Abstract
Two full-length cDNAs, HvNRT2.3 and HvNRT2.4, were isolated from roots of barley (Hordeum vulgare), using reverse transcriptase-PCR and RACE-PCR. The corresponding polypeptides, consisting of 507 amino acids (molecular masses of 54.6 kD), belong to the major facilitator superfamily (MFS), and are closely related (>87% identity) to those encoded by HvNRT2.1 and HvNRT2.2 (formerly BCH1 and BCH2, respectively) from roots of barley. The latter are considered to encode inducible high-affinity NO3− transporters (Trueman et al., 1996). HvNRT2 transcripts were undetectable in NO3−-deprived plants. Following exposure to either NO3− or NO2−, transcript abundance and 13NO3− influx increased to a maximum by 6 to 12 h, then declined in HvNRT2.1, HvNRT2.2, and HvNRT2.3. The pattern of HvNRT2.4 transcript abundance was different, remaining high after achieving peak abundance. When external NO3− concentrations were varied from 0 to 500 μm under steady-state conditions of NO3− supply, HvNRT2 transcript accumulation and 13NO3− influx were highest in 50 μm NO3− -grown plants. When NH4+ was provided together with NO3−, transcript accumulation during the first 2 h was similar to that due to NO3− alone, but by 4 h the transcript level was significantly reduced. HvNRT2 transcript was undetectable in leaf tissues.
The absorption of NO3− by root cells is mediated by at least three kinetically distinct and thermodynamically active transport systems that are localized in the plasma membranes of root cells. The three transport systems were first characterized on the basis of their different responses to external NO3− concentrations and by their different NO3− inducibility (for review, see Glass and Siddiqi, 1995; Crawford and Glass, 1998). The constitutive high-affinity transport system (CHATS) is a low-capacity, high-affinity transporter that is expressed without the necessity of prior exposure to NO3− (Behl et al., 1988; Siddiqi et al., 1990; Aslam et al., 1992). This transporter represents the main pathway for NO3− entry into roots from low external NO3− on first exposure to NO3−, and is therefore critical for the induction of the high-capacity, high-affinity inducible transport system (IHATS). This transport system can be induced by either NO3− or NO2− (Siddiqi et al., 1992; Aslam et al., 1996). A notable feature of the time course of this induction is that it is typically followed by down-regulation of NO3− influx to a much lower steady-state level. While it is very evident that the induction of the IHATS is mediated by NO3−, the subsequent down-regulation has been attributed to NO3−, NH4+, and/or various amino acids (Ingemarsson et al., 1987; Siddiqi et al., 1990; Lee et al., 1992; Muller and Touraine, 1992).
At high external NO3− concentrations (>200 μm), a low-affinity transport system (LATS) becomes apparent. This system, like CHATS, is expressed in barley (Hordeum vulgare) plants grown in the complete absence of NO3− and shows no evidence of saturation, even at NO3− concentrations as high as 50 mm (Siddiqi et al., 1990). All three transporter systems bring about a rapid depolarization of the membrane electrical potential difference when exposed to exogenous NO3−, which is consistent with the hypothesis that the free energy for active transport of NO3− is provided by the proton motive force via a 2H+:1NO3− symport (Ullrich and Novacky, 1981; McClure et al., 1990; Glass et al., 1992; Meharg and Blatt, 1995; Wang and Crawford, 1996). At present, the only genetic information on the CHATS comes from the isolation of a chlorate-resistant mutant (chl8) from Arabidopsis (Wang and Crawford, 1996). Physiological studies of this mutant showed that plants grown in submerged cultures without NO3− failed to show the typical pattern of NO3− uptake or depolarization of membrane electrical potential differences when exposed to NO3−, suggesting an absence of normal CHATS expression. By contrast, the IHATS and LATS activities of this mutant were normal.
Genes that are thought to encode the LATS and IHATS transporters have been isolated from various higher plants. In the case of LATS, this was accomplished by screening Arabidopsis mutants with chlorate (a toxic analog of NO3−). This resulted in the isolation and characterization of chlorate-resistant mutants (Doddema and Telkamp, 1979). Of these, only one (B1) was affected in NO3− transport (the other mutants were defective in the synthesis of molybdate cofactor and NO3− reductase). Using the same screening method, Tsay et al. (1993) isolated a T-DNA-tagged mutant that mapped to the same locus as B1. A T-DNA tagged CHL1 gene and the wild-type homolog were subsequently isolated. The expression of CHL1 cDNA in Xenopus oocytes resulted in the accumulation of NO3− and the subsequent depolarization of the oocyte membrane upon exposure to NO3−. These results provide more direct evidence that the protein encoded by the CHL1 gene is capable of NO3− transport (Tsay et al., 1993). Physiological analysis of the chl1 deletion mutant by Touraine and Glass (1997) and Arabidopsis transformed with chl1 under the control of the 35S promoter (Huang et al., 1996) was interpreted to indicate the existence of two low-affinity NO3− transport systems in Arabidopsis. These transport systems and/or their respective genes appear to be differentially regulated by ammonium or products of ammonium assimilation.
The first gene encoding an inducible high-affinity NO3− transporter from eukaryotic organisms was cloned from Aspergillus nidulans (Johnstone et al., 1990; Unkles et al., 1991). The crnA mutant (Tomsett and Cove, 1979) was defective in NO3− uptake in conidiospores and young mycelia (Brownlee and Arst, 1983). The crnA gene, which was able to restore NO3− uptake in this mutant (Johnstone et al., 1990; Unkles et al., 1991), encodes a protein that is 507 amino acids long and contains 12 membrane-spanning regions. This protein is a member of the major facilitator superfamily (MFS) (Trueman et al., 1996), a superfamily of membrane proteins that contains the conserved amino acid motif of (D/N) RXGR(R/K) and IX2RX3GX3G between membrane spanning domains 2 and 3 (Henderson, 1991).
A number of genes that are homologous with crnA has been cloned from other eukaryotes. These include YNT1 from Hansenula polymorpha (Perez et al., 1997), CrNRT2.1 and CrNRT2.2 from Chlamydomonas reinhardtii (Quesada et al., 1994), HvNRT2.1 and HvNRT2.2 (formerly BCH1 and BCH2) from barley (Trueman et al., 1996), NpNRT2.1 from Nicotiana plumbaginifolia (Quesada et al., 1997), GmNRT2 from soybean (Glycine max) (Amarashinghe et al., 1998), and AtNRT2.1 and AtNRT2.2 from Arabidopsis (Zhuo et al., 1999). All of the above are thought to encode high-affinity NO3− transporters and belong to the MFS.
In barley the isolation of a BCRNA fragment by PCR using oligonucleotides directed at a conserved MFS motif led to the isolation of the first HvNRT2.1 and HvNRT2.2 genes encoding putative NO3−-inducible high-affinity transporters in higher plants (Trueman et al., 1996). Northern-blot analysis of nitrogen-starved barley plants showed that the HvNRT2 transcript accumulated rapidly in roots following provision of NO3− (Trueman et al., 1996). This is in agreement with physiological data showing that NO3− influx can increase up to 30-fold in the high-affinity range upon NO3− treatment (Siddiqi et al., 1989). Subsequently, in both N. plumbaginifolia and Arabidopsis, levels of NpNRT2.1 and AtNRT2.1 transcripts decreased when the NO3− supply was maintained beyond the period of peak induction (Krapp et al., 1998; Zhuo et al., 1999). This pattern of expression correlates with the overshoot of high-affinity NO3− transport and the subsequent decline to a lower steady-state level referred to above (Siddiqi et al., 1989). Reduced nitrogen forms such as NH4+ or Gln, which are known to diminish NO3− uptake when applied in the presence of NO3−, decreased NpNRT2.1 and AtNRT2.1 transcript levels in roots of N. plumbaginifolia and Arabidopsis, respectively (Quesada et al., 1997; Krapp et al., 1998; Zhuo et al., 1999). By use of metabolic inhibitors, particularly Met sulfoximine and azaserine, which block the enzymes Gln synthetase and GOGAT, respectively, it was suggested that both NH4+ and Gln were active in the down-regulation of AtNRT2.1 expression (Zhuo et al., 1999).
In barley, the genome organization may allow for the presence of as many as seven to 10 members of the HvNRT2 gene family (Trueman et al., 1996). Due to the considerable physiological data available for this species, barley is an important model system in which to investigate the mechanism of transcriptional regulation of NO3− transport. In this report, we describe the isolation of two new cDNAs, HvNRT2.3 and HvNRT2.4, which are closely related to HvNRT2.1 and HvNRT2.2, and the isolation of the 5′ upstream region of HvNRT2.1, HvNRT2.2, and HvNRT2.3. We have also characterized the expression pattern of the HvNRT2 family of genes and 13NO3− influx in response to the provision of various nitrogen sources in parallel experiments.
MATERIALS AND METHODS
Plant Material
Seven-day-old seedlings of barley (Hordeum vulgare cv Klondike) were used in all experiments. Seeds were surface-sterilized with 20% (v/v) commercial bleach solution and rinsed with de-ionized water. The seeds were placed on a nylon mesh (pore size, 4 mm) fixed onto 20-mm (8 seeds) or 60-mm (25 seeds) plexiglass discs, depending on the experiment. The discs were placed in moist sand in the dark, and the seeds were covered to a depth of 10 mm. After 3 d the seedlings were transferred to 40-L hydroponic tanks and grown in nitrogen-free one-tenth-strength modified Johnson's solution (Siddiqi et al., 1989) for 4 d more. Depending on the experiment, nitrogen was supplied in the form of NO3−, NO2−, or NH4+. The K+ concentration was monitored daily and the concentrations of K+ and other nutrients were restored by the addition of a concentrated stock solution to maintain them at constant levels. The pH of the solutions was maintained at 6.2 ± 0.3 by the addition of excess CaCO3 powder. Plants were maintained in a controlled environment chamber with a 16-h/8-h light/dark cycle at 20°C ± 2°C and 70% relative humidity. Light (photon flux density at plant level of approximately 300 μmol m−2 s−1) was provided by fluorescent tubes with a spectral composition similar to sunlight.
RNA and DNA Isolation
Total RNA was isolated using Trizol Reagent (Life Technologies/Gibco-BRL, Cleveland) with two modifications. First, after the tissue was ground in a mortar and Trizol reagent was added at a ratio of 0.2 g of tissue per milliliter of Trizol, the homogenate was centrifuged at 8,000g for 30 min to remove cellular debris. Second, after the total RNA was isolated, it was again extracted with phenol:chloroform:iso-amyl alcohol (25:24:1) and precipitated with sodium acetate (final concentration 0.3 m) and 2 volumes of ethanol. An mRNA isolation kit (FastTrack, Invitrogen, Carlsbad, CA) was used according to the manufacturer's instructions. Genomic DNA was isolated as described by Asubel et al. (1995).
cDNA and Genomic Library
Messenger RNAs isolated from roots of 7-d-old barley seedlings treated for 2 and 6 h with 10 mm KNO3 were used as the template for cDNA synthesis. A cDNA synthesis kit (Marathon, CLONTECH Laboratories, Palo Alto, CA) was used for the construction of a cDNA library. A DNA walking kit (PromoterFinder, CLONTECH Laboratories) was used for the construction of a barley genomic DNA library.
Northern-Blot Analysis
Total RNA was separated on a 1.2% (w/v) agarose gel containing 1× MOPS buffer (20 mm 3-[N-morpholino] propanesulfonic acid, 8 mm sodium acetate and 1 mm EDTA) with 2.2 m formaldehyde, at 60 V for 3.5 h, then washed twice in water, and the RNA was transferred by capillary action to nylon membrane (Hybond N+, Amersham-Pharmacia Biotech, Uppsala). The membrane was baked for 2 h at 80°C to fix the RNA, and was then placed in prehybridization solution for 1 or 4 h (random labeled probe or oligonucleotide probe, respectively). Membranes were next exposed to hybridization solution with 32P-labeled probe for 12 to 16 h. For random-labeled probes, prehybridization and hybridization solutions were 6× SSC, 5× Denhardt's solution, 0.5% (w/v) SDS, and 20 μg mL−1 sonicated herring sperm DNA, respectively. Random-labeled probes were made with a kit (Prime-A-Gene, Promega, Madison, WI) using an internal fragment from the HvNRT2.3 gene from plasmid pBCH3 by digestion with EcoRV and AflIII.
Control levels of total RNA were probed with a fragment of the 25S gene on plasmid pV25S by digestion with XhoI. Membranes were washed according to the manufacturer's instructions with 0.25 SSC and 0.1% (w/v) SDS at 42°C for 15 min for the final wash. Oligonucleotide probing, prehybridization solution, and hybridization solution consisted of 50% (v/v) formamide, 6× SSC, 0.01% (w/v) SDS, and 0.05 mg mL−1 salmon sperm DNA. Prehybridization was at 37°C for 4 h, while hybridization was for 12 to 16 h. Washing of the membrane consisted of 2× 15-min washes at room temperature with 2× SSC and 0.05% (w/v) SDS. The oligonucleotides used as probes were DX 46: 5′CTGTAGTTCAGTACTTGTACATAGG for the HvNRT2.1 gene; DX48: 5′CACTGTACGTGTACACAGGTAAAG for the HvNRT2.2 gene; BCH3: 5,GGTCCAAATGGAGGTGGAGG for the HvNRT2.3 gene; and BCH4: 5′ CAAAATTTGAAACTTATACGTGTAGG for the HvNRT2.4 gene. T4 DNA kinase (Life Technologies, Gaithersburg, MD) and [α-32P]ATP (Amersham-Pharmacia Biotech) were used for end-labeling of oligonucleotides. G-25 spin columns (Pharamacia, Montreal, Canada) were used to separate unincorporated [32Pα]ATP from reaction mixtures.
Isolation and Screening of HvNRT2.3 and HvNRT2.4 cDNAs by Reverse Transcriptase-PCR and RACE-PCR
The isolation of HvNRT2.3 was by 5′- and 3′-RACE-PCR. Oligonucleotide BCRNA-7 (5′GTATGGGTGTGCCTTCCT) was used for the 3′ prime race, while for 5′-RACE-PCR and isolation of a full-length cDNA, BCH3 (5′TGCCTTATACCTGCTGCTGGGGTG) was used. The cDNA template was fabricated using the cDNA synthesis kit. 5′- and 3′-RACE-PCR conditions were 7 min at 94°C then 35 cycles of 94°C for 45 s, 62°C for 45 s, with a 5-min extension period at 72°C.
The isolation of HvNRT2.4 was by 5′- and 3′-RACE-PCR. The oligonucleotide DZ44: 5′ GGACTAGCAGCGGGT was used in the initial 3′-RACE-PCR. RACE-PCR conditions were 94°C for 7 min, then 35 cycles of 94°C for 45 s, 50°C for 45 s, and 72°C for 4 min. The PCR reaction products were purified and then separated on a 1.2% (w/v) agarose gel. The digested DNA was then transferred to nylon membrane (Hybond N+, Amersham-Pharmacia Biotech) for Southern analysis. Positive PCR products were cloned into pCR2.1 (Invitrogen) and sequenced. Oligonucleotide BCH4 (5′ CAAAATTTGAAACTTATACGTGTAGG) was used for the isolation of 5′-RACE-PCR product.
Full-length clones for HvNRT2.3 and HvNRT2.4 were generated by PCR using a high-fidelity PCR system (Expand Long, Boehringer Mannheim/Roche, Basel). For HvNRT2.4, oligonucleotides used were BCH3-5, 5′GGTCCAAATGGAGGTGGAGG and BCH3, while for HvNRT2.4, oligonucleotides BCH4-5, 5′CTCAGTAGATATGGAGGTGAGGC and BCH4 were used. The full-length PCR products were subcloned into pCR2.1 (Invitrogen). After restriction endonuclease analysis of the resulting cDNAs, resulting overlapping fragments were subcloned into pBlueScript I KS+ (Stratagene, La Jolla, CA). Sequences were determined on both strands using M13 forward and reverse primers. For regions where subclones could not be generated, specific oligonucleotides were designed for sequencing.
Isolation of Upstream Region of HvNRT2.1, HvNRT2.2, and HvNRT2.3
A DNA walking kit (PromoterFinder, CLONTECH Laboratories) was used for the isolation of the upstream region of HvNRT2.1, HvNRT2.2, and HvNRT2.3. Specific oligonucleotides were designed to hybridize with the 5′-untranslated region (UTR). For HvNRT2.1, the oligonucleotides were GB1 (5′CAACAACTAGAAGCAGCTAATGGTGGC) and GB1-2 (5′GTTGCAGCTCTTGAGCTTGGCTTGCAA); for HvNRT2.2, GB2 (5′TCGAGCTAGCTAGCTTAGTCGCACTGG) and GB2-2 (5′GTGTGTCTTTAATGGTGGTTGCTGCTG); and for HvNRT2.3, GB3 (5′ GGACCTTGCTTGATCGAGCTAGTCTCC) and GB3-2 (5′ GGAGCTAGC- TTGCTTGATCAGCTGCAG). All PCR reactions used a high-fidelity PCR system (Boehringer Mannheim/Roche). For the first round of PCR the oligonucleotide AP1 was used (PromoterFinder DNA walking kit) with GB1, GB2, or GB3, at 92°C for 3 min, then 30 cycles of 92°C for 25 s, 65°C for 30 s, and 68°C for 10 min. The amplicon was diluted one-tenth, and then a second round of PCR (nested) was conducted using AP2 (PromoterFinder DNA walking kit) and GB1-2, GB2-2, or GB3-3. The resulting PCR products were purified using the Gene Clean Kit (BIO101, Vista, CA), and cloned into pCR2.1.
NO3− Influx
NO3− influx experiments were carried out essentially as described by Siddiqi et al. (1989). Barley plants were grown on sand for 3 d, transferred to hydroponic tanks for 4 d, and then exposed to various media according to the experimental design. Plants were then transferred to 0.5-L vessels containing unlabeled uptake solution for 5 min so as to bring the root epidermal and cortical apoplasm to the same NO3− concentration as was used for the influx determination. After this pretreatment they were transferred to 0.5-L vessels containing a 50 μm NO3− influx solution labeled with 13NO3− for a period of 5 min. Thereafter, plants were transferred back to the 0.5-L vessel of unlabeled nutrient solution for 3 min to remove tracer from the cell wall. Roots and shoots were harvested separately and placed into 20-mL scintillation vials for counting in a γ-counter (Minaxi δ Auto-γ 5000 series, Packard Instruments, Meriden, CT). 13NO3− was produced as described by Kronzucker et al. (1995).
RESULTS
Isolation of HvNRT2.3 and HvNRT2.4 cDNAs
The isolation of HvNRT2.3 (accession no. AF091115) was accomplished by RACE-PCR. Oligonucleotides directed to the BCRNA fragment (Trueman et al., 1996) were designed and subsequently used for 5′- and 3′-RACE PCR. The sequencing of the 3′-RACE-PCR product indicated a new member of the HvNRT2 family of genes in barley, which we designated as the 3BCH3 fragment. The 5′-RACE-PCR product was a contaminant of HvNRT2.1. Therefore, a new oligonucleotide was designed based on sequence data from the 3′-UTR of the 3BCH3 fragment. 5′-RACE-PCR resulted in the isolation of a full-length cDNA. The cDNA sequence revealed that it was a new member of the HvNRT2 family of genes in barley. This cDNA, which is 1,822 bp in length, was designated HvNRT2.3.
A different strategy was used in the isolation of the HvNRT2.4 (accession no. AF091116) cDNA. We designed an oligonucleotide encoding protein consensus sequences found in the CrNRT2.1, CrNRT2.2, HvNRT2.1, and HvNRT2.2 cDNAs. This consensus sequence represents amino acid positions 166 to 174 of the HvNRT2.1 protein. This amino acid motif has the sequence GLAAGWGNM, which is conserved among the NO3−/NO2− subgroup of the MFS (Trueman et al., 1996). The cDNA library used for 3′-RACE-PCR was digested with a number of restriction endonucleases that digest within HvNRT2.1 and HvNRT2.2 cDNAs. This was done to rule out the possibility that HvNRT2.1 and HvNRT2.2 cDNAs would be amplified. The 3′-RACE-PCR products were transferred to nylon membrane and probed with HvNRT2.1 cDNA at medium stringency. The resulting RACE-PCR products that hybridized with the HvNRT2.1 probe were cloned and sequenced. The sequencing data indicated that one of the RACE-PCR products was another member of the HvNRT2 gene family in barley. This fragment is designated 3′HvNRT2.4. An oligonucleotide specific to the 3′-UTR of this gene was designed and synthesized. 5′-RACE-PCR resulted in the isolation and cloning of HvNRT2.4 cDNA. This cDNA is 1,705 bp in length.
Protein Structure, Genetic Analysis, Comparison of the Nucleotide, and Protein Sequences
The predicted HvNRT2.3 and HvNRT2.4 proteins are 507 amino acids in length, with molecular masses of 54.6 kD. The pIs of the HvNRT2.3 and HvNRT2.4 proteins are 8.21 and 8.54, respectively. Both predicted proteins contain 12 membrane spanning regions and have the MFS conserved sequence of (D/N) RXGR(R/K) and IX2RX3GX3G (Henderson, 1991; Marger and Saier, 1993). We analyzed the predicted protein sequences of HvNRT2 proteins with PROSITE program (Bairoch et al., 1997) and found possible sites for protein phosphorylation (see asterisks, Fig. 1). HvNRT2.3 and HvNRT2.4 proteins have three possible protein kinase C phosphorylation sites (Woodgett et al., 1986; Kikkawa et al., 1988) at positions 28 to 30, 381 to 383, and 484 to 486, with residue compositions of SFR, SRR, and SER, respectively. HvNRT2.3 and HvNRT2.4 proteins also have three casein kinase II sites (Pinna, 1990) at positions 453 to 456, 463 to 466, and 482 to 485, with residue compositions of TEEE, SEEE, and SRSE, respectively. The phosphorylation sites are all on the predicted cytoplasmic face of the NRT2 proteins. These phosphorylation sites were also present in HvNRT2.1. The predicted localization of HvNRT2.3 and HvNRT2.4 proteins using the PSORT program (Nakai and Kanehisa, 1992) was the plasma membrane.
Figure 1.
Predicted amino acid sequence of HvNRT2.3 (accession no. AF091115) and HvNRT2.4 (accession no. AF091116) with alignment of eight full-length sequences representing the following other inducible high-affinity NO3− transporters: HvNRT2.1 (accession no. U34198), HvNRT2.2 (accession no. U34290), NpNRT2.1 (accession no. Y08210), AtNRT2.1 (accession no. Z97058), GmNRT2.1 (accession no. AF047718), CRNA (accession no. U34382), YNT1 (accession no. Z69783), and CrNRT2.1 (accession no. Z25438). The alignment was made using Multialign (Smith et al., 1996; BCM launcher, Baylor College of Medicine, Houston). Possible sites for protein phosphorylation are indicated by asterisks.
We analyzed and compared the protein and nucleotide sequences of the four known members of the HvNRT2 multigene family, and compared them with each other and with other NO3− transporters. Figure 1 shows the alignment of the predicted protein sequences, while Figure 2 shows the phylogenetic relationships of the four predicted HvNRT2 proteins compared with the NO3− transporters NpNRT2.1, AtNRT2.1, CrNRT2.1, GmNRT2, YNT1, and CRNA. The amino acid sequences of the predicted proteins of the HvNRT2 family shows >87% identity, with HvNRT2.2 being the most divergent. The highest level of sequence divergence was observed at 5′- and 3′-UTRs of all four cDNAs. In comparing the predicted protein sequence of HvNRT2 genes from barley with the other plant NRT2 genes, we found a stretch of 21 amino acids near the amino terminus of the NRT2 proteins (GmNRT2, AtNRT2.1, and NpNRT2.1) that was not present in the HvNRT2 proteins. Analysis of this sequence predicts a possible protein kinase C phosphorylation site (TGR), and/or a casein kinase II phosphorylation site (TGRE) (Woodgett et al., 1986; Kikkawa et al., 1988; Pinna, 1990).
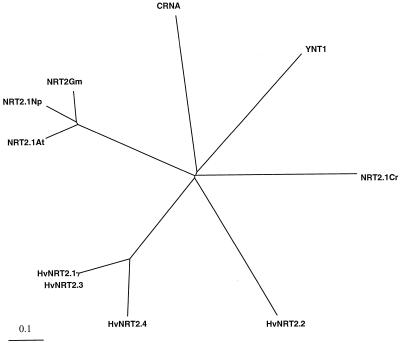
Figure 2.
Phylogeny of predicted amino acid sequence of inducible high-affinity NO3− transporters. The phylogeny was obtained with the PileUp program (Genetics Computer Group) with HvNRT2.1, HvNRT2.2, HvNRT2.3, HvNRT2.4, AtNRT2.1, NpNRT2.1, GmNRT2.1, CRNA, YNT1, and CrNRT2.1.
Isolation, Analysis, and Comparison of 5′ Upstream Regions for HvNRT2.1, HvNRT2.2, and HvNRT2.3
We isolated the 5′ upstream region of HvNRT2.1 (accession no. AF189727), HvNRT2.2 (accession no. AF189728), and HvNRT2.3 (accession no. AF189729) (DNA fragments 535, 635, and 1,436 bp in length, respectively) by the use of a DNA walking kit. The TATA boxes were located at −48, −37, and −45 for HvNRT2.1, HvNRT2.2, and HvNRT2.3, respectively. In comparing the promoter sequences of HvNRT2.1, HvNRT2.2, and HvNRT2.3, we found that HvNRT2.1 had 65% homology with HvNRT2.3 and 58% homology with HvNRT2.2. By comparison, HvNRT2.2 had 53.2% homology with HvNRT2.3. We found one stretch of DNA present in the promoter sequence of HvNRT2.1, HvNRT2.2, and HvNRT2.3, which was highly homologous, (domain I) 16/19 (84.2% identity) with the consensus sequence TGATTCCGTNNGNTGCAAT. If we specifically compared the areas adjacent to domain I of HvNRT2.1 and HvNRT2.3, this domain increased both in DNA size and homology 29/33 (87.8% homology). We also found another stretch of DNA, domain II, with 62/69 identical nucleotides (89.8% homology). In Arabidopsis a putative cis-acting NO3−-inducible element (NIE) containing a core sequence (A[G/C] TCA) preceded by an AT-rich region is considered to be involved in the induction of the NO3− reductase genes by NO3− (Hwang et al., 1997). In barley the HvNRT2 promoters were found to contain this core sequence. HvNRT2.1 has one copy of the core sequence at −430, HvNRT2.2 has two copies at −120 and −299, and HvNRT2.3 has four copies at −684, −764, −994, and −1,267 from the transcription start site.
Time Profile of NO3−-Induced mRNA of the HvNRT2 Multigene Family and 13NO3− Influx
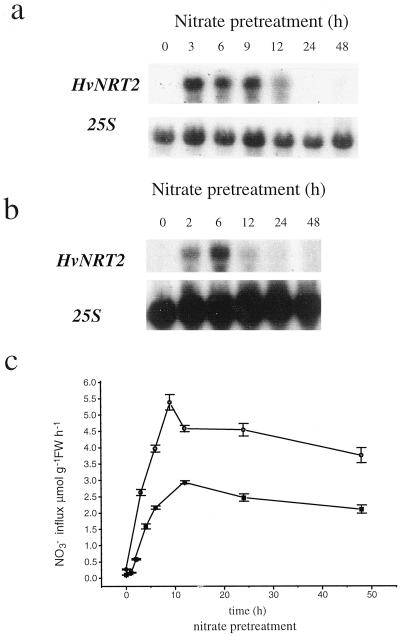
The effects of two NO3− concentrations (1 and 10 mm) on the expression of HvNRT2 multigene family and NO3− influx was investigated for various pretreatment times (0–48 h). Northern-blot analysis using an internal fragment of HvNRT2.3 (which was able to recognize all members of the HvNRT2 family), showed that 1 mm NO3− induced the accumulation of HvNRT2 transcript in roots to their highest level within 3 h of treatment. Thereafter, transcript levels steadily decreased to undetectable levels by 24 h (Fig. 3a). The same overall pattern was observed for the 10 mm NO3− treatment. HvNRT2 mRNA accumulation peaked at 6 h, and then decreased to undetectable levels by 24 h (Fig. 3b). In short-term experiments in which plants were supplied with 10 mm NO3−, HvNRT2 transcript accumulation was observed within 30 min of the onset of NO3− treatment (data not shown). In parallel experiments, using 50 μm external NO3− to measure high-affinity NO3− influx (Fig. 3c), influx increased 20-fold from the onset of 1 mm NO3− pretreatment to a maximum value at 9 h, then decreased. Nevertheless, even at 48 h, influx remained relatively high, despite the fact that transcript abundance had decreased to undetectable levels. This may indicate the participation of other transport systems in the measured influx. The pattern of response to pretreatment with 10 mm NO3− was essentially similar to the 1 mm pretreatment (Fig. 3c).
Figure 3.
Time course effects of NO3− pretreatment on root HvNRT2 transcript abundance and 13NO3− influx. a, Northern blot from roots of plants exposed to 1 mm KNO3 for the times shown. b, Northern blot from roots of plants exposed to 10 mm KNO3 for the times shown. Twenty micrograms of total RNA was introduced into each lane and washed at medium stringency. Northern blots were probed with the 25S ribosomal subunit to ensure equal loading of RNA. c, 13NO3− influx values measured at 50 μm NO3− for plants pretreated with 1 (●) or 10 (▪) mm NO3− for the times shown.
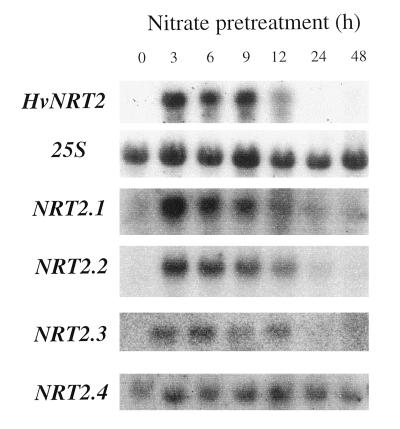
To investigate the expression of each of the known HvNRT2 cDNAs, oligonucleotide probes directed to the 3′-UTR were designed and used in northern-blot analysis. Figure 4 shows the accumulation of specific HvNRT2 mRNAs in nitrogen-starved plants following exposure to 1 mm NO3− for 0, 3, 6, 9, 12, 24, and 48 h. Accumulation of HvNRT2.1, HvNRT2.2, and HvNRT2.3 transcripts in roots peaked at 3 to 6 h, then declined to undetectable levels by 12 to 24 h. By contrast, HvNRT2.4 transcript levels, which increased within 3 h of NO3− feeding, remained at elevated levels for the duration of the experiment (48 h). To visualize transcript abundance of each of the different HvNRT2 homologs, x-ray film was exposed to HvNRT2 northern blots for the following times: HvNRT2.1, 12 h; HvNRT2.2, 48 h; HvNRT2.3, 16 h; and HvNRT2.4, 96 h.
Figure 4.
Northern-blot analysis of members of the HvNRT2 family of genes. Time course effects of pretreating nitrogen-starved plants with 1 mm NO3− for 0, 3, 6, 9, 12, 24, and 48 h. Twenty micrograms of total RNA was introduced into each lane, probed with HvNRT2.3 internal fragment (which recognizes all known HvNRT2 homologs), and then washed at medium stringency (panel 1). Northern blots were probed with the 25S ribosomal subunit to ensure equal loading of RNA. Northern blots were also probed with specific oligonucleotides directed to HvNRT2.1, HvNRT2.2, HvNRT2.3, and HvNRT2.4 transcripts, and washed at high stringency.
Effect of Various External NO3− Concentrations on HvNRT2 Transcript Levels and NO3− Influx
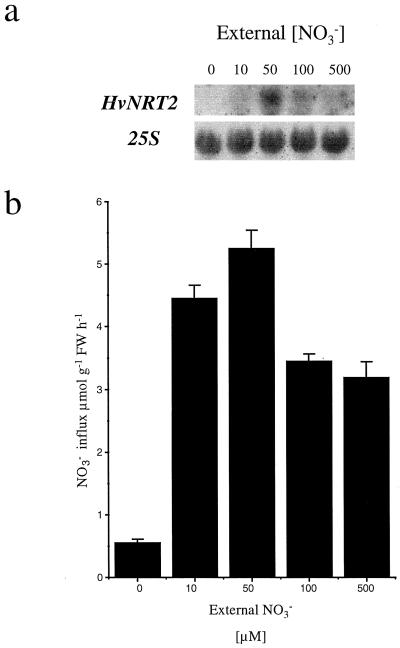
To investigate the effect of external NO3− concentrations on HvNRT2 transcript levels and NO3− influx under steady-state culture conditions, barley plants were grown in hydroponic solutions containing 0, 10, 50, 100, and 500 μm NO3− for 4 d. Both HvNRT2 transcript levels (northern-blot analysis, Fig. 5a) and 13NO3− influx from 50 μm NO3− (Fig. 5b) were subsequently monitored in roots of these plants. NO3− influx varied from 0.54 μmol g−1 fresh weight h−1 in plants maintained at 0 external NO3− to its maximum rate (5.32 μmol g−1 fresh weight h−1) for plants maintained at 50 μm NO3−. At higher NO3− concentrations, influx decreased to 3.15 μmol g−1 fresh weight h−1 in plants maintained at the 500 μm level. The abundance of HvNRT2 transcript followed the same pattern.
Figure 5.
Effect of external NO3− supply on transcript abundance of HvNRT2 genes and 13NO3− influx in roots of barley plants grown on 0, 10, 50, 100, and 500 μm NO3−. a, Northern-blot analysis of HvNRT2 transcript abundance; 20 μg of total RNA was introduced into each lane and washed at medium stringency. To ensure equal loading into the lanes, RNA was probed with the 25S ribosomal subunit and washed at high stringency. b, NO3− influx was measured at 50 μm NO3− after 4 d of growth at the NO3− concentrations shown.
Effect of NO2− and NH4+ on Transcript Levels of the HvNRT2 Multigene Family
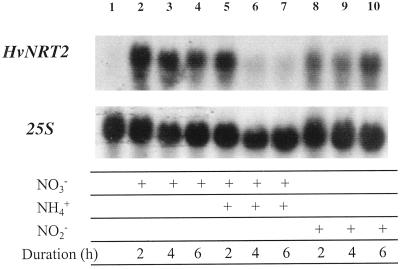
Figure 6 shows the effect of simultaneously providing 10 mm NH4+ on the accumulation of HvNRT2 transcript during induction of NO3− transport by 10 mm NO3−. Two hours after providing NH4+ and NO3− together, no effect on HvNRT2 transcript accumulation was apparent, compared with plants supplied with NO3− only. However, by 4 and 6 h, respectively, transcript abundance decreased dramatically. In parallel experiments, we measured 13NO3− influx at 50 μm NO3− in plants supplied with both NO3− and NH4+ for 6 h. 13NO3− influx in these plants and in nitrogen-starved plants remained at low levels.
Figure 6.
Effects of the duration of NO3−, NO3− plus NH4+, and NO2− pretreatment on HvNRT2 transcript abundance. Lane 1, Plants grown in nitrogen-free medium; lanes 2 through 4, 10 mm NO3−-supplied plants; lanes 5 through 7, 10 mm NH4+ and 10 mm NO3−-co-supplied plants; and lanes 8 through 10, 10 mm NO2−-supplied plants for times shown. Twenty micrograms of total RNA was introduced into each lane and probed with HvNRT2.3 internal fragment and washed at medium stringency. Northern blot was probed with the 25S ribosomal subunit to ensure equal loading of RNA.
The capacity of exogenously supplied NO2− to induce HvNRT2 transcript accumulation in nitrogen-starved barley seedlings was also investigated (Fig. 6). Supplying 10 mm NO2− increased the accumulation of HvNRT2 transcripts in roots of barley seedlings, but at a slower rate than in the NO3− treatment.
DISCUSSION
Characterization of HvNRT2 cDNAs and 5′ Upstream Regions
In this study, reverse transcriptase-PCR and RACE-PCR were employed for the isolation of two new putative high-affinity NO3− transporter genes (HvNRT2.3 and HvNRT2.4) from barley. The coding regions of these genes are highly conserved with respect to each other and to HvNRT2.1 and HvNRT2.2 (Trueman et al., 1996), with greater than 87% homology at the protein level. Both HvNRT2.3 and HvNRT2.4 have the conserved motif of the major facilitator superfamily, and, possibly, protein kinase C and casein kinase II phosphorylation sites. The location and number of sites are constant for all of the known NRT2 proteins, and the sites appear to be located on the cytoplasmic face of each protein. The major difference between the NRT2 proteins from barley and those from N. plumbaginifolia, Arabidopsis, and soybean is a deletion of 21 amino acids in the NRT2 proteins at the amino terminus. In this deletion are located protein kinase C and casein kinase II phosphorylation sites. This may indicate that there are differences in post-translation modification of the barley NRT2 proteins. The role of these phosphorylation sites was not assessed in this study, but it is interesting that protein kinase C and casein kinase II sites were present in barley NRT2 proteins, as well as in the N terminus (21 amino acid sequence) of the other NRT2 proteins. In barley, Southern-blot analysis demonstrated that there are seven to 10 NRT2 homologs (Trueman et al., 1996), while in N. plumbaginifolia, soybean, and Arabidopsis, there appear to be only two copies of the gene (Quesada et al., 1997; Amarashinghe et al., 1998; Zhuo et al., 1999). Why barley should possess seven to 10 copies of this gene family is unknown.
Analysis of the 5′ upstream region of HvNRT2.1, HvNRT2.2, and HvNRT2.3 revealed that these sequences are less conserved (>53% homology). One region of DNA (domain I) was conserved in the promoter sequences of HvNRT2.1, HvNRT2.2, and HvNRT2.3. Also present was domain II, a region of DNA comprising 69 bp with a homology of 89%. The roles of these sequences are unknown, but they may function in the regulation of the NRT2 genes. The NIE core elements, demonstrated to be present in variable copy number in the HvNRT2 promoter (see “Results” section), may participate in NO3− induction of these genes (Hwang et al., 1997). Unlike the core sequences of Arabidopsis, the barley HvNRT2 and NR (NAR1 and NAR7) promoters are not preceded by an AT-rich region.
Time Profile of HvNRT2 Transcript Levels and NO3− Transport
The substantial increase of high-affinity NO3− uptake following first exposure to NO3− has been referred to as NO3− induction (Jackson et al., 1973; Goyal and Huffaker, 1986). This process typically increases rates of net NO3− uptake several-fold (Warner and Huffaker, 1989). Using the Klondike barley and 13NO3− to measure plasma membrane influx, Siddiqi et al. (1989) demonstrated a 28-fold increase. It is evident that the increase of influx associated with induction is governed by the constitutive value of influx associated with CHATS activity and the extent of the IHATS flux. In Steptoe barley, the CHATS activity was already substantially higher than in Klondike, and therefore the increase of influx associated with induction was substantially less in Steptoe than in Klondike (King et al., 1993). In the present experiments using 1 mm or 10 mm NO3− treatments, both influx and HvNRT2 transcript levels increased with the same time dependence. Peak activities of both transcript and influx occurred during the first 6 to 12 h, after which they declined. Plants pretreated at 10 mm NO3− had lower values of influx than those treated with 1 mm NO3−, and this probably reflects a greater down-regulation by larger internal nitrogen pools in the 10 mm NO3−-treated plants. This decline of HvNRT2 transcript has been shown for other cDNAs encoding putative NO3− transporters when plants were pretreated with NO3− for >12 h as in N. plumbaginifolia (Quesada et al., 1997) and Arabidopsis (Zhuo et al., 1999).
The investigation of the effects of NO3− treatment on individual members of the HvNRT2 family of genes revealed that these genes are coordinately induced in root tissue with approximately the same pattern in time as observed using a single HvNRT2 (HvNRT2.3) probe, which recognized all members of this family. Nevertheless, the transcript abundances varied among the HvNRT2 homologs in the following order: HvNRT2.1 = HvNRT2.3 > HvNRT2.2 > HvNRT2.4. This result does not preclude differential expression by different cell types of the root (e.g. endodermis, stele, root hairs, etc.). In tomato, expression of the LeNRT1.1 gene, which encodes a low-affinity NO3− transporter, is predominately localized in the root cylinder, while LeNRT1.2 is predominately localized in root hairs (Lauter et al., 1996). NpNRT2.1 has also been shown to be highly expressed in epidermal and endodermal cells at the root tip, and in lateral root primordia and epidermis of mature roots (Krapp et al., 1998). Likewise, there was a differential expression of the AtNRT1 gene in Arabidopsis, the gene being expressed in outer layers of the root tip and in progressively deeper layers with increasing distance from the root tip (Huang et al., 1996). Clearly, a significant question remains as to the functional roles of the multiple representatives of the HvNRT2 family of genes.
The long-term effect of exposure to external NO3− (0, 10, 50, 100, or 500 μm) prior to influx measurements revealed that both HvNRT2 transcript levels and influx were highest in plants grown with 50 μm NO3− (Fig. 5). The reported Km of IHATS, 25 to 100 μm, varies with genotype and as a function of NO3− pretreatment (Siddiqi et al., 1990). The present correlation between NRT2 transcript levels and influx values confirms earlier observations to this effect in Arabidopsis (Zhuo et al., 1999) and provides further evidence that the NRT2 genes encode components of the inducible high-affinity transport systems for NO3− influx.
Effects of External NO2− or NH4+ on NO3− Influx and HvNRT2 Transcript Accumulation
To investigate whether nitrogen signals other than NO3− may be involved in the induction of the HvNRT2 genes, NO2− was supplied in place of NO3− during the standard induction treatment. Furthermore, to determine whether NH4+ might impact upon the induction process, 10 mm NH4+ was provided together with 10 mm NO3− in separate experiments (Fig. 6). Although NO2− is rarely present in soil solution, under laboratory conditions it is able to induce both NO2− and NO3− uptake (Siddiqi et al., 1992; King et al., 1993; Aslam et al., 1996), albeit at a slower rate. NO2− at 10 mm increased HvNRT2 transcript levels within 2 h of treatment (Fig. 6). It may be that the slower response to NO2− is due to a slower transduction of the NO2− signal or to lower rates of NO2− uptake. Alternatively, perhaps NO2− was responsible for toxic effects at the concentration provided.
When NH4+ was provided together with NO3−, HvNRT2 transcript accumulation was unaffected for the first 2 h of the treatment, but by 4 to 6 h, there was virtually no transcript present. It is thought that NH4+ may: (a) inhibit NO3− influx directly at the transport step through effects at the plasma membrane (Glass et al., 1985; Lee and Drew 1989; King et al., 1993; Aslam et al., 1996), and (b) function as a signal resulting in repression of IHATS transcription either through effects of NH4+ itself or after its conversion to amino acids. Earlier physiological studies suggested that NH4+ effects on NO3− uptake resulted from downstream assimilation products of NH4+ (Breteler and Siegerist, 1984; Lee and Drew, 1989; Muller and Touraine, 1992). Strong support for this hypothesis comes from treatments with Met sulfoximine, which have resulted in a release from the inhibitory effects of exogenous NH4+ (Breteler and Siegerist, 1984; Lee and Drew, 1989). However, other studies have demonstrated a failure of Met sulfoximine to relieve the inhibitory effects of NH4+ during application of Met sulfoximine (King et al., 1993; Aslam et al., 1996). In the present experiments high levels of NO3− pretreatment (10 mm) were employed to ensure that NO3− would enter the root by both the high- and low-affinity transport systems, notwithstanding an inhibitory effect of NH4+ on high-affinity NO3− uptake. This would distinguish between low levels of induction of NO3− influx arising from inadequate internal [NO3−] and low levels of induction resulting from down-regulation of HvNRT2 expression by NH4+ or its assimilation products.
The present results (Fig. 6) appear to show that induction by 10 mm NO3− proceeded normally in the first 2 h of NH4+ treatment despite the presence of exogenous NH4+. It may be that there is insufficient buildup of cytosolic NH4+ or products of its assimilation to cause any negative effects on induction in the first hours of exposure to NH4+. In a study of AMT1 expression in Arabidopsis, 9 h of exposure to 5 mm NH4NO3 were required before AMT1 expression was reduced to approximately 20% of its original value (Rawat et al., 1999). By this time root Gln concentrations had increased approximately 5-fold, and this nitrogen fraction was responsible for the largest change of root nitrogen composition under these conditions. Likewise, in experiments examining the expression of AtNRT2.1 in roots of Arabidopsis, Zhuo et al. (1999) demonstrated that transcript abundance was reduced to levels corresponding to those of uninduced plants after 3 h of azaserine treatment. Azaserine blocks the activity of Glu synthase, reducing Glu and increasing Gln concentrations. This observation is consistent with a potent effect of Gln on AtNRT2.1 transcript abundance. However, the same study revealed that treatments with Met sulfoximine reduced transcript abundance to approximately 10% of control (induced) plants, indicating that NH4+ itself may also act with negative effects at the level of transcription.
In summary, transcripts of members of the HvNRT2 gene family are induced by both NO3− and NO2−. The promoter sequence contains a core NIE domain, which has been implicated in the induction pathway of NR genes in Arabidopsis. Also present in the promoter sequences of HvNRT2.1, HvNRT2.2, and HvNRT2.3 genes was a common domain I whose function is presently unknown. The presence of NH4+ during the first hours of induction appears not to affect HvNRT2 transcript accumulation, suggesting that cytosolic [NH4+] may need to build to a sufficiently high level (by 4–6 h) before transcript abundance is affected. Alternatively, the result may indicate that downstream metabolites such as Gln are more important regulators of NRT2 expression. Transcript levels of all known members of the HvNRT2 gene family (HvNRT2.1, HvNRT2.2, HvNRT2.3, and HvNRT2.4) at first increased following provision of NO3−, then, as is the case for NO3− influx, down-regulation occurred (with the exception of HvNRT2.4, which remained constant for the duration of the experiment). The strong correlation between patterns of HvNRT2 expression and high-affinity NO3− influx provides further support for the identification of the HvNRT2 genes as participants in high-affinity NO3− influx in barley roots.
ACKNOWLEDGMENTS
We thank Dr. H.J. Kronzucker, Dr. Jan K. Schjoerring, and D. Brito for their help with influx experiments, and Dr. B. Touraine for critical reading of the manuscript. Dr. Brian Forde is thanked for provision of the pBCH1 and pBCH2 plasmids.
Footnotes
This work was supported by Natural Sciences and Engineering Research Council of Canada Strategic and Research grants to A.D.M.G.
LITERATURE CITED
- Amarashinghe BHRR, De Bruxelles G, Braddon M, Onyeocha I, Forde BG, Udvardi MK. Regulation of GmNRT2 expression and nitrate transport activity in roots of soybean (Glycine max) Planta. 1998;206:44–52. doi: 10.1007/s004250050372. [DOI] [PubMed] [Google Scholar]
- Aslam M, Travis R, Rains D, Huffaker R. Effect of ammonium on the regulation of nitrate and nitrite transport systems in roots of intact barley (Hordeum vulgareL.) seedlings. Planta. 1996;200:58–63. [Google Scholar]
- Aslam M, Travis RL, Huffaker RC. Comparative kinetics and reciprocal inhibition of nitrate and nitrite uptake in roots of uninduced and induced barley (Hordeum vulgareL.) seedlings. Plant Physiol. 1992;99:1124–1133. doi: 10.1104/pp.99.3.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1995. Preparation and analysis of DNA; pp. 2.0.1–2.9.10. [Google Scholar]
- Bairoch A, Bucher P, Hofmann K. The PROSITE database, its status in 1997. Nucleic Acids Res. 1997;25:217–221. doi: 10.1093/nar/25.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behl R, Tischner R, Raschke K. Induction of high-capacity nitrate-uptake mechanism in barley roots prompted by nitrate uptake through a constitutive low-capacity mechanism. Planta. 1988;176:235–240. doi: 10.1007/BF00392450. [DOI] [PubMed] [Google Scholar]
- Breteler H, Siegerist M. Effect of ammonium on nitrate utilization by roots of dwarf bean. Plant Physiol. 1984;75:1099–1103. doi: 10.1104/pp.75.4.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlee A, Arst HJ. Nitrate uptake in Aspergillus nidulansand involvement of the third gene of the nitrate assimilation gene cluster. J Bacteriol. 1983;155:1136–1146. doi: 10.1128/jb.155.3.1138-1146.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford NM, Glass ADM. Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci. 1998;3:389–395. [Google Scholar]
- Doddema H, Telkamp GP. Uptake of nitrate by mutants of Arabidopsis thaliana, disturbed in uptake or reduction of nitrate: II. Kinetics Physiol Plant. 1979;45:332–338. [Google Scholar]
- Glass ADM, Bordeleau L, Thompson RG. Regulation of NO3− influx: studies with 13NO3−. Plant Physiol. 1985;77:379–381. doi: 10.1104/pp.77.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass ADM, Shaff JE, Kochian LV. Studies of the uptake of nitrate in barley: IV. Electrophysiology. Plant Physiol. 1992;99:456–463. doi: 10.1104/pp.99.2.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass ADM, Siddiqi MY. Nitrogen absorption by plant roots. In: Srivastava HS, Singh RP, editors. Nitrogen Nutrition in Higher Plants. New Delhi, India: Associated Publishers; 1995. pp. 21–56. [Google Scholar]
- Goyal S, Huffaker R. The uptake of NO3−, NO2− and NH4+ by intact wheat (Triticum aestivum) seedlings: I. Induction and kinetics of transport systems. Plant Physiol. 1986;82:1051–1056. doi: 10.1104/pp.82.4.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson P. Sugar transport proteins. Curr Opin Struct Biol. 1991;1:590–601. [Google Scholar]
- Huang NC, Chiang CS, Crawford NM, Tsay YF. CHL1 encodes a component of the low affinity nitrate uptake system in Arabidopsisand shows cell type-specific expression in roots. Plant Cell. 1996;8:2183–2191. doi: 10.1105/tpc.8.12.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang CF, Lin Y, D'Souza T, Cheng CL. Sequences necessary for nitrate-dependent transcription of Arabidopsis nitrate reductase genes. Plant Physiol. 1997;113:853–862. doi: 10.1104/pp.113.3.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingemarsson B, Oscarson P, Af Ugglas M, Larsson CM. Nitrogen utilization in Lemna: II. Studies of nitrate uptake using 13NO3−. Plant Physiol. 1987;85:860–864. doi: 10.1104/pp.85.3.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson WA, Flesher D, Hageman RH. Nitrate uptake by dark-grown corn seedlings: some characteristics of apparent induction. Plant Physiol. 1973;51:120–127. doi: 10.1104/pp.51.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone I, McCabe P, Grieve P, Guur S, Cole G, Brow M, Unkles SE, Kinghorn JR, Innis M. Isolation and characterization of the crnA-niiA-niaD gene cluster for nitrate assimilation in Aspergillus nidulans. Gene. 1990;90:181–192. doi: 10.1016/0378-1119(90)90178-t. [DOI] [PubMed] [Google Scholar]
- Kikkawa U, Ogita K, Shearman MS, Ase K, Sekiguchi K, Naor Z, Kishimoto A, Nishizuka Y, Saito N, Tanaka C. The family of protein kinase C: its molecular heterogeneity and differential expression. Cold Spring Harbor Symp Quant Biol. 1988;53:97–102. doi: 10.1101/sqb.1988.053.01.015. [DOI] [PubMed] [Google Scholar]
- King BJ, Siddiqi MY, Ruth TJ, Warner RL, Glass ADM. Feedback regulation of nitrate influx in barley roots by nitrate, nitrite, and ammonium. Plant Physiol. 1993;102:1279–1286. doi: 10.1104/pp.102.4.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp A, Fraisier V, Scheible WR, Quesada A, Gojon A, Stitt M, Caboche M, Daniel-Vedele F. Expression studies of Nrt2:1Np,a putative high affinity nitrate transporter: evidence for its role in nitrate uptake. Plant J. 1998;14:723–731. [Google Scholar]
- Kronzucker HJ, Siddiqi MY, Glass ADM. Compartmentation and flux characteristics of nitrate in spruce. Planta. 1995;196:674–682. [Google Scholar]
- Lauter FR, Ninnemann O, Bucher M, Riesmeier JW, Frommer WB. Preferential expression of an ammonium transporter and of two putative nitrate transporters in root hairs of tomato. Proc Natl Acad Sci USA. 1996;93:8139–8144. doi: 10.1073/pnas.93.15.8139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RB, Drew MC. Rapid reversible inhibition of nitrate influx in barley by ammonium. J Exp Bot. 1989;40:741–752. [Google Scholar]
- Lee RB, Purves J, Ratcliffe R, Saker L. Nitrogen assimilation and the control of ammonium and nitrate absorption by maize roots. J Exp Bot. 1992;43:1385–1396. [Google Scholar]
- Marger M, Saier M. A major superfamily of transmembrane facilitators that catalyse uniport, symport and antiport. Trends Biochem Sci. 1993;18:13–20. doi: 10.1016/0968-0004(93)90081-w. [DOI] [PubMed] [Google Scholar]
- McClure PR, Kochian LV, Spanswick RM, Shaff JE. Evidence for co-transport of nitrate and protons in maize roots: I. Effects of nitrate on the membrane potential. Plant Physiol. 1990;93:281–289. doi: 10.1104/pp.93.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meharg AA, Blatt MR. NO3− transport across the plasma membrane of Arabidopsis thalianaroot hairs: kinetic control by pH and membrane voltage. J Membr Biol. 1995;145:49–66. doi: 10.1007/BF00233306. [DOI] [PubMed] [Google Scholar]
- Muller B, Touraine B. Inhibition of NO3−uptake by various phloem-translocated amino acids in soybean seedlings. J Exp Bot. 1992;43:617–623. [Google Scholar]
- Nakai K, Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez MD, Gonzalez C, Avila J, Brito N, Siverio JM. The YNT1 gene encoding the nitrate transporter in the yeast Hansenula polymorpha is clustered with genes YNI1 and YNR1encoding nitrite and nitrate reductase and its disruption causes inability to grow in nitrate. Biochem J. 1997;321:397–403. doi: 10.1042/bj3210397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinna LA. Casein kinase 2: an ‘eminence grise’ in cellular regulation? Biochim Biophys Acta. 1990;1054:267–284. doi: 10.1016/0167-4889(90)90098-x. [DOI] [PubMed] [Google Scholar]
- Quesada A, Galvan A, Fernandez E. Identification of nitrate transporter genes in Chlamydomonas reinhardtii. Plant J. 1994;5:407–419. doi: 10.1111/j.1365-313x.1994.00407.x. [DOI] [PubMed] [Google Scholar]
- Quesada A, Krapp A, Trueman LJ, Daniel-Vedele F, Fernandez E, Forde BG, Caboche M. PCR-identification of a Nicotiana plumbaginifolia cDNA homologous to the high-affinity nitrate transporters of the crnAfamily. Plant Mol Biol. 1997;34:265–274. doi: 10.1023/a:1005872816881. [DOI] [PubMed] [Google Scholar]
- Rawat SR, Silim SN, Kronkucker HJ, Siddiqi MY, Glass ADM. AtAMT1 expression and NH4+ uptake in roots of Arabidopsis thaliana: evidence for regulation by root glutamine levels. Plant J. 1999;19:143–152. doi: 10.1046/j.1365-313x.1999.00505.x. [DOI] [PubMed] [Google Scholar]
- Siddiqi MY, Glass ADM, Ruth TJ, Fernando M. Studies of the regulation of nitrate influx by barley seedlings using 13NO3−. Plant Physiol. 1989;90:806–813. doi: 10.1104/pp.90.3.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqi MY, Glass ADM, Ruth TJ, Rufty T. Studies of the uptake of nitrate in barley: I. Kinetics of 13NO3−influx. Plant Physiol. 1990;93:1426–1432. doi: 10.1104/pp.93.4.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqi MY, King BJ, Glass ADM. Effects of nitrite, chlorate and chlorite on nitrate uptake and nitrate reductase activity. Plant Physiol. 1992;100:644–650. doi: 10.1104/pp.100.2.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R, Wiese B, Wojzynski M, Davison D, Woeley K. BCM search launcher: an integrated interface to molecular biology data base search and analysis services available on the World Wide Web. Genome Res. 1996;6:454–462. doi: 10.1101/gr.6.5.454. [DOI] [PubMed] [Google Scholar]
- Tomsett A, Cove D. Deletion mapping of the niiA niaD region of Aspergillus nidulans. Genet Res. 1979;34:19–32. doi: 10.1017/s001667230001925x. [DOI] [PubMed] [Google Scholar]
- Touraine B, Glass ADM. NO3− and ClO3− fluxes in the chl1-5 mutant of Arabidopsis thaliana: does the CHL1-5 gene encode a low-affinity NO3− transporter? Plant Physiol. 1997;114:137–144. doi: 10.1104/pp.114.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trueman LJ, Richardson A, Forde BG. Molecular cloning of higher plant homologues of the high-affinity nitrate transporters of Chlamydomonas reinhardtii and Aspergillus nidulans. Gene. 1996;175:223–231. doi: 10.1016/0378-1119(96)00154-0. [DOI] [PubMed] [Google Scholar]
- Tsay YF, Schroeder JI, Feldman A, Crawford NM. The herbicide sensitivity gene CHL1 of Arabidopsisencodes a nitrate-inducible nitrate transporter. Cell. 1993;72:705–713. doi: 10.1016/0092-8674(93)90399-b. [DOI] [PubMed] [Google Scholar]
- Ullrich W, Novacky A. Nitrate-dependent membrane potential changes and their induction in Lemna gibbaG1. Plant Sci Lett. 1981;22:211–217. [Google Scholar]
- Unkles SE, Hawker KL, Grieve C, Campbell EI, VanMontague P, Kinghorn JR. crnA encodes a nitrate transporter in Aspergillus nidulans. Proc Natl Acad Sci USA. 1991;88:204–208. doi: 10.1073/pnas.88.1.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Crawford NM. Genetic identification of a gene involved in constitutive, high-affinity nitrate transport in higher plants. Proc Natl Acad Sci USA. 1996;93:9297–9301. doi: 10.1073/pnas.93.17.9297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner R, Huffaker R. Nitrate transport is independent of NADH and NAD(P)H nitrate reductases in barley seedlings. Plant Physiol. 1989;91:947–953. doi: 10.1104/pp.91.3.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodgett JR, Gould KL, Hunter T. Substrate specificity of protein kinase C: use of synthetic peptides corresponding to physiological sites as probes for substrate recognition requirements. Eur J Biochem. 1986;161:177–184. doi: 10.1111/j.1432-1033.1986.tb10139.x. [DOI] [PubMed] [Google Scholar]
- Zhuo D, Okamoto M, Vidmar JJ, Glass ADM. Regulation of putative high-affinity nitrate transporter (Nrt2;1At) in roots of Arabidopsis thaliana. Plant J. 1999;17:563–569. doi: 10.1046/j.1365-313x.1999.00396.x. [DOI] [PubMed] [Google Scholar]