, Breurec S, Engstrand L, Perets TT, Megraud F, Lehours P. Dormant phages of Helicobacter pylori reveal distinct populations in Europe. Scientific reports. 2015;5:14333. doi: 10.1038/srep14333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahara K, Furuta Y, Oshima K, Yoshida M, Azuma T, Hattori M, Uchiyama I, Kobayashi I. Chromosome painting in silico in a bacterial species reveals fine population structure. Molecular biology and evolution. 2013;30:1454–1464. doi: 10.1093/molbev/mst055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahiro K, Hirayama T, Moss J, Noda M. Helicobacter pylori VacA toxin causes cell death by inducing accumulation of cytoplasmic connexin 43. Cell death & disease. 2015;6:e1971. doi: 10.1038/cddis.2015.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaoka Y. Mechanisms of disease: Helicobacter pylori virulence factors. Nature reviews. Gastroenterology & hepatology. 2010;7:629–641. doi: 10.1038/nrgastro.2010.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaoka Y, Kwon DH, Graham DY. A M(r) 34,000 proinflammatory outer membrane protein (oipA) of Helicobacter pylori. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:7533–7538. doi: 10.1073/pnas.130079797. [DOI] [PMC free article] [PubMed] [Google Scholar]