Abstract
Sequence-specific recognition of peptides is of enormous importance to many chemical and biological applications, but has been difficult to achieve due to the minute differences in the side chains of amino acids. Acidic peptides are known to play important roles in cell growth and gene expression. In this work, we report molecularly imprinted micelles coded with molecular recognition information for the acidic and hydrophobic side chains of acidic peptides. The imprinted receptors could distinguish acidic amino acids from other polar and nonpolar amino acids, with dissociation constants of tens of nanomolar for biologically active peptides containing up to 18 amino acids.
Keywords: peptide binding, molecular recognition, receptor, molecular imprinting, micelle
Graphical Abstract
We describe a general method to construct synthetic receptors with high sequence specificity for the acidic and hydrophobic residues of peptides. The receptors could bind biological peptides with tens of nanomolar of binding affinity and were prepared straightforwardly through molecular imprinting in less than 2 days.
Introduction
Using 20 amino acids, nature constructs countless numbers of peptides and proteins with diverse structures and functions. These amino acids provide the functional “codes” that define the hydrophobic, hydrophilic, acidic, and basic side chains at specific positions along the peptide chain. The primary sequence is important to the conformational preference of the peptide and its secondary/tertiary structures under a given condition. Although not a direct determinant of the final function, the primary “code” of amino acids does contain all the information on the functional groups of a protein, including those involved in the binding, catalysis, or other molecular tasks the protein performs.
Chemists have long been interested in the molecular recognition of peptides.[1] In addition to their fundamental importance to supramolecular chemistry, these receptors have practical applications. With many peptides serving as neurotransmitters, neuromodulators, and hormones in biology,[2] synthetic receptors could be used to alter or inhibit these interactions. If effective strategies can be developed to recognize peptides in a sequence-selective manner, it might be possible to extend the method to recognize proteins by their characteristic amino acids on the surface.[3]
In principle, to prepare a strong and selective receptor for a peptide, all one needs to do is to create a “supramolecular code” on the receptor, complementary to the amino acid “code” of the peptide, in hydrophobicity, hydrogen bonds, and acidic/basic functionalities. The challenge, however, lies in the technicalities. If donor/acceptor motifs can be clearly identified and used in hydrogen-bond-based molecular recognition, doing so for a peptide chain with a complex combination of functionalities is a completely different matter. When the binding takes place in water, additional challenges exist because hydrogen bonds, arguably the best tool available for selective molecular recognition, are often ineffective in aqueous solution.[4]
In the last several decades, chemists have constructed peptide receptors using many different materials including macrocycles[1a, 1c] such as cyclodextrin[1b, 1d] and cucurbituril,[1h, 1j, 1l] amide oligomers[1a, 1f, 1g, 1i, 1k] and self-assembled nanocages.[1e] Nonetheless, most of these receptors can only bind particular types of peptides, usually fairly short ones. This is understandable. Because numerous binding interactions need to work together to achieve selective binding of a peptide even moderate in length, the difficulty in the design and synthesis of its receptor increases exponentially as the peptide gets longer, especially if the receptor is molecular in nature.
Molecular imprinting is a distinctively different concept for constructing guest-tailored receptors.[5] Instead of building discrete molecular structures and then fitting them with guests, researchers perform polymerization/cross-linking directly around the guests (i.e., template molecules). Removal of the templates leaves behind imprinted binding sites in the polymer matrix. The technique has advanced greatly in the last decades and found applications in separation, sensing, and enzymatic catalysis, including in peptide recognition.[6] In addition to traditional polymers and surfaces, imprinting could occur unimolecularly within discrete structures such as dendrimers.[7]
One of the most difficult challenges in peptide recognition is derived from the subtle structural difference of amino acids: leucine and isoleucine differ by the position of a single methyl group; aspartic acid and glutamic acid by the addition of one methylene. Such minute differences demand an extremely high level of precision in the molecular recognition. As a result, despite decades of research, a general method for sequence-specific binding of peptides remains elusive.[3, 8]
We recently reported our first step toward a general method for peptide recognition[9] via molecular imprinting within cross-linked micelles.[10] However, since these receptors rely heavily on hydrophobic interactions to operate, the method is expected to work poorly for peptides rich in hydrophilic amino acids, excluding a large number of biological peptides. Acidic peptides are known to play important roles in cell growth and gene expression.[11] Aspartic and glutamic acids are key to the aqueous solubility of many proteins and play important roles in the catalysis of numerous enzymes. In this work, we describe a method to encode the receptors with molecular recognition information for acidic groups on the side chain and at the C-terminus. The discovery expands the supramolecular code of our molecularly imprinted nanoparticles (MINPs) and significantly broadens the scope of peptides to be recognized. Given the importance of peptide recognition in many fundamental and applied research fields, we expect these easily synthesized, protein-sized, water-soluble receptors for peptides to be very useful in biology and chemistry.
Results and Discussion
Design and Synthesis
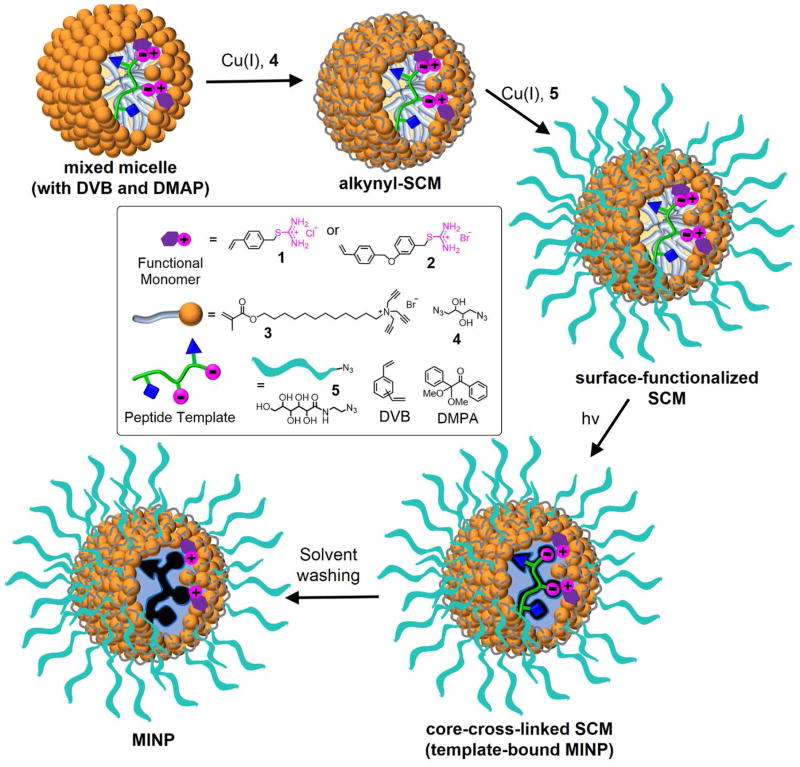
To recognize carboxylic acids on a peptide, we synthesized thiouronium derivatives 1 and 2 (Scheme 1) as the functional monomers (FMs) for the imprinting. Their design was based on the ion-pairing interaction between carboxylic acid and guanidinium or related cationic salts.[12] The two compounds differ in hydrophobicity but have the same binding group. Their vinyl group allows the compounds to be polymerized by radical polymerization. To be useful in peptide recognition, the thiouronium FM needs to be highly effective at binding carboxylic acid but not overwhelming in strength to cause nonspecific binding of acidic peptides. In addition, it needs to help the MINP differentiate the two acidic amino acids, which can be very challenging given their similarity.
Scheme 1.
Preparation of peptide-binding MINP with imbedded functional monomers (FMs).
As shown in Scheme 1, the preparation of the MINP started with solubilizing the peptide template within the micelle of cross-linkable surfactant 3. Click-cross-linking of the micelle on the surface using diazide 4 yielded surface-cross-linked micelle (SCM).[13] The SCM had multiple residual alkyne groups because of the 1:1.2 ratio between the tripropargylated cross-linkable surfactant and the diazide cross-linker. Ligand 5 was installed by another round of click reaction to cover the cross-linked micelle with a layer of hydrophilic groups, allowing us to recover the final MINPs by precipitation into acetone and washing with organic solvents.[10] Right in the beginning, the micelles contained DVB, DMPA (a photoinitiator), and functional monomer (FM) 1 or 2. UV irradiation initiated free radical polymerization/cross-linking of the micellar core, yielding the core-cross-linked SCM, with the template still bound in the binding site and the FMs covalently attached to the core. The template was removed by repeated solvent washing, yielding MINP with the vacant binding site. Usually, the entire preparation and purification were complete in <2 days once the starting materials were available.
We did not attempt to control the pH of the solution during MINP preparation because we did not want the micellization process to be complicated by buffer molecules. The relatively high pKa of S-alkylthiouronium (~9.8)[14] and low pKa of the acidic side chains of peptides (~4) suggest that the hydrogen bond-reinforced thiouronium–carboxylate salt bridge could dominate in quite broad a range of pH near neutral.[15] The notion was supported by our binding studies (vide infra).
Synthesis and characterization of the materials followed previously reported procedures[16] and the details are reported in the Supporting Information. Generally, the reaction progress was monitored by 1H NMR spectroscopy. Dynamic light scattering (DLS) afforded the size and molecular weight of the MINP. The nanoparticles were typically 4–5 nm in diameter, with an estimated M.W. of 50,000–56,000. The size was confirmed by transmission electron microscopy (Figure S1 in Supporting Information).
Optimization of the MINP preparation
Our first template was WDW. The tryptophan-aspartic acid-tryptophan tripeptide has two hydrophobic residues and one acidic one, making the total number of acids two including the C-terminal one. The fluorescent W allowed us to study the binding by both isothermal titration calorimetry (ITC) and fluorescence titration. Its hydrophobicity enabled us to study the interplay between hydrophobicity and the thiouronium–carboxylate salt bridge in the imprinting and binding of the MINP.
In general, ITC and fluorescence titrations showed excellent agreement (Table 1). ITC has the advantage of affording the number of binding sites per particle (N), in addition to other thermodynamic binding parameters such as the binding constant (Ka), enthalpy (ΔH), and entropy (ΔS).[17] However, since fluorescence titration was faster and easier to perform, we used it whenever it was possible (i.e., when the peptide contained fluorescent tryptophan). Because the binding affinity changed little (<10%) from water to 25 mM HEPES buffer (pH 7.4), with or without FMs (entries 2 and 7), we performed the majority of the bindings in water (we will come back to this point near the end of the paper).
Table 1.
Binding data for WDW by MINP(WDW).[a]
| entry | FM | surfactant | FM/template | Ka (× 105 M−1) | N | −ΔG (kcal/mol) | ΔH (kcal/mol) | TΔS (kcal/mol) |
|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 3 | 0 | 15.2 ± 1.2 (15.7 ± 1.4) | 0.80 ± 0.03 | 8.4 | −76.7 ± 3.3 | −68.3 |
| 2 | 1 | 3 | 0 | (17.4 ± 0.1)[b] | -- | 8.5 | -- | -- |
| 3 | 1 | 3 | 3:1 | 33.4 ± 1.5 (34.3 ± 1.8) | 1.08 ± 0.01 | 8.9 | −59.8 ± 0.4 | −50.9 |
| 4 | 2 | 3 | 1:1 | 28.8 ± 2.9 (28.8 ± 1.2) | 0.77 ± 0.01 | 8.8 | −56.5 ± 1.2 | −47.7 |
| 5 | 2 | 3 | 2:1 | 54.4 ± 5.6 (53.6 ± 4.9) | 0.80 ± 0.01 | 9.2 | −75.2 ± 1.1 | −66.0 |
| 6 | 2 | 3 | 3:1 | 82.7 ± 4.9 (80.8 ± 7.2) | 0.83 ± 0.02 | 9.4 | −119.8 ± 3.3 | −110.4 |
| 7 | 2 | 3 | 3:1 | (76.5 ± 0.5)[b] | -- | 9.4 | -- | -- |
| 8 | 2 | 3 | 4:1 | 57.9 ± 3.8 (58.7 ± 3.3) | 1.26 ± 0.01 | 9.2 | −101.6 ± 0.8 | −92.4 |
| 9 | 2 | 3 | 5:1 | 45.4 ± 2.3 (39.1 ± 1.0) | 1.15 ± 0.01 | 9.1 | −103.9 ± 0.8 | −94.8 |
| 10 | 2 | 6 | 0 | (10.1 ± 1.6) | -- | 8.2 | -- | -- |
| 11 | 2 | 6 | 3:1 | (76.7 ± 1.9) | -- | 9.4 | -- | -- |
| 12 | 2 | 7 | 0 | (14.3 ± 3.4) | -- | 8.4 | -- | -- |
| 13 | 2 | 7 | 3:1 | (63.3 ± 6.0) | -- | 9.3 | -- | -- |
The titrations were performed in Millipore water unless indicated otherwise. The Ka values in the parentheses were determined by fluorescence titration and those without the parentheses by ITC.
The binding was measured in 25 mM HEPES buffer (pH 7.4).
As shown in Table 1, MINP(WDW) prepared with the template but without any FM bound the peptide with Ka = 15.3 × 105 M−1 in water (entry 1). This binding constant reflects the effectiveness of “hydrophobic encoding” of the imprinted receptor, as polymerization/cross-linking around the indole side chain of tryptophan is expected to create complementary pockets in the nonpolar region of the cross-linked micelle.[9] Although the two carboxylates of WDW were expected to form salt bridges with the cross-linked ammonium surfactant,[10] we consider such electrostatic interactions as background in the presence of functional monomers such as 1 and 2, which are expected to form specific hydrogen bond-reinforced ion pairs.
The binding constant above was somewhat weaker than what was obtained for WGWG by MINP(WGWG) under similar conditions (Ka = 56.2 × 105 M−1).[9] Unless the last glycine (G) contributes significantly to the binding of WGWG, the weaker binding in WDW (by its own MINP) seems to suggest that the aspartic acid negatively impacts the imprinting/binding of the peptide in the absence of specific FMs. Ionic groups such as carboxylate have a strong tendency to stay on the surface of the micelle, to be solvated by water. Hydrophobic binding between the tryptophans and the MINP, on the other hand, is stronger when the binding pocket is relatively deep in the hydrophobic core [18]. Although we could not pin down the exact reason for the weakened binding of WDW (in the absence of FMs), the above two requirements do seem to contradict each other. (We will come back to this point again later in this paper.)
FM 1 and 2 both enhanced the binding, indicating that the hydrogen bond-reinforced thiouronium–carboxylate salt bridges were effective. At the same FM/template ratio (3:1), the more hydrophobic 2 strengthened the binding more than the less hydrophobic 1 (entries 3 and 6). We attributed the difference to their different incorporation into the micelle: being cationic, both 1 and 2 are repelled by the cationic micelle. The more hydrophobic 2 is anticipated to have a stronger tendency to stay in the hydrophobic microenvironment of the micelle, whereas 1 may migrate into the aqueous phase due to its water solubility. For the imprinting to work, the FM clearly needs to stay with the template within the micelle during polymerization.
Our data also show that the optimal FM/template ratio was 3:1 for FM 2 (Table 1, entries 4–9). Since WDW has two carboxylates in the structure, the ratio suggests a 50% excess of the thiouronium was needed for the best result. As will be shown by additional studies, this ratio was fairly constant over different templates (vide infra).
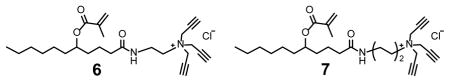
We recently reported amide-containing cross-linkable surfactants 6 and 7.[18] The hydrogen-bonding capabilities of the amide were found to enhance the imprinting and binding of hydrogen bond-containing templates significantly, including oligosaccharides.[16f] However, with or without the thiouronium FM, surfactant 6 or 7 afforded MINPs with weaker binding than those prepared with 1 (entries 10–13). Thus, even if hydrogen bonds could be formed between the cross-linked surfactants and the peptide template, they brought no net benefit.
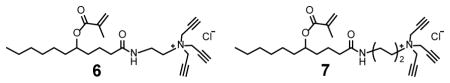
The behavior of the amide-containing MINPs might be puzzling, given that they enhanced the imprinting and binding of many templates with hydrogen-bonding functionalities.[16f, 18] However, our previous work also showed that location of the hydrogen-bonding group in the template is highly important to the performance of such MINPs [18]. The templates that benefit most from the amide generally need to have their hydrophobic group penetrate through the amide layer, reaching the hydrophobic core of the micelle in order to maximize the hydrophobic interactions. In addition to many hydrogen-bonding groups in the main chain, a peptide has ionic ammonium/carboxylate groups. Their solvation demands the peptide to stay near the surface of the micelle during imprinting and binding. Quite likely, such a configuration would not allow the relatively short hydrophobic side chains (even that of tryptophan) to penetrate deep into the hydrophobic core when an amide layer exists in the MINP.
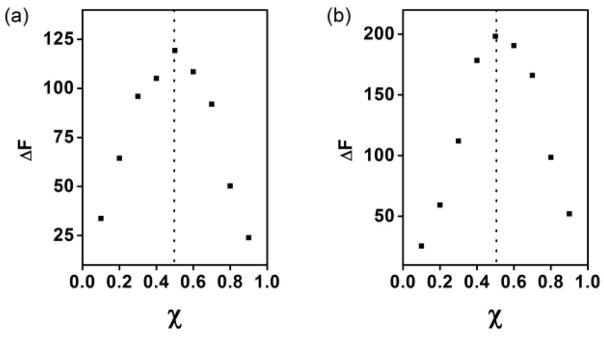
One of the most important properties of the MINP is its controllable number of binding sites.[10] Usually, we keep the surfactant/template ratio to 50:1. With each MINP containing ~50 (cross-linked) surfactants, the ratio gives one binding site per nanoparticle on average. The number was confirmed by the ITC studies, as shown in Table 1. The 1:1 binding stoichiometry was also evident from the Job plots, for the MINP prepared with or without FM 2 (Figure 1). Our previous work shows that, by changing the surfactant/template ratio, the number of binding sites can be easily controlled.[10]
Figure 1.
Job’s plots for WDW with (a) MINP(WDW) prepared (a) without FM (a) and with 3 equiv FM 2 (b). The total concentration of MINP and the guest was 10 μM. χ = [Host]/{[Host] + [Guest]}.
Effects of hydrophobicity and the number of salt bridges
The two tryptophans afforded significant hydrophobic driving force to the binding (Table 1, entry 1).[19] FM 2 was able to increase the binding constant by 5 times (compare entries 1 and 6). This is good news to us but how about other peptides? Is the salt bridge equally effective for peptides with different hydrophobicity?
Table 2 addresses these questions. All the peptides have two carboxylic acids, one from the aspartic acid side chain and the other from the carboxy terminal. The number of tryptophan increased from 0 to 1 to 2 from GDG to GDW to WDW. Note that the positions of the carboxylic acids stay the same within the series.
Table 2.
Effects of peptide hydrophobicity on imprinting and binding using FM 2.[a]
| entry | template | FM/template | Ka (× 105 M−1) | Krel | N | −ΔG (kcal/mol) | ΔH (kcal/mol) | TΔS (kcal/mol) |
|---|---|---|---|---|---|---|---|---|
| 1 | GDG | 0:1 | 0.53 ± 0.1 | 0.69 ± 0.36 | 6.4 | −42.2 ± 5.4 | −35.8 | |
| 2 | GDG | 3:1 | 4.6 ± 0.3 | 8.7 | 1.03 ± 0.02 | 7.7 | −20.1 ± 0.9 | −12.4 |
| 3 | GDW | 0:1 | 6.4 ± 0.7 | 0.83 ± 0.03 | 7.9 | −96.5 ± 5.0 | −88.6 | |
| 4 | GDW | 3:1 | 19.8 ± 1.0 | 3.1 | 1.02 ± 0.06 | 8.6 | −88.6 ± 2.0 | −80.0 |
| 5 | WDW | 0:1 | 15.2 ± 1.2 | 0.80 ± 0.03 | 8.4 | −76.7 ± 3.3 | −68.3 | |
| 6 | WDW | 3:1 | 82.7 ± 4.9 | 5.4 | 0.83 ± 0.02 | 9.4 | −119.8 ± 3.3 | −110.4 |
The Ka values were determined by fluorescence titrations in Millipore water. Krel is the binding constant of the MINP with the thiouronium FM relative to that without.
We were pleased to see that even the most hydrophilic peptide (GDG) showed significant binding toward its MINP, with Ka = 0.53 × 105 M−1 in water (entry 1). Since this peptide does not have any significant hydrophobicity, the 6.4 kcal/mol binding free energy (−ΔG) is quite impressive. The “background” electrostatic interactions between the oppositely charged MINP and template, the hydrogen bonding interactions formed between the peptide and the MINP (e.g., with triazole on the cross-linked surfactant or hydroxyl groups on the cross-linker or surface ligand), and any desolvation during the binding probably all contributed to the binding.
The addition of the first tryptophan increased the binding by 1.5 kcal/mol (entry 3) and the second addition by 0.5 kcal/mol (entry 5). Note that these numbers represent the binding between each peptide and its own MINP, not different peptides to the same receptor. The energy difference, thus, reflects the effectiveness of the imprinting, as well as that of the binding.
Importantly, in all three peptides, the thiouronium FM clearly enhanced the binding, shown by the Krel value, i.e., the binding constant of the MINP with the thiouronium FM relative to that without. Meanwhile, as the number of hydrophobic residues increased, the enthalpic driving force became stronger but were offset by an increasingly larger (unfavorable) entropic term (Table 2). Thus, all the bindings were enthalpically driven, including those shown in Table 1.
We then studied the effect of the (hydrogen bond-reinforced) salt bridge on the imprinting and binding (Table 3). In this series, we kept the number (= 2) and the position of the tryptophan the same and increased the number of aspartic acid from 1 to 3. As a result, the salt bridges that could be potentially formed between the peptide and its MINP increased from 2 to 4.
Table 3.
Effects of the number of salt bridges on imprinting and binding using FM 2.[a]
| Entry | template | FM/template | Ka (×105 M−1) | Krel | −ΔG (kcal/mol |
|---|---|---|---|---|---|
| 1 | WDW | 0:1 | 15.7 ± 1.4 | 8.45 | |
| 2 | WDW | 3:1 | 80.8 ± 7.2 | 5.1 | 9.42 |
| 3 | WDWD | 0:1 | 11.4 ± 0.8 | 8.26 | |
| 4 | WDWD | 3:1 | 78.8 ± 12.7 | 6.9 | 9.41 |
| 5 | WDWD | 4.5:1 | 116.5 ± 17.2 | 10.2 | 9.64 |
| 6 | WDWD | 6:1 | 94.7 ± 8.1 | 8.3 | 9.52 |
| 7 | WDWDD | 0:1 | 9.3 ± 0.8 | 8.14 | |
| 8 | WDWDD | 6:1 | 134.8 ± 10.5 | 14.5 | 9.73 |
The Ka values were determined by fluorescence titrations in Millipore water. Krel is the binding constant of the MINP with the thiouronium FM relative to that without.
Our data indicate that, as the number of carboxylate increases, the binding became weaker for the MINP prepared without FM (Table 3, entries 1, 3, and 7). Although the effect was moderate, the trend corroborated with our earlier results to support that, without proper binding groups for the carboxylates, their strong solvation by water makes the peptide stay closer to the surface of the micelle and interferes with the hydrophobic imprinting that requires the hydrophobic residues to move deeper into the micellar core.
Encouragingly, Krel increased steadily, from 5.1 for WDW to 10.2 for WDWD, and to 14.5 for WDWDD at the same FM/carboxylate ratio (1.5:1). As shown by entries 4–6, the 50% excess FM per carboxylate was optimal for the imprinting of WDWD, the same as found before.
The results so far clearly demonstrated the effectiveness of the thiouronium–carboxylate salt bridge in the supramolecular coding of the MINP receptors: they worked for both hydrophilic peptides and hydrophobic ones, and continued to enhance the binding as the number of salt bridges increased.
In Table 4, we varied both the hydrophobicity and the number of salt bridges. In these examples, we kept the FM/carboxylate ratio at 1.5:1, the optimal number confirmed by two studies shown above.
Table 4.
Interplay of hydrophobicity and salt bridges.[a]
| Entry | template | FM | # of salt bridge[b] | Ka (×105 M−1) | Krel | −ΔG (kcal/mol) |
|---|---|---|---|---|---|---|
| 1 | WNW | none | 0 | 35.7 ± 5.1 | 8.94 | |
| 2 | WNW | 2 | 1 | 59.1 ± 5.9 | 1.7 | 9.24 |
| 3 | WWW | none | 0 | 123.5 ± 9.9 | 9.67 | |
| 4 | WWW | 2 | 1 | 159.6 ± 6.5 | 1.3 | 9.83 |
| 5 | WDW | none | 0 | 15.7 ± 1.4 | 8.45 | |
| 6 | WDW | 2 | 2 | 80.8 ± 7.2 | 5.1 | 9.42 |
| 7 | WEW | none | 0 | 16.2 ± 1.5 | 8.47 | |
| 8 | WEW | 2 | 2 | 92.7 ± 9.2 | 5.7 | 9.50 |
The Ka values were determined by fluorescence titrations in Millipore water. When FM 2 was used, the FM/carboxylate ratio was kept 1.5:1 in all cases. Krel is the binding constant of the MINP with the thiouronium FM relative to that without.
The salt bridge refers to the hydrogen bond-reinforced thiouronium–carboxylate salt bridge.
For the two very hydrophobic peptides (WNW and WWW), the hydrophobic imprinting apparently was highly efficient, affording nearly 9 kcal/mol in binding free energy without any FMs. With one thiouronium–carboxylate salt bridge (at the carboxy terminal), the enhancement (Krel) was expectedly low, only 1.3–1.7 (Table 4, entries 2 and 4). The data in entries 5 and 6 were the same as in Tables 1–3, and are listed here for comparison purposes. As shown by entries 7 and 8, the thiouronium FM also worked well for peptides containing glutamic acid (E), enhancing the binding constant by a similar degree as for aspartic acid (D).
Binding selectivity of small peptides
Molecular imprinting has the benefit of supramolecularly encoding the receptor with molecular recognition information for the entire peptide template, main chain and side chains at the same time. Since the hydrophobic residues on a peptide give us excellent sequence-selectivity according,[9] we focused on several closely related amino acids in this study, knowing that the hydrophobic residues will continue to impart selectivity to the MINP receptor.
Figure 2 compares the binding affinity of different peptides toward the MINP prepared from WDW, normalized to the binding affinity of the template itself. We included data for MINP(WDW) prepared with and without FM 2. In the absence of FM, replacing one of the tryptophans with glycine (in GDW) or removing it (in DW) weakened the binding significantly. Switching the position(s) of the tryptophan (in DWW or WWD), as well as adding a tryptophan (in WWW) weakened the binding even more. All of these results are fully consistent with the rules found in the hydrophobic imprinting.[9] Thus, although hydrophobic effect is nonspecific by itself, imprinted hydrophobic binding pockets with specific shape and size can be highly discriminating in their binding.
Figure 2.
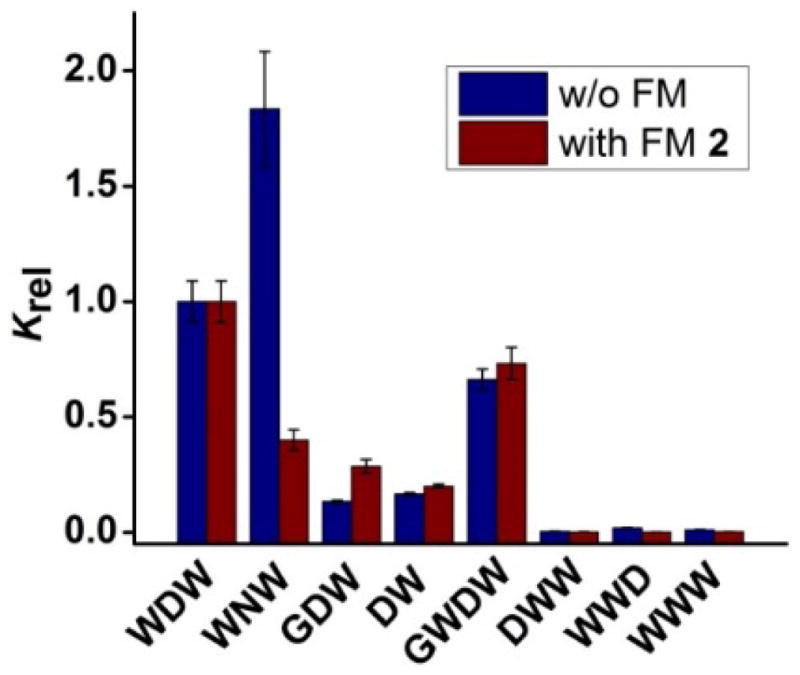
Binding selectivity of MINP(WDW) prepared without (blue bars) and with FM 2 (red bars). The binding data are reported in Table S1 in the Supporting Information. Krel is the binding affinity of different peptides toward the MINP prepared from WDW, normalized to the binding affinity of the template itself.
For the binding of MINP without FM, the most unusual result was that WNW showed stronger binding for MINP(WDW) than the templating peptide WDW (Figure 2). This would be normally interpreted as failure in molecular imprinting. However, asparagine (N) and aspartic acid (D) differ subtly: the two have the same number of carbons and both contain a carbonyl group, except one (carbonyl) from a primary amide and the other form a carboxylic acid. If we consider our earlier evidence for the interference of hydrophobic binding by the ionic carboxylates, the stronger binding of WNW may not seem as strange. Essentially, the imprinting and binding of WDW have to deal with the conflictory requirements of the ionic and hydrophobic side chains for optimal interactions, as discussed earlier. Once the aspartic acid is replaced with asparagine, the dilemma no longer exists. Being extremely similar in structure to the template (i.e., WDW), WNW should be able to occupy the binding site created after WDW. Not only so, its tryptophans can now optimize the hydrophobic interactions with the binding pockets, without the constraint set by the aspartic acid ionic side chain—a feature beneficial to the binding.
When MINP(WDW) was encoded with the carboxylate-binding thiouronium groups, the binding affinity improved as shown by our earlier data, as well as Table S1. Importantly, the binding selectivity was maintained in most cases (Figure 2). Most notably, WNW, which showed the unusually stronger binding than the template itself toward the MINP prepared without FM, now behaved normally by displaying a weaker binding than the templating peptide. Thus, once the aspartic acid side chain was made to participate in the specific carboxylate–thiouronium salt bridge, the residue becomes an important contributor to the binding selectivity (as well as to binding affinity).
Table 5 shows the (absolute) binding constants of MINP(WDW) and MINP(WEW) for peptide WDW and WEW. The comparison shows the binding selectivity of the MINPs for the two subtly different acidic amino acids. In all cases, the MINP always preferred its own template over the minutely different competitor, indicative of effective molecular imprinting. Encouragingly, both the MINPs prepared with and without FM 2 displayed moderate selectively: with the cross-reactivity (i.e., Krel) ranging from 0.54 to 0.72 for the non-matching peptide. Although the selectivity might have improved slightly with MINP(WDW) functionalized with the thiouronium binding group (entry 6), the improvement was rather limited. The binding studies also demonstrated that the thiouronium FM strengthened the binding, without comprising the binding selectivity.
Table 5.
Differentiation of aspartic acid (D) and glutamic acid (E).[a]
| Entry | template | Guest | FM | Ka (×105 M−1) | Krel |
|---|---|---|---|---|---|
| 1 | WDW | WDW | none | 15.7 ± 1.4 | |
| 2 | WDW | WEW | none | 10.7 ± 0.7 | 0.68 |
| 3 | WEW | WEW | none | 16.2 ± 1.5 | |
| 4 | WEW | WDW | none | 11.7 ± 1.2 | 0.72 |
| 5 | WDW | WDW | 2 | 80.8 ± 7.2 | |
| 6 | WDW | WEW | 2 | 44.0 ± 4.2 | 0.54 |
| 7 | WEW | WEW | 2 | 92.7 ± 9.2 | |
| 8 | WEW | WDW | 2 | 66.1 ± 5.1 | 0.71 |
The Ka values were determined by fluorescence titrations in Millipore water. Krel is the binding constant of the MINP with the thiouronium FM relative to that without. The FM/carboxylate ratio was kept 1.5:1 in all cases.
Imprinting and binding of biological peptides
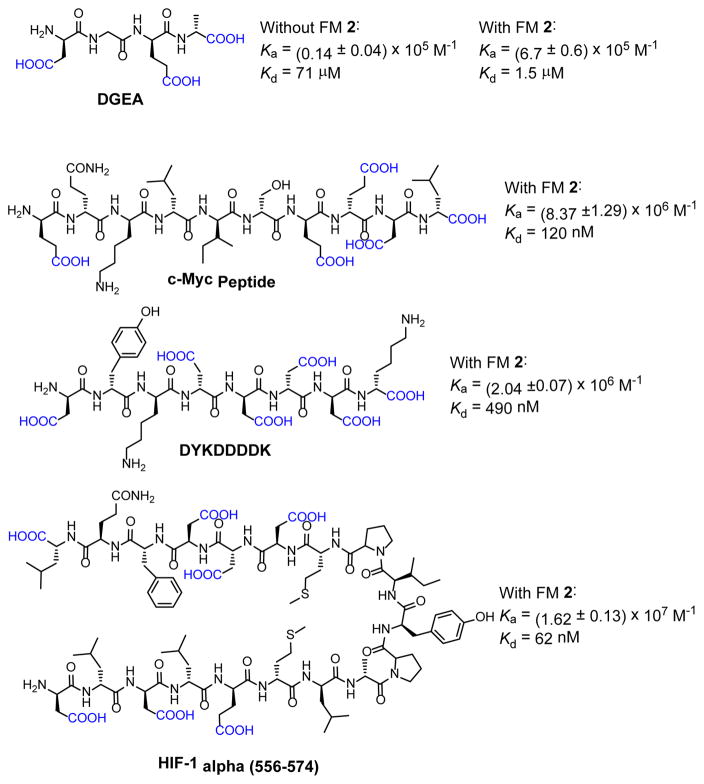
Having confirmed that thiouronium FM 2 was able to effectively encode the MINP receptor for aspartic acid and glutamic acid, we moved on to study a number of biological peptides bearing acidic side chains (Figure 3). The 1:1.5 carboxylate/FM 2 was maintained in all cases.
Figure 3.
Structures of biological peptides studied and the binding constants (Ka) and dissociation constants (Kd) obtained.
For the simplest DGEA, a tetrapeptide in collagen critical to the binding of the alpha 2 beta 1 integrin receptor,[20] FM2 enhanced the binding impressively, by nearly 50 times. As we have observed previously for the hydrophobic peptides, the benefit of the molecular imprinting is that, the longer the peptide, the more hydrophobic and now also acidic residues would contribute to the binding and the stronger the binding will be. Impressively, for HIFF-1 alpha, a peptide made of 18 amino acids and the longest we have imprinted so far, a dissociation constant of 62 nM was obtained.
pH effect on the binding of biological peptides
We performed the majority of the binding studies in Millipore water because our initial studies showed that the binding of WDW by its MINP showed little change (<10%) from water to HEPES buffer (pH 7.4), with or without FMs (Table 1, entries 2 and 7). Negligible pH effects in MINP binding has also been observed previously for other peptides (e.g., WWGG).[9] Nonetheless, the biological acidic peptides contain many ionizable groups. It is important to understand how the MINP receptors behave under different pH conditions.
Our model peptide was DYKDDDDK, the FLAG-tag frequently used for protein purification and for studying proteins in living cells.[21] Its abundant acidic and basic groups suggest that its binding could be quite sensitive to solution pHs. As shown in Table 6, its binding constant was 20.4 × 105 M−1 in water, without any pH adjustment. The binding constant dropped approximately by half in 10 mM HEPES buffer. At first appearance, there seemed to be a quite significant pH effect. However, when the pH was changed to 6.0 and 9.0 using two other buffers, the binding constant stayed nearly the same, suggesting otherwise. Suspecting that the sulfonate and carboxylate groups of the these buffer molecules might have competed with the carboxylate groups of the peptide for the thiouronium binding groups, we performed the binding in Tris-HCl buffer at both pH 7.4 and 9.0. Indeed, in the absence of competing binding functionalities, the binding became stronger, similar to that in water.
Thus, the binding of DYKDDDDK showed little dependence on solution pH (6–9), despite its many ionizable groups (Table 6). This feature makes the binding more predictable in different environments and could be quite useful in biological applications of the MINPs. Cationic micelles are known to be slightly basic on their surface due to their electrical potential, even when the bulk solution is neutral.[22] MINP, being a polycation, should do the same.[23] Since the acidic side chains of aspartic acid and glutamic acids have a pKa of ~4, they should be readily deprotonated near the surface of the MINP during the binding. This could be the possible reason for the lack of pH effect at 6–9 for the MINP binding.
Table 6.
Effects of pH and buffer on the binding of DYKDDDDK.[a]
| Entry | buffer | pH | Ka (×105 M−1) |
|---|---|---|---|
| 1 | none | unadjusted | 20.4 ± 0.7 |
| 2 | 10 mM HEPES | 7.4 | 11.7 ± 0.6 |
| 3 | 10 mM MES | 6.0 | 11.2 ± 0.5 |
| 4 | 10 mM Bicine | 9.0 | 12.6 ± 1.0 |
| 5 | 10 mM Tris-HCl | 7.4 | 25.9 ± 2.0 |
| 6 | 10 mM Tris-HCl | 9.0 | 24.2 ± 3.1 |
The Ka values were determined by ITC. The FM/carboxylate ratio was kept 1.5:1 in all cases.
Conclusions
Many imprinted polymers have been used for peptide recognition, ranging from materials useful for peptide extraction and separation to lightly cross-linked nanogels with improved biocompatibility.[6] In comparison to these materials, our cross-linked micelles generate protein-sized water-soluble nanoparticles with a controllable number of binding sites. This work has expanded the “supramolecular codes” of our imprinted micellar receptors significantly. Thiouronium FM 2 turned out highly effective in the molecular imprinting of peptides containing acidic side chains. In the presence of the FM (typically used at 1.5 equiv to the carboxylic acids), both hydrophobic residues and the acidic side chains contribute strongly to the binding of the imprinted receptors. The enhancement of the binding by the thiouronium depends on the hydrophobicity of the peptides, and generally becomes more significant when the peptides become less hydrophobic. This feature is highly desirable because we already have a very effective imprinting for hydrophobic peptides whose hydrophobic groups alone could afford enormous driving forces to the binding.
These imprinted receptors in general have excellent selectivities in peptide recognition. In this work, the thiouronium FM 2 further improved the selectivity, particularly those that cannot be distinguished by the hydrophobic imprinting alone (i.e., aspartic acid and asparagine). A nearly two-fold selectivity between aspartic and glutamic acids in the tripeptides was impressive, given their extreme similarities. Importantly, for an imprinted receptor, all residues and the peptide backbone contribute to the binding, including glycine. Thus, even relatively small single-residue-selectivity would be magnified as the chain gets longer.
Several lines of evidence support that, during the molecular imprinting of the peptides without the thiouronium FM, the ionic side chains could interfere with the imprinting of the hydrophobic residues, as the carboxylate groups and the hydrophobic side chains prefer different locations in the micelle. The result is somewhat weaker binding for peptides containing acidic side chains. In the presence of FM 2, when specific hydrogen bond-reinforced carboxylate–thiouronium salt bridges exist, the acidic side chains strengthened the binding while improving the overall binding selectivity. One clear example is shown in Table 4: WDW, for example, was bound more strongly by its MINP than WNW (by its own) in the presence of FM 2; in the absence of the FM, the opposite trend was observed.
Supplementary Material
Acknowledgments
We thank the National Institute of General Medical Sciences of the National Institutes of Health (R01GM113883) for financial support of the research.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supporting information for this article is given via a link at the end of the document.
References
- 1.a) Hong JI, Namgoong SK, Bernardi A, Still WC. J Am Chem Soc. 1991;113:5111–5112. [Google Scholar]; b) Breslow R, Yang Z, Ching R, Trojandt G, Odobel F. J Am Chem Soc. 1998;120:3536–3537. [Google Scholar]; c) Hossain MA, Schneider HJ. J Am Chem Soc. 1998;120:11208–11209. [Google Scholar]; d) Yamamura H, Rekharsky MV, Ishihara Y, Kawai M, Inoue Y. J Am Chem Soc. 2004;126:14224–14233. doi: 10.1021/ja046612r. [DOI] [PubMed] [Google Scholar]; e) Tashiro S, Tominaga M, Kawano M, Therrien B, Ozeki T, Fujita M. J Am Chem Soc. 2005;127:4546–4547. doi: 10.1021/ja044782y. [DOI] [PubMed] [Google Scholar]; f) Wright AT, Anslyn EV, McDevitt JT. J Am Chem Soc. 2005;127:17405–17411. doi: 10.1021/ja055696g. [DOI] [PubMed] [Google Scholar]; g) Schmuck C, Wich P. Angew Chem Int Ed. 2006;45:4277–4281. doi: 10.1002/anie.200601046. [DOI] [PubMed] [Google Scholar]; h) Reczek JJ, Kennedy AA, Halbert BT, Urbach AR. J Am Chem Soc. 2009;131:2408–2415. doi: 10.1021/ja808936y. [DOI] [PubMed] [Google Scholar]; i) Niebling S, Kuchelmeister HY, Schmuck C, Schlucker S. Chem Sci. 2012;3:3371–3377. [Google Scholar]; j) Smith LC, Leach DG, Blaylock BE, Ali OA, Urbach AR. J Am Chem Soc. 2015;137:3663–3669. doi: 10.1021/jacs.5b00718. [DOI] [PubMed] [Google Scholar]; k) Faggi E, Vicent C, Luis SV, Alfonso I. Org Biomol Chem. 2015;13:11721–11731. doi: 10.1039/c5ob01889g. [DOI] [PubMed] [Google Scholar]; l) Sonzini S, Marcozzi A, Gubeli RJ, van der Walle CF, Ravn P, Herrmann A, Scherman OA. Angew Chem Int Ed. 2016;55:14000–14004. doi: 10.1002/anie.201606763. [DOI] [PubMed] [Google Scholar]
- 2.Sewald N, Jakubke H-D. Peptides: chemistry and biology. Wiley-VCH; Weinheim: 2002. [Google Scholar]
- 3.Peczuh MW, Hamilton AD. Chem Rev. 2000;100:2479–2494. doi: 10.1021/cr9900026. [DOI] [PubMed] [Google Scholar]
- 4.a) Oshovsky GV, Reinhoudt DN, Verboom W. Angew Chem Int Ed. 2007;46:2366–2393. doi: 10.1002/anie.200602815. [DOI] [PubMed] [Google Scholar]; b) Kataev EA, Müller C. Tetrahedron. 2014;70:137–167. [Google Scholar]
- 5.a) Wulff G. Angew Chem Int Ed Engl. 1995;34:1812–1832. [Google Scholar]; b) Wulff G. Chem Rev. 2001;102:1–28. doi: 10.1021/cr980039a. [DOI] [PubMed] [Google Scholar]; c) Haupt K, Mosbach K. Chem Rev. 2000;100:2495–2504. doi: 10.1021/cr990099w. [DOI] [PubMed] [Google Scholar]; d) Ye L, Mosbach K. Chem Mater. 2008;20:859–868. [Google Scholar]; e) Shea KJ. Trends Polym Sci. 1994;2:166–173. [Google Scholar]; f) Sellergren B. Techniques and instrumentation in analytical chemistry. Vol. 23. Elsevier; Amsterdam: 2001. [Google Scholar]; g) Komiyama M. Molecular imprinting: from fundamentals to applications. Wiley-VCH; Weinheim: 2003. [Google Scholar]; h) Yan M, Ramström O. Molecularly imprinted materials: science and technology. Marcel Dekker; New York: 2005. [Google Scholar]; i) Alexander C, Andersson HS, Andersson LI, Ansell RJ, Kirsch N, Nicholls IA, O’Mahony J, Whitcombe MJ. J Mol Recognit. 2006;19:106–180. doi: 10.1002/jmr.760. [DOI] [PubMed] [Google Scholar]; j) Sellergren B, Hall AJ. In: Supramolecular Chemistry: From Molecules to Nanomaterials. Steed JW, Gale PA, editors. Wiley; 2012. Online. [Google Scholar]; k) Haupt K. Topics in current chemistry. Springer; Heidelberg ; New York: 2012. [Google Scholar]
- 6.a) Klein JU, Whitcombe MJ, Mulholland F, Vulfson EN. Angew Chem Int Ed. 1999;38:2057–2060. doi: 10.1002/(SICI)1521-3773(19990712)38:13/14<2057::AID-ANIE2057>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]; b) Nishino H, Huang CS, Shea KJ. Angew Chem Int Ed. 2006;45:2392–2396. doi: 10.1002/anie.200503760. [DOI] [PubMed] [Google Scholar]; c) Hoshino Y, Kodama T, Okahata Y, Shea KJ. J Am Chem Soc. 2008;130:15242–15243. doi: 10.1021/ja8062875. [DOI] [PubMed] [Google Scholar]; d) Hoshino Y, Koide H, Urakami T, Kanazawa H, Kodama T, Oku N, Shea KJ. J Am Chem Soc. 2010;132:6644–6645. doi: 10.1021/ja102148f. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Urraca JL, Aureliano CSA, Schillinger E, Esselmann H, Wiltfang J, Sellergren B. J Am Chem Soc. 2011;133:9220–9223. doi: 10.1021/ja202908z. [DOI] [PubMed] [Google Scholar]; f) Banerjee S, König B. J Am Chem Soc. 2013;135:2967–2970. doi: 10.1021/ja4001568. [DOI] [PubMed] [Google Scholar]; g) Qader AA, Urraca J, Torsetnes SB, Tønnesen F, Reubsaet L, Sellergren B. J Chromatogr A. 2014;1370:56–62. doi: 10.1016/j.chroma.2014.10.023. [DOI] [PubMed] [Google Scholar]; h) Zhang Y, Deng C, Liu S, Wu J, Chen Z, Li C, Lu W. Angew Chem Int Ed. 2015;54:5157–5160. doi: 10.1002/anie.201412114. [DOI] [PubMed] [Google Scholar]; i) Schwark S, Sun W, Stute J, Lutkemeyer D, Ulbricht M, Sellergren B. RSC Adv. 2016;6:53162–53169. [Google Scholar]
- 7.a) Zimmerman SC, Wendland MS, Rakow NA, Zharov I, Suslick KS. Nature. 2002;418:399–403. doi: 10.1038/nature00877. [DOI] [PubMed] [Google Scholar]; b) Zimmerman SC, Zharov I, Wendland MS, Rakow NA, Suslick KS. J Am Chem Soc. 2003;125:13504–13518. doi: 10.1021/ja0357240. [DOI] [PubMed] [Google Scholar]
- 8.Maity D, Schmuck C. Synthetic Receptors for Biomolecules: Design Principles and Applications. The Royal Society of Chemistry; 2015. pp. 326–368. [Google Scholar]
- 9.Awino JK, Gunasekara RW, Zhao Y. J Am Chem Soc. 2017;139:2188–2191. doi: 10.1021/jacs.6b12949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Awino JK, Zhao Y. J Am Chem Soc. 2013;135:12552–12555. doi: 10.1021/ja406089c. [DOI] [PubMed] [Google Scholar]
- 11.a) Mancinelli L, Chillemi F, Cardellini E, Marsili V, Giavarini F, De Angelis L, Lugaro G, Gianfranceschi GL. Biol Chem. 1999;380:31–40. doi: 10.1515/BC.1999.004. [DOI] [PubMed] [Google Scholar]; b) Castigli E, Mancinelli L, Mariggio MA, Gianfranceschi GL. Am J Physiol, Cell Physiol. 1993;265:C1220–C1223. doi: 10.1152/ajpcell.1993.265.5.C1220. [DOI] [PubMed] [Google Scholar]; c) Huff T, Müller CSG, Otto AM, Netzker R, Hannappel E. Int J Biochem Cell Biol. 2001;33:205–220. doi: 10.1016/s1357-2725(00)00087-x. [DOI] [PubMed] [Google Scholar]
- 12.a) Ariga K, Kunitake T. Acc Chem Res. 1998;31:371–378. [Google Scholar]; b) Hannon CL, Anslyn EV. In: Bioorganic Chemistry Frontiers. Dugas H, Schmidtchen FP, editors. Springer; Heidelberg: 1993. pp. 193–255. [Google Scholar]; c) Orner BP, Hamilton AD. J Inclusion Phenom Macrocyc Chem. 2001;41:141–147. [Google Scholar]; d) Schmidtchen FP, Berger M. Chem Rev. 1997;97:1609–1646. doi: 10.1021/cr9603845. [DOI] [PubMed] [Google Scholar]
- 13.a) Zhang S, Zhao Y. Macromolecules. 2010;43:4020–4022. [Google Scholar]; b) Zhao Y. Langmuir. 2016;32:5703–5713. doi: 10.1021/acs.langmuir.6b01162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Albert A, Goldacre R, Phillips J. J Chem Soc. 1948:2240–2249. [Google Scholar]
- 15.Although S-alkylthiouronium salts can hydrolyze in water, the hydrolysis usually requires a strongly basic solution (e.g., 2.5 M NaOH) at refluxing temperatures. For details, see: Urquhart GG, Gates JW, Connor R. Org Syn. 1941;21:36–38.
- 16.a) Awino JK, Zhao Y. Chem Commun. 2014;50:5752–5755. doi: 10.1039/c4cc01516a. [DOI] [PubMed] [Google Scholar]; b) Awino JK, Zhao Y. Chem-Eur J. 2015;21:655–661. doi: 10.1002/chem.201404919. [DOI] [PubMed] [Google Scholar]; c) Awino JK, Hu L, Zhao Y. Org Lett. 2016;18:1650–1653. doi: 10.1021/acs.orglett.6b00527. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Awino JK, Zhao Y. ACS Biomater Sci Eng. 2015;1:425–430. doi: 10.1021/acsbiomaterials.5b00042. [DOI] [PubMed] [Google Scholar]; e) Awino JK, Gunasekara RW, Zhao Y. J Am Chem Soc. 2016;138:9759–9762. doi: 10.1021/jacs.6b04613. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Gunasekara RW, Zhao Y. J Am Chem Soc. 2017;139:829–835. doi: 10.1021/jacs.6b10773. [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Awino JK, Zhao Y. Org Biomol Chem. 2017;15:4851–4858. doi: 10.1039/c7ob00764g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidtchen FP. In: Supramolecular Chemistry: From Molecules to Nanomaterials. Steed JW, Gale PA, editors. Wiley; 2012. Online. [Google Scholar]
- 18.Arifuzzaman MD, Zhao Y. J Org Chem. 2016;81:7518–7526. doi: 10.1021/acs.joc.6b01191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tryptophan is not considered highly hydrophobic by the GRAVY index. However, it can create a significantly larger binding pocket in the hydrophobic core of MINP than other amino acids, providing a strong hydrophobic driving force for binding as a result.
- 20.Staatz WD, Fok KF, Zutter MM, Adams SP, Rodriguez BA, Santoro SA. J Biol Chem. 1991;266:7363–7367. [PubMed] [Google Scholar]
- 21.Hopp TP, Prickett KS, Price VL, Libby RT, March CJ, Pat Cerretti D, Urdal DL, Conlon PJ. Bio/Technology. 1988;6:1204–1210. [Google Scholar]
- 22.Roy D, Karmakar R, Mondal SK, Sahu K, Bhattacharyya K. Chem Phys Lett. 2004;399:147–151. [Google Scholar]
- 23.Chadha G, Zhao Y. J Colloid Interface Sci. 2013;390:151–157. doi: 10.1016/j.jcis.2012.09.042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.