Abstract
The world-renowned Thai Hom Mali Rice has been the most important aromatic rice originating in Thailand. The aromatic variety was collected from Chachoengsao, a central province, and after pure-line selection, it was officially named as Khao Dawk Mali 105, (KDML105). Because of its superb fragrance and cooking quality, KDML105 has been a model variety for studying genes controlling grain quality and aroma. The aromatic gene was cloned in KDML105, as an amino aldehyde dehydrogenase (AMADH) or better known as BADH2 located on chromosome 8. Later on, all other aromatic rice genes were discovered as allelic to the AMADH. As a selection of local landrace variety found in rainfed areas, the Thai Jasmine rice showed adaptive advantages over improved irrigated rice in less fertile lowland rainfed conditions. Because KDML105 was susceptible to most diseases and insect pests, marker-assisted backcross selection (MABC) was used for the genetic improvement since 2000. After nearly 17 years of MABC for integrating new traits into KDML105, a new generation of KDML105, designated HM84, was developed which maintains the cooking quality and fragrance, and has gained advantages during flash flooding, disease, and insect outbreak.
Electronic supplementary material
The online version of this article (10.1186/s12284-018-0212-7) contains supplementary material, which is available to authorized users.
Keywords: Thai Hom Mali, Oryza sativa L, Marker-assisted backcross selection, Gene pyramiding, Amino aldehyde dehydrogenase, Flooding tolerance, Blast resistance, Bacterial blight resistance, Brown planthopper tolerance
Origin and distribution
Thai Jasmine rice is officially known as Thai Hom Mali rice. Hom Mali local landrace varieties were widely distributed throughout Thailand under a variety of names including ‘Khao Hom’, ‘Hom Mali’, and ‘Khao Mali’. In 1945, the best Hom Mali local variety was discovered by a farmer in Lam Pradoo district, Chonburi province, and in 1951, 199 panicles of the local variety were selected from a nearby district of Chachearngsoa province for pure line selection (Bureau of Rice Research and Development 2010). Based on its superior physical appearance, cooking quality, and grain aroma, row ‘105’ was selected and officially named “Khao Dawk Mali 4–2-105” or “Khao Dawk Mali 105” or “KDML105” for short. KDML105 was released to farmers in 1959. Thai Jasmine rice was produced annually at approximately 9.4 M ton of paddy rice from 4.3 M ha (Department of Foreign Trade 2014; Varinruk 2017).
KDML105 has a tall, lanky, floppy architecture. The average plant height at harvest is approximately 140 cm. KDML105 has a photoperiod sensitivity that determines its flowering date around October. Specifically, the critical induction signal is the day/night time duration. When day length is shorter than 11:52 h, KDML105 will initiate its floral organ (Kumboonreang 2011). The critical day length for photoperiod sensitivity can be met at the end of September when rainfall is normally heavy. Therefore, the full bloom is normally expected in the last two weeks of October, and harvesting will start in the second week of November every year. This photoperiod sensitivity is considered an adaptive advantage for lowland rainfed rice to survive and produce without irrigation water.
Review
In traditional lowland rainfed rice cultivation, farmers ask the rice spirit to protect their crop from flooding or drought during cultivation. KDML105 and its mutagenized counterparts, RD15 and RD6, have been widely cultivated in such a stressful mega-ecosystem which occupies 70% of the rice area in Thailand (Jongdee et al. 2006). Characterized by fluctuation in rainfall distribution and poor soil fertility, the lowland rainfed area produces the highest quality Jasmine rice. Adaptation to the infertile lowland rainfed area of the northeast is the benefit of this high-quality rice. It is mildly tolerant to drought, salinity, and acid sulfate soil (Bureau of Rice Research and Development 2010). KDML105 is not a well-known drought-tolerant variety, but its adaptability in a rainfed lowland ecosystem was ascribed to the plasticity of the root system under abiotic stresses (O’Toole and Bland 1987). Plasticity describes the ability of roots to respond to soil moisture fluctuation (Banco et al. 2000). Particularly, in a paddy field with hardpan, KDML105 consistently expresses its root growth adaptability to water stress via greater root branching (Kano-Nakata et al. 2013). Expression of root-branching ability in a shallow soil layer plays an important role in capturing water for rapid recovery from drought stresses and is the key adaptive trait of KDML105 in lowland rainfed conditions (Kano-Nakata et al. 2013).
Most rice is very sensitive to salinity, at a salinity level from 3 dS m− 1 (USDA 2013). The salinity at such level affects rice growth by reducing germination rate, plant height, tillering, root growth, and spikelet fertility. At the ECe as low as 3.5 dSm− 1, grain yield loss can be at about 10% depending on varietal difference (Rice Knowledge Bank, International Rice Research Institute, IRRI). Under a salt-affected area in the NE of Thailand, KDML105 was adaptive to salt-affected area in the NE, where 1.84 M ha or 16% of the lowland rainfed area is classified as an inland saline area ranging from 11 to 35 dS m− 1 (Im-Erb et al. 2013; Arunin and Pongwichian 2015). The saline soil in this area is high in sodium and chloride content and has a sandy soil texture, low organic matter, and minimal fertility (Arunin 1984). Cultivation of KDML105 in this area was located on slightly to moderately salt-affected soils, with ECe of 2–8 dS m− 1 (Im-Erb et al. 2013; Arunin and Pongwichian 2015). However, KDML105 is less productive in high (8–16 dS m− 1) and very high saline soil (> 16 dS m− 1). Therefore, increasing KDML105 salinity tolerance can help poor farmers to grow this fragrant rice in high-salinity areas.
The plasticity of KDML105 seems to be inherited to its irradiated mutants, RD15 and RD6. These three cultivars occupied 5.5 M ha of the mega-lowland rainfed area mostly in the Northeast of Thailand (Fig. 1). On the other hand, the three mega-varieties, KDML105, RD15, and RD6, were not successful in the central plain due to the widespread occurrence of various diseases and insect pests in irrigated areas. This implies that the three Jasmine rice varieties are more adaptable to abiotic than biotic stresses. Therefore, breeding for resistance to biotic and abiotic stresses is the most sensible approach to strengthen KDML105 in lowland rainfed and irrigated areas.
Fig. 1.
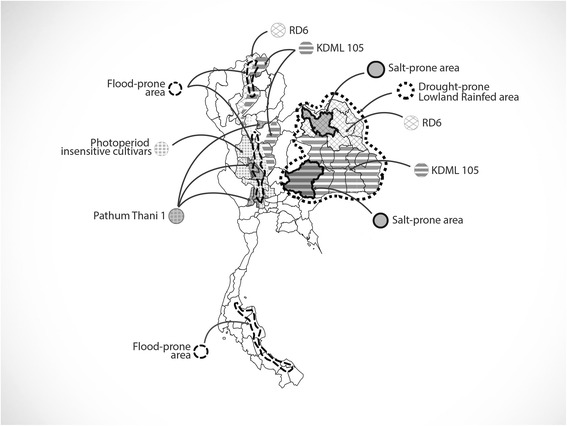
Geographical distribution of Thai Hom Mali rice, RD6, photoperiod insensitive aromatic and non-aromatic rice cultivars, in relation to abiotic stresses drought, salinity, and flooding, in the lowland rainfed area in Thailand
Cooking quality, fragrance, and interaction with environment
The superb cooking quality of KDML105 is characterized by its soft-texture, aromatic, long-slender white rice. Low amylose content, a low degree of gelatinization, and soft gel consistency are biochemical characteristics of the cooked KDML105 rice. When grown in its optimal environment, the physical appearance of polished grains is pearly, clear, very long and slender, with a glossy exterior. The dimension of brown rice is 2.1 × 7.5 × 1.8 mm (width x length x thickness), and the length/width ratio is 3.42. Its head rice yield is 58–60%.
Cooking and eating quality
Eating quality is determined by consumer preference and perception of cooked rice under an optimal cooking protocol. Cooking quality can be determined as soft-texture based on physicochemical properties including amylose content (AC), amylopectin content (AP), gelatinization temperature (GT), and gel consistency (GC) (Juliano and Perez 1984; Lanceras et al. 2000; Hsu et al. 2014). QTL mapping of the cooking quality of KDML105 was reported (Lanceras et al. 2000) in a 141 F8 recombinant inbred line (RIL) population from a cross between KDML105 and CT9993–5–10-1-M, a drought tolerant, japonica, upland rice from the Center of International Tropical Agriculture (CIAT). The major QTL on chromosome 6, known as the Waxy locus, with modifier genes on chromosomes 3, 4, and 7, determined 14–16% AC of KDML105. AC and GC were linked on QTLch6. The waxy allele of KDML105 was classified as Wxb or Wxg1 (Lanceras et al. 2000; Teng et al. 2012). Three QTLs for GT were mapped on chromosome 6 as the single major QTL, and the two minor QTLs were mapped on chromosome 2. The major QTLch6 for GT is known as the alk locus encoding soluble starch synthase IIa (SSIIa) (Umemoto et al. 2002). Grain aroma is controlled by a single recessive gene located on chromosome 8 (Singh et al. 2007; Tragoonrung et al. 1996). QTL analysis of grain aroma identified one major QTL controlling grain aroma mapped close to RG28 on chromosome 8 (Ahn et al. 1992) in Azucena (Lorieux et al. 1996), Basmati (Singh et al. 2007) and KDML rice (Tragoonrung et al. 1996). Because sensory evaluation of grain aroma is inaccurate, the discovery of genes responsible for grain aroma for MAS is the key to improving KDML105.
Discovery of the aromatic gene
The content of 2-acetyl-1-pyrroline (2AP) is the major determinant of grain aroma, as the 2AP content and the QTL of grain aroma were coincidentally mapped on the same locus on chromosome 8 (Lorieux et al. 1996). Map-based cloning of the aromatic genes in KDML105 was initiated on Os2AP isogenic lines differing in 2AP content and developed from a KDML cross (Vanavichit et al. 2004; Vanavichit et al. 2005). The responsible gene, named Os2AP, was identified as a member of gamma-aminobutylaldehyde dehydrogenase (AMADH) and functions as the metabolic switch between γ-aminobutylaldehyde (GBAL) and gamma-aminobutyric acid (GABA) in non-aromatic isogenic lines (Vanavichit et al. 2005). The crystallization of the 503-amino acid Os2AP protein from the non-aromatic isogenic line was reported (Kuaprasert et al. 2011). The structure was predicted to contain three binding domains: an NAD+ binding, an oligomerization, and a substrate binding domain where an aldehyde dehydrogenase cysteine subdomain was located (Chen et al. 2008; Wongpanya et al. 2011). In the aromatic isogenic lines and KDML105, the specific 8-bp deletion was identified within exon 7 of the AMADH of KDML105, causing a premature stop codon, non-sense-mediated decay, and non-functioning of Os2AP, respectively (Vanavichit et al. 2005). RNAi against the Os2AP in the non-aromatic Japonica rice cv. Nipponbare resulted in the suppression of AMADH expression, resulting in the accumulation of 2AP at the level of KDML105 under the same growing condition (Vanavichit et al. 2005). Similar conclusions for the gene responsible for emerging aromatic rice and its aromatic compound biosynthesis switch were reported by most of the research groups as BADH2 (Bradbury et al. 2005; Chen et al. 2008; Sun et al. 2008; Shi et al. 2008; Bradbury et al. 2008). Comparing the aromatic isogenic line and RNAi in the N15 dilution experiment and inclusion of the inhibitors of enzymes in polyamine biosynthesis revealed ornithine as the initial amino acid precursor of N, and polyamine is a major biosynthetic pathway leading to 2AP accumulation (Vanavichit et al. 2005; Vanavichit and Yoshihashi 2010).
Variation in aromatic quality
Among major aromatic rice cultivars, although they carry the same aromatic allele of AMADH, considerable variation in 2AP content was reported, for example, in Basmati rice (0.34 ppm), Jasmine rice (0.81 ppm) and Texmati rice (0.53 ppm) (Goufo et al. 2010; Gaur et al. 2016). Environmental factors will always have a strong influence on the synthesis and degradation of 2AP content, resulting in quantitative variation of 2AP in rice grains. Additionally, other volatile compounds may add flavors to the pandan-like aroma. Analysis of volatile aromatic compounds during grain ripening identified odor-active compounds (OAC), including 2AP, decanal, pentanal, phenylacetaldehyde, hexanal, (E)-2-nonenal, nonanal, heptanal, 1-octanol, 1-octen-3-ol, and 2-pentylfuran as contributors toward the unique sensory of specific rice fragrance (Hinge et al. 2016). Also, terpenoids, another class of volatile aromatic compounds found mostly in herbs, were identified in rice bran from purple, red, and brown rice varieties (Chumpolsri et al. 2015). Also, in the rice bran extracts of KDML105, major terpenoid odorants were found, such as limonene, trans-b-ocimene, b-cymene, and linalool (Chumpolsri et al. 2015). This finding may help explain the difference in the sensory evaluation of several aromatic rice varieties before and after cooking.
Geographical dependency
The accumulation of 2AP and grain qualities of aromatic, low-amylose rice is influenced by season and production area. The first report came from seven KDML rice samples collected between 2000 and 2001 from the north (N), central (C), and northeast (NE) regions of Thailand, which were quantified for grain 2AP content (Yoshihashi et al. 2004). The outcomes showed that the lowland rainfed area in the NE was the optimal environment for the accumulation of 2AP in rice grain (Yoshihashi et al. 2004). In particular, the NE had the best rice quality regarding physical appearance and cooking qualities (Anun et al. 2000). Environmental variation of cultivated areas affects the nitrogen, lipid, starch, fiber, and ash of polished rice (McCall et al. 1953), and low-amylose rice was particularly sensitive (Asaoka et al. 1989). Based on physical and cooking quality evaluation, the production area of KDML105 was classified as most favorable, favorable, or less favorable (Anun et al. 2000). Based on pasting properties rapid visco analysis (RVA) and the colorimetric properties of rice starch (Differential Scanning Colorimetry analysis), KDML105 environmental quality was classified into five groups: most favorable (one group), favorable (one group), and less favorable (three sub-groups) (Pitiphunpong and Suwannaporn 2009). In the most favorable area, “Tung-kula-rong-hai,” which comprises three provinces in central NE Thailand, was praised for the geographical distinction for overall Jasmine rice qualities, in particular, 2AP (Yoshihashi et al. 2004). Poor fertility, sandy and mildly saline soil, and a cool, dry, sunny atmosphere during the ripening stage are distinctive characteristics of the area that affect both grain quality and grain 2AP content (Yoshihashi et al. 2004).
Susceptibility to biotic stresses
KDML105 and its induced-mutation RD15 and RD6, are highly susceptible to bacterial leaf blight (BLB), leaf/neck blast and brown planthopper/whiteback hopper because of the lack of R genes (Toojinda et al. 2005). Overall, the 3.2 M ha production area for KDML105 and RD15 is considered the largest single varietal type in a lowland rainfed ecosystem and is at high risk of the BLB, BL, and BPH outbreak. In lowland rainfed conditions, blast disease caused by Pyricularia grisea has threatened KDML, RD15, and RD6 productivity at every stage of growth, but particularly during the ripening stage, called neck blast. Of all rice growing countries, Thailand has the most diversified pathotypes in the world (Sreewongchai 2008; Chaipanya et al. 2017), as the fungus mutates quickly. In October 1993, 200,000 ha of Jasmine rice was devastated during the ripening stage, when farmers have no option for mitigation (Rice Today January-March 2006). Breeding for durable blast resistance from various donors with conventional and marker-assisted selection has reduced farmers’ risk of devastation by disease and ensured the grain yield and quality of Thai Jasmine Rice. Lacking major R genes, KDML and RD15 are highly susceptible to BPH (Nilaparvata lugens Stål) in every stage of plant development (Jairin et al. 2005). Also, releasing aromatic compound 2AP from all plant parts making KDML even more risk of being devastated as 2AP may act as a possible chemical attractant to brown planthopper (BPH) (Kamolsukyunyong et al. 2013). Therefore, breeding for durable resistance to BLB, BL, and BPH of KDML is the most practical mitigation strategy to prevent crop loss.
New generation of jasmine Rice: Plus one
At the initial stage, mutation breeding utilizing gamma rays generated two important progenies: RD6, waxy Jasmine rice, and RD15, early-maturing Jasmine rice, which were released in 1977 and 1978, respectively (Rice Department). The first blast tolerant variety with a Jasmine-like quality was initiated in 1999 using IR77924–62–71-1-2 as the donor for two backcrosses to KDML, followed by pedigree selection (Supapoj et al. 2009). The elite progeny RD33, photoperiod-insensitive Jasmine rice, was released in 2006, as an alternative aromatic variety with blast resistance for NE and N Thailand (Rice Today Jan-March 2006). Marker-assisted backcrossing was comprehensively developed to add more resistance genes for diseases, insects, and abiotic stresses to KDML105 (Toojinda et al. 2005). These new generations of KDML progeny possess new traits, such as tolerance to submergence, bacterial leaf blight, and brown planthopper (Table 1). Background selection was performed using 65 well-distributed SSR markers to preserve most of the KDML105 genetic background for the adaptability and market quality of the Thai Jasmine rice (Table 2).
Table 1.
KASP SNP markers for specific gene target used for MABC for improving KDML105
| Traits | Marker Name | Chr. | QTL/gene | LGC codef |
|---|---|---|---|---|
| Aroma | Aroma_2-3e | 8 | OsBADH2 | 002–0829.1 |
| Amylose content (AC) | wx_5UTR_G/Ta | 6 | GBSS | 002–0052.1 |
| Gelatinization temperature (GT) | ALK_ex8_SNP_GC/TTa | 6 | SSIIa | 002–0049.1 |
| Bacterial leaf blight (BLB) | xa5a | 5 | xa5 | 002–0775.1 |
| Bacterial leaf blight (BLB) | SNP_P100 Xa21e | 11 | Xa21 | 002–0998.1 |
| Brown planthopper (BPH) | OsSTPS2_21bp_delb | 4 | OsSTPS2 | 002–0120.1 |
| Brown planthopper (BPH) | OsLecRK3_QBPHRe | 4 | OsLecRK3 | 002–0263.1 |
| Brown planthopper (BPH) | Bph32_2_1223332e | 6 | BPH 32 | |
| Blast (BL) | BLch11-Pikm-2c,d | 11 | NB-ARC domain containing protein | 002–0754.1 |
| Blast (BL) | TBGI055578_TC_Chr1a | 1 | secretory carrier-associated membrane protein | 002–0768.1 |
| Blast (BL) | TBGI055841a | 1 | hypothetical protein | 002–0819.1 |
| Blast (BL) | TBGI453126a | 11 | NBS-LRR disease resistance protein | 002–0820.1 |
| Blast (BL) | TBGI453598a | 11 | retrotransposon protein | 002–0821.1 |
| Blast (BL) | TBGI454069a | 11 | receptor kinase-like protein | 002–0822.1 |
| Blast (BL) | TBGI454717a | 11 | C2H2 zinc finger protein | 002–0823.1 |
| Blast (BL) | TBGI454800a | 11 | zinc finger, C3HC4 type | 002–0824.1 |
| Submergence (SUB) | Sub1A_SNP1e | 9 | Sub1A | 002–0152.1 |
| Submergence (SUB) | Sub1C_loci5a | 9 | Sub1C | 002–0995.1 |
Table 2.
Pyramiding nine target genes/QTLs into the KDML105 background by marker-assisted backcrossing illustrated in Fig. 2. Background selection with markers was conducted only in the Plus-1 and Plus-4 BILs. X= The gene/QTLs were added to the new varieties by backcrossing
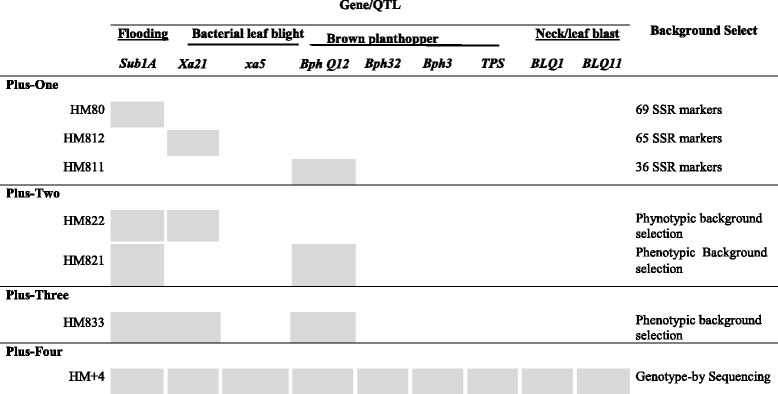
TPS Terpene synthase (OsSTPS2)
KDML-Sub1
The first marker-assisted selection line generated, HM80, was the first Sub1- Jasmine rice with flash flooding tolerance using IR49830–7–1-2-2 as the donor (Siangliw et al. 2003). This line was officially released in 2013 by the Rice Department. After the worst flooding in decades that affected Thailand in 2011, RD51 (HM80) was released for farmers in 2013. The planting area of the RD51 in 2013–14 was only 3000 ha in a flood-prone area or only 1% of the Thai Jasmine rice (KDML105 + RD15) (Table 3). Later, the HM80 was used as the general flooding donor to develop advance backcross pyramid lines (Fig. 2, Table 4).
Table 3.
Planting area of the Thai Jasmine rice (KDML105 + RD15), KDML-derived varieties, RD6, RD51, RD33, and Pathumthani 1 (PTT1), in the wet and dry seasons, 2013–14 (Unpublished Rice Department 2014)
| Varieties | Area (ha) | Total (ha) | |
|---|---|---|---|
| Wet season | Dry season | ||
| KDML105 | 3,870,560 | 3630 | 3,874,190 |
| RD15 | 238,470 | 1580 | 240,050 |
| RD6 | 1,407,250 | 390 | 1,407,640 |
| RD51 | 3170 | 290 | 3460 |
| RD33 | 110 | 2950 | 3060 |
| PTT1 | 8900 | 46,110 | 55,010 |
| Total (ha) | 5,528,460 | 54,950 | 5,583,410 |
Fig. 2.
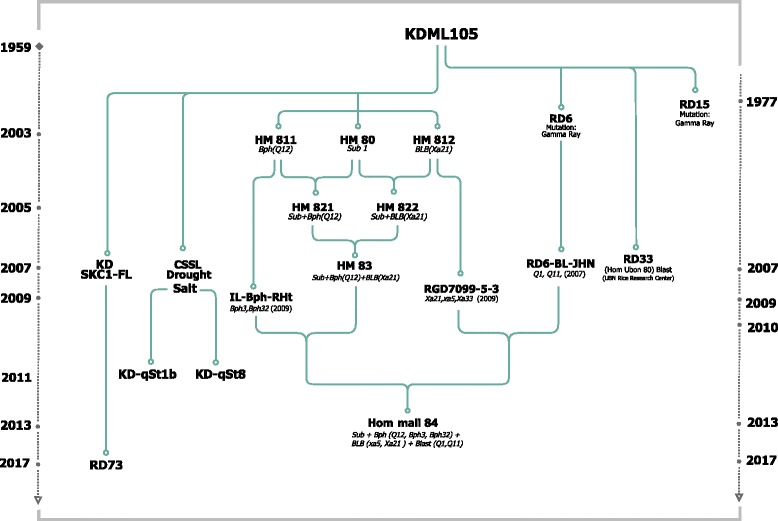
Schematic development of KDML105 from a well-adapted, local aromatic rice cultivar to new KDML-like rice varieties with resistance to submergence, bacterial leaf blight, leaf/neck blast, and brown planthopper using comprehensive marker-assisted gene/QTL pyramiding backcross breeding
Table 4.
Trait evaluation, agronomic characteristics, grain quality, and cooking quality traits of the four selected HomMali 84, KDML105, and KD EDVs (For trait evaluation methods, see Additional file 1)
| Name | HM84 progenies | KDML105 | KD EDV | ||||||
|---|---|---|---|---|---|---|---|---|---|
| KD-BLB | Plus III | RD6-Blast | KD-Rathu | ||||||
| 6_14G08 | 24_16D06 | 2_14A03 | 4_14B09 | ||||||
| aSub | 84.67 | 55.00 | 49.00 | 63.67 | 7.00 | 5.3 | 75.7 | 33.3 | 6.5 |
| BLB(4 isolates) | R | R | R | R | S | R | R | S | S |
| BPH (2 biotype) | R | R | R | MR | S | S | MR | S | R |
| Blast (8 Mixed) | R | R | R | R | S | S | S | R | S |
| bDM | 126.67 | 125.33 | 129.00 | 120.33 | 120.67 | 120.33 | 122.00 | 121.67 | 127.30 |
| NTP | 13.00 | 10.00 | 12.33 | 13.67 | 14.33 | 9.00 | 13.67 | 8.33 | 12.67 |
| PH(cm) | 112.80 | 114.77 | 112.10 | 113.97 | 98.23 | 101.33 | 113.33 | 110.23 | 99.77 |
| PSF(%) | 72.33 | 72.27 | 71.07 | 68.57 | 68.10 | 60.27 | 77.27 | 55.07 | 74.33 |
| TGW(g) | 30.03 | 29.93 | 27.80 | 27.90 | 29.93 | 29.65 | 28.89 | 26.33 | 27.97 |
| GY(kg/ha) | 2766.69 | 2887.50 | 2266.69 | 2833.31 | 2554.19 | 2360.44 | 3130.31 | 2795.81 | 2921.06 |
| cBR1/ (%) | 76.06 | 73.39 | 68.10 | 73.03 | 70.40 | 70.64 | 69.48 | 72.54 | 71.88 |
| HR1/ (%) | 62.08 | 61.26 | 55.02 | 49.00 | 57.84 | 50.44 | 51.49 | 58.14 | 58.85 |
| GL/W1/ | 3.4 | 3.16 | 3.2 | 3.29 | 3.42 | 3.24 | 3.22 | 3.11 | 3.15 |
| PRL1/ (cm) | 0.75 | 0.73 | 0.75 | 0.78 | 0.72 | 0.7 | 0.71 | 0.7 | 0.67 |
| ASV1/ | 6.0 | 6.1 | 6.0 | 6.0 | 6.0 | 2.3 | 6 | 6.1 | 6.2 |
| Amylose(%) | 16.42 | 17.7 | 14.14 | 14.55 | 17 | 16.58 | 17.21 | 8.2 | 16.94 |
a/Trait evaluation
Sub = Average % plant survival (%PS) after 15 days of flash flooding
BLB = average lesion length in centimeters of the damage caused by the BB isolate including TXO152, TXO85, TXO155 and TXO156
BPH = Severity scores with UBN biotype at 9 DAI when TN1, the susceptible control died
Blast = Average blast injury score when attacked by 8 mixed blast isolates from Thailand
b/Agronomic characteristics
DM (days to maturity), NTP (number of tillers per plant), PH (plant height from the soil surface to the neck of the panicle), PSF (percent spikelet fertility), TGW (1000-grain weight) and GY (grain yield)
c/Grain quality and cooking quality
BR (brown rice), HR (head rice), GL/W (grain length-width ratio), PRL (polished rice length), AC (amylose content; Julaino, 1971), ASV (alkaline spreading value)
EDV = Essentially-derived variety
KDML-Xa21
The introduction of the Xa21 gene from the highly resistant donor line, IR1188, into KDML105 successfully generated the first Jasmine-like BLB resistant line, HM812 (Fig. 2, Table 4). Because of the limitations of Xa21 against multiple strains of Xanthomonas oryzae (Xo) found in Thailand, xa5, xa33 (t), xa34(t), and qBB11 were introduced into the KDML105 background by MABC (Korinsak, 2009).
KDML-BphQTLCh12
More than 30 BPH R genes/quantitative trait loci (QTL) were mapped (Ling and Weilin 2016). Eight BPH R genes have been identified, including BPH14 (Du et al. 2009), BPH26 (Tamura et al. 2014), BPH3 (Liu et al. 2014), BPH29 (Wang et al. 2015), BPH9 (Zhao et al. 2016), BPH18 (Ji et al. 2016), BPH32 (Ren et al. 2016), and BPH31 (Prahalada et al. 2017). After the rigorous screening of the rice germplasm, two broad-spectrum resistant donors for BPH were identified. Abhaya (Aba), a gall midge-resistant variety from India (Kalode et al. 1993; Rao and Kandalkar 1992), and Rathu Heenati (RHt), a broad-spectrum BPH-resistant variety from Sri Lanka (Lakshminarayana and Khush 1977), were used as donors in MABC to introduce durable BPH resistance into KDML. At the initial phase, the MABC program successfully introduced the BphQTLCh12 donated from Abhaya into HM811 (Table 4).
New generation of jasmine Rice: Plus-two and plus-three
Based on HM80 (Sub1) and HM811 (BPHch12), the major QTL for flooding tolerance, Sub1, was pyramided into HM811 using MABC with extreme phenotypic selection (Korinsak et al. 2016). The first pyramid line, HM821, gained Jasmine-like physical, fragrant and cooking qualities with enhanced submergence tolerance and BPH resistance (Fig. 2). The second Plus-2 line, HM822 (Sub1A and Xa21), was combined by backcrossing HM80 to HM812 (Xa21). Following the similar MABC platform, HM821 was backcrossed to HM822 to generate the Plus-three line, HM83, in 2007 (Fig. 2).
New generation of jasmine Rice: Plus-four
Significant improvements to HM84 from the HM83 generation included introduction of new BPH, BLB, and Blast (BL) resistance into the HM83 background. This was the first generation into which the two QTLs for resistance to leaf and neck blast were introgressed into the Plus-3 background. In total, nine foreground genes/QTLs were introgressed into four Plus-4 BILs (Tables 2 and 4).
Integration of durable RHt-BPH
HM811 was utilized in several pyramiding breeding experiments until the Abhaya-derived BphQTLch12 was no longer effective against the emerging biotypes of BPH. To overcome such rapid evolution, a more durable, tolerant donor, Rathu Heenati (RHt), was used for MABC in the second phase. QTL mapping analysis of RHt revealed BphQTL chr4 (Sun et al. 2005) and BphQTLchr6 (Jairin et al. 2007), which are later referred to as Bph3 (Liu et al. 2014) and Bph32 (Ren et al. 2016), respectively. Introgression of the QTL from RHt, a high-amylose rice, into the soft, susceptible Jasmine rice background was a challenge because of the tight linkage between BphQTLch6 and the waxy gene (GBSS). The genetic distance between RM589 (BphQTLch6) and RM190 (Waxy) is 1.6 cM (Jairin et al. 2007) or 383 Kb (Jairin et al. 2009). The first step to break the tight linkage between the two loci was to generate a large F2 population between KDML105 (waxy b, bphQTLch6) and RHt (Waxy A, BphQTLch6), followed by rigorous MAS for the (waxy b, BphQTLch6) haplotype (Jairin et al. 2007). The recombinants were backcrossed to KDML by MABC with ฺBPH screening using a broad spectrum of Thai BPH biotypes (Jairin et al. 2007). For BphQTLch4, one resistant IL-BPH carried a functional sesquiterpene synthase (STPS) gene located on the long arm of chromosome 4. STPS was up-regulated by BPH infestation in the IL-BPH but with no expression in infested KDML, which carries a large mutation in its promoter region (Kamolsukyunyong et al. 2013). Analysis of monoterpenoid profiling released upon infestation from the IL-BPH and KDML revealed the differential accumulation of (E)-citral, citronellal, (E)-geraniol, β-citronellol, citronellyl acetate, and geranyl acetate in the BPH resistant IL (Pitija et al. 2014). Also, during BPH infestation, KDML105 activated the amino acid-mediated pathway, while IL-BPH-RHt activated nucleotide biosynthesis by the purine and pyrimidine compound-mediated salvage pathway (Uawisetwathana et al. 2015). Such targeted release of specific terpenoids and amino acids upon insect infestation formed the basis of antibiosis and antixenosis of the interaction between rice and BPH.
Broad-spectrum blast resistance
Leaf and neck blast QTLs were genetically mapped using two recombinant inbred line (RIL) populations derived from KDML x JHN (Noenplab et al. 2006) and JHN x IR64 (Sreewongchai et al. 2010). The two broad-spectrum resistant QTLs derived from JHN, BLQTLch1_JHN and BLQTLch11_JHN, contributed 35–43% percent variance explained (PVE) using three blast isolates. They were backcrossed using markers into RD6 (RD6-BL-JHN) (Wongsaprom et al. 2010) and were used as the donor for HM83. Gene cloning of the two QTLs revealed Pish_J and Pi7_J as BLQTLch1_JHN and BLQTLch11, respectively (Chaipanya et al. 2017). Four BLQTLs were combined into RD6 (RGD7005–164) (Noenplab et al. 2006; Wongsaprom et al. 2010; Sreewongchai et al. 2010; Korinsak 2010).
Strengthening of BLB resistance
With the rapid evolution of Xo pathotypes in recent years, screening of R genes for broad-spectrum resistance against the pathotypes in Thailand identified xa5 and Xa33 as stronger R genes compared to Xa21 (Korinsak 2009). By using five effective Xo isolates, pyramided backcross inbred lines (BIL) with the combination of xa5, Xa21, xa33(t), Xa34(t), and qBB11 on an indica variety were more effective than BIL with single R genes. Among single R genes, xa5 showed the broadest resistance followed by Xa34(t) > xa33(t), > Xa21 (Korinsak 2009). The addition of xa5 to HM83 significantly improved resistance to BLB in the lowland rainfed and irrigated areas of Thailand. Therefore, deploying the best combination of R genes may prolong the productivity of newly-developed BLB resistant KDML105 in irrigated area.
Successful pyramiding
HM84, the outcome of genetic integration of the three KDML plus-one lines, delivered stronger BPH, BLB, and BL resistance into HM83 (Fig. 2 and Table 4). Nine foreground genes/QTLs were incorporated by MABC into KDML105 in a stepwise approach, from Plus-1, Plus-2, Plus-3, and Plus-4 (Table 2). Four sub-lines of HM84 derived after background selection showed tolerance to submergence, BPH, BLB, and BL, while good cooking quality and aroma were maintained (Table 4). Under pest-free conditions, HM84 on average yielded almost equal to KDML105 (Table 4). Under flooding, BLB, BL, and BPH epidemics, HM84 showed clear advantages over the susceptible KDML105 due to the addition of resistance genes/QTLs (data not shown).
KDML-CSSL as a platform for improving drought tolerance
The limitation of map-based cloning of QTL had been the driving factor in the development of KDML105 chromosome segment substitution lines (CSSLs). The process of map-based cloning requires the development of near-isogenic lines for fine-scale mapping followed by QTL cloning. CSSL is a novel mapping population that carries a specific chromosomal segment from a donor line in the genetic background of the recurrent line. Association of QTL with a particular chromosomal segment can be performed by genetic analysis using a CSSL population; at the same time, CSSLs can be developed quickly as NIL-containing target regions/QTLs of interest. This is a good strategy to accurately study genetic regions for complex traits such as drought resistance.
A population of chromosomal segment substitution lines with a KDML105 background was developed by crossing KDML105 with IR68586-F2-CA-31 (also known as DH103) and IR68586-F2-CA-143 (DH212), which are doubled haploid lines derived from a cross between CT9993–510–1-M (CT9993), an upland japonica rice, and IR62266–42–6-2 (IR62266), an irrigated indica rice (Toojinda et al. 2011). The QTL segment in the CSSLs was originally mapped from the doubled haploid population of CT9993 and IR62266. These populations differ in potential yield, osmotic adjustment (OA), and root characteristics such as a deep, thick rooting system. This population was developed at CIAT, Colombia, and the IRRI, Philippines. Several research institutes have collaborated and used this population for the genetic study of traits associated with drought tolerance (Blum et al. 1999; Tripathy et al. 2000; Zhang et al. 2001; Babu et al. 2003; Robin et al. 2003; Nguyen et al. 2004).
The QTL study was conducted by phenotyping the 220 DH lines and parents with yield, yield components and agronomic traits under control and drought conditions. A population of 154 DH lines was randomly selected from the full set used in developing the genetic map (Zhang et al. 2001), which was used to identify the QTL-controlling traits above. Five chromosomal segments, namely, chromosomes 1, 3, 4, 8 and 9, were identified carrying several overlapping QTL for drought resistance traits covering 49, 15, 53, 60 and 30 cM of the chromosomes, respectively (Lanceras et al. 2004). These regions were introgressed into KDML105 by MABC and selfing until BC3F3, and lines carrying the QTL segments were selected using SSR markers in every generation (Siangliw et al. 2007). Further backcrossing was advanced into BC5, and purification for homozygosity at the QTL regions was accomplished in the F2, F3, and F4 generations. In total, 135 KDML105 CSSLs were developed, comprised of 30, 22, 41, 31 and 15 lines carrying QTL in chromosomes 1, 3, 4, 8 and 9, respectively (Toojinda et al. 2011; Kanjoo 2012). These isoQTL lines can be used to develop KDML Plus-5 for greater drought tolerance shortly.
Adapting KDML to high-saline soil
The SKC1 gene on chromosome 1, an HKT-type transporter (OsHKT1;5), was identified as a controller of shoot K content related to salinity tolerance in rice (Gregorio et al. 2002, Lin et al. 2004; Ren et al. 2005; Thomson et al. 2010). Introgression of SKC1 from FL496 (IR66946-3R-196-1-1) or FL530 (IR66496-3R-230-1-1) into KDML105 was accomplished using MABC to generate KD-SKC1-FL (Fig. 2). The improved KD-SKC1-FL showed greater tolerance, with a lower Na/K ratio and higher yield under salt stress at 10–12 ds m− 1 (Punyawaew et al. 2016). One elite line was released to farmers under the name “RD73” by the Thailand Rice Department in 2017. In addition to SKC1, qSt1b, located on the lower region of chromosome 1, significantly improved photosynthesis efficiency with less injury under salinity stress (Siangliw et al. 2014; Kanjoo et al. 2011; Thomson et al. 2010; Chutimanukul et al. 2013). The third QTL, qSt8, identified on chromosome 8, protected rice plants from salinity stress. KD-CSSL-qST1/8 carrying qSt1b and qSt8 shows more tolerance under salt stress at 10–14 dS m− 1 (Kanjoo et al. 2011, Chutimanukul et al. 2013, Nounjan et al. 2016). Therefore, pyramiding of SKC1, qSt1, and qSt8 may enhance productivity under salt stress.
Conclusions
The 58 years-old KDML105, selected from a widely adapted aromatic landrace found in the lowland rainfed area, has become an iconic rice cultivar since 1959. To continue its productivity into the next century, KDML was rigorously and carefully enriched for new traits using mutation, conventional breeding, and MABC while conserving its superb cooking quality and adaptive advantages in the lowland rainfed area. After 13 years of MABC, new generations of KDML backcross inbred lines were carefully developed by pyramiding six QTLs with six gene-specific alleles into KDML105. Salt tolerance KDML Plus-1 and Drought tolerance CSSLs are integrating into the HM84. This newly emerging HM84 will have advantages over KDML105 in that affected area where bacterial leaf blight, blast, brown planthopper, and flooding are problems. With these adaptive advantages and superb cooking quality, the innovative KDML 105 will be more productive in the subsistence and pesticide-free lowland rainfed area.
Additional file
Evaluation of abiotic and biotic stress traits and recording of important agronomic traits method. (DOCX 22 kb)
Acknowledgements
This work was supported by the National Center for Genetic Engineering and Biotechnology (BIOTEC) and National Science and Technology Development Agency (NSTDA), NSTDA Research Chair (Grant No. P12-01898), Agriculture Research Development Agency (ARDA) (Grant No. P12/2552), Rice Department and Kasetsart University.
Funding
National Center for Genetic Engineering and Biotechnology (BIOTEC) and National. Science and Technology Development Agency (NSTDA)(NSTDA Research Chair (Grant No. P12–01898)). Agriculture Research Development Agency (ARDA) (Grant No. P12/2552).
Availability of data and materials
Not applicable
Abbreviations
- 2AP
2-acetyl-1-pyrroline
- AC
amylose content
- AP
amylopectin content
- BL
blast
- BLB
bacterial leaf blight
- BPH
brown planthopper
- C
Central
- CSSLs
chromosome segment substitution lines
- EDV
Essentially-derived variety
- GC
gel consistency
- GT
gelatinization temperature
- HM
Hom Mali
- KDML105
Khao Dawk Mali 105
- MABC
marker-assisted backcross selection
- N
North
- NE
North-East
- PVE
percent variance explained
- RHt
Rathu Heenati
- RIL
recombinant inbred line
- RVA
rapid visco analysis
- Xo
Xanthomonas oryzae
Authors’ contributions
Conceived and writer: AP. Review BPH: WK. Review BLB, BL and MABC: SR and EP. Review drought and salinity: JLS and MS. Field data collection: SuT. Performed bioinformatics and statistical analysis: CS and EC. MABC design: TT. Review KDML history and statistics: SoT. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12284-018-0212-7) contains supplementary material, which is available to authorized users.
Contributor Information
Apichart Vanavichit, Email: vanavichit@gmail.com.
Wintai Kamolsukyeunyong, Email: wtkmsyy@gmail.com.
Meechai Siangliw, Email: meechai.siangliw@gmail.com.
Jonaliza L. Siangliw, Email: jsiangliw@gmail.com
Suniyom Traprab, Email: suniyomt@yahoo.com.
Siriphat Ruengphayak, Email: phatsterster@gmail.com.
Ekawat Chaichoompu, Email: ekawatun@gmail.com.
Chatree Saensuk, Email: chatsaen@gmail.com.
Ekapol Phuvanartnarubal, Email: banky31@gmail.com.
Theerayut Toojinda, Email: toojindatheerayut@gmail.com.
Somvong Tragoonrung, Email: somvong.tra@gmail.com.
References
- Ahn SN, Bollich CN, Tanksley SD. RFLP tagging of a gene for aroma in rice. Theor Appl Genet. 1992;84:825–828. doi: 10.1007/BF00227391. [DOI] [PubMed] [Google Scholar]
- Anun D, Suthep L, Boriboon S, Vassana V, Ngamchuen K. Research of Rice and Rice products research program, Department of Agriculture, Bangkok. 2000. Characterization of Khao Dawk Mali 105 and RD15 rice cultivated area in Thailand. [Google Scholar]
- Arunin S. Ecology and management of problem soils in Asia. Taipei, Rep. of China: Food and Fertilizer Technology Center for the Asian and Pacific Region; 1984. Characteristics and management of salt-affected soil in the northeast of Thailand. [Google Scholar]
- Arunin S, Pongwichian P. Salt-affected soils and management in Thailand. Bull Soc Sea Water Sci Jpn. 2015;69(5):319–325. [Google Scholar]
- Asaoka M, Okuno K, Hara K, Oba M, Fuwa H. Effects of environmental temperature at the early developmental stage of seeds on the characteristics of endosperm starches of rice (Oryza sativa L.) Denpun Kagaku. 1989;36:1–8. [Google Scholar]
- Ashikawa I, Hayashi N, Abe F, Wu J, Matsumoto T. Characterization of the rice blast resistance gene Pik cloned from Kanto 51. Mol Breed. 2012;30:485–494. doi: 10.1007/s11032-011-9638-y. [DOI] [Google Scholar]
- Babu RC, Nguyen BD, Chamarerk V, Shanmugasundaram P, Chezhian P, Jeyaprakash P, Ganesh SK, Palchamy A, Sadasivam S, Sarkarung S, Wade LJ, Nguyen HT. Genetic analysis of drought resistance in rice by molecular markers: association between secondary traits and field performance. Crop Sci. 2003;43:1457–1469. doi: 10.2135/cropsci2003.1457. [DOI] [Google Scholar]
- Banco DM, Yamauchi A, Kamoshita A, Wade LJ, Pardales JR., Jr Dry matter production and root system development of rice cultivars under fluctuating soil moisture. Plant Prod Sci. 2000;3(2):197–207. doi: 10.1626/pps.3.197. [DOI] [Google Scholar]
- Blum A, Mayer J, Golan G, Sinmena B. Drought tolerance of a doubled-haploid line population if rice in the field. In: Ito O, O’Toole J, Hardy B, editors. Genetic improvement of rice for water-limited environments. Los Banos, Philippines: International Rice Research Institute; 1999. pp. 319–330. [Google Scholar]
- Bradbury LMT, Fitzgerald TL, Henry RJ, Jin Q, Waters DL. The gene for fragrance in rice. Plant Biotechnol J. 2005;3:363–370. doi: 10.1111/j.1467-7652.2005.00131.x. [DOI] [PubMed] [Google Scholar]
- Bradbury LMT, Gillies SA, Brushett DJ, Waters DLE, Henry RJ. Inactivation of an aminoaldehyde dehydrogenase is responsible for fragrance in rice. Plant Mol Biol. 2008;68:439–449. doi: 10.1007/s11103-008-9381-x. [DOI] [PubMed] [Google Scholar]
- Bureau of Rice Research and Development (2010) Khao Dawk Mali 105. Rice Department, Bangkok. ISBN: 978–974–403-692-6
- Chaipanya C, Jeanie M, Yanoria T, Quime B, Longya A, Korinsak SP, Korinsak S, Toojinda T, Vanavichit A, Jantasuriyara C, Zhou B. Dissection of broad-spectrum resistance of the Thai rice variety Jao Hom Nin conferred by two resistance genes against rice blast. Rice. 2017;10:18. doi: 10.1186/s12284-017-0159-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Yang Y, Shi W, Ji Q, He F, Zhang Z, Cheng Z, Liu X, Xu M. Badh2, encoding betaine aldehyde dehydrogenase, inhibits the biosynthesis of 2-acetyl-1-pyrroline, a major component in rice fragrance. Plant Cell. 2008;20:1850–1861. doi: 10.1105/tpc.108.058917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumpolsri W, Wijit N, Boontakham P, Nimmanpipug N, Sookwong P, Luangkamin S, Wongpornchai S. Variation of terpenoid flavor odorants in the bran of some black and white rice varieties analyzed by GC×GC-MS. J Food Nutr Res. 2015;3(2):114–120. doi: 10.12691/jfnr-3-2-7. [DOI] [Google Scholar]
- Chutimanukul P, Chaidee A, Buaboocha T, Siangliw M, Toojinda T, Chadchawan S, Kositsup B (2013) Effect of salt stress on photosynthesis and growth in salt-tolerant rice lines obtained from a CSSL population. Thai J Genet S 1:276–279
- Department of Foreign Trade (2014) Rice in Thailand: Agriculture, history what makes jasmine rice so special. [cited 2017 Oct 18]. Available from: http://factsanddetails.com/southeast-asia/Thailand/sub5_8h/entry-3321.html
- Du B, Zhang W, Liu B, Hu J, Wei Z, Shi Z, He R, Zhu L, Chen R, Han B, He G. Identification and characterization of Bph14, a gene conferring resistance to brown planthopper in rice. Proc Natl Acad Sci U S A. 2009;106:22163–22168. doi: 10.1073/pnas.0912139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur A, Shabir Wani H, Pandita D, Bharti N, Malav A, Shikari A, Bhat A. Understanding the fragrance in rice. J Rice Res. 2016;4:e125. doi: 10.4172/2375-4338.1000e125. [DOI] [Google Scholar]
- Goufo P, Duan M, Wongpornchai S, Tang X. Some factors affecting the concentration of the aroma compound 2-acetyl-1-pyrroline in two fragrant rice cultivars grown in South China. Front Agric China. 2010;4:1–9. doi: 10.1007/s11703-009-0087-x. [DOI] [Google Scholar]
- Gregorio GB, Senadhira D, Mendoza RD, Manigbas NL, Roxas JP, Guerta CQ. Progress in breeding for salinity tolerance and associated abiotic stresses in rice. Field Crops Res. 2002;76:91–101. doi: 10.1016/S0378-4290(02)00031-X. [DOI] [Google Scholar]
- Hinge VR, Patil HB, Nadaf AB. Aroma volatile analyses and 2AP characterization at various developmental stages in basmati and non-basmati scented rice (Oryza sativa L.) cultivars. Rice. 2016;9:38. doi: 10.1186/s12284-016-0113-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YC, Tseng MC, Wu YP, Lin MY, Wei FJ, Hwu KK, Hsing YI, Lin YR. Genetic factors responsible for eating and cooking qualities of rice grains in a recombinant inbred population of an inter-subspecific cross. Mol Breed. 2014;34:655–673. doi: 10.1007/s11032-014-0065-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im-Erb R, Neawsuparb K, Sombatpanit S. Soil salinization assessment and monitoring at Boe Klue District, Nan Province, northern Thailand. In: Shahid S, Abdelfattah M, Taha F, editors. Developments in soil salinity assessment and reclamation. Dordrecht: Springer; 2013. pp. 75–86. [Google Scholar]
- Jairin J, Phengrat K, Teangdeerith S, Vanavichit A, Toojinda T. Mapping of a broad-spectrum brown planthopper resistance gene, Bph3, on rice chromosome 6. Mol Breed. 2007;19:35–44. doi: 10.1007/s11032-006-9040-3. [DOI] [Google Scholar]
- Jairin J, Teangdeerith S, Leelagud P, Kothcharerk J, Sansen K, Yi M, Vanavichit A, Toojinda T. Development of rice introgression lines with brown planthopper resistance and KDML105 grain quality characteristics through marker-assisted selection. Field Crops Res. 2009;110:263–271. doi: 10.1016/j.fcr.2008.09.009. [DOI] [Google Scholar]
- Jairin J, Toojinda T, Tragoonrung S, Tayapat S, Vanavichit A. Multiple genes determining brown planthopper (Nilaparvata lugens Stål) resistance in backcross introgressed lines of Thai jasmine rice ‘KDML105’. ScienceAsia. 2005;31:129–135. doi: 10.2306/scienceasia1513-1874.2005.31.129. [DOI] [Google Scholar]
- Ji H, Kim S-R, Kim Y-H, Suh J-P, Park H-M, Sreenivasulu N, Misra G, Kim S-M, Hechanova S-L, Kim H, Lee G-S, Yoon U-H, Kim T-H, Lim H, Suh S-C, Yang J, An G, Jena KK. Map-based cloning and characterization of the bph18 gene from wild rice conferring resistance to brown planthopper (bph) insect pest. Sci Rep. 2016;6:34376. doi: 10.1038/srep34376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongdee B, Pantuwan G, Fukai S, Fischer K. Improving drought tolerance in rainfed lowland rice: an example from Thailand. Agric Water Manag. 2006;80:225–240. doi: 10.1016/j.agwat.2005.07.015. [DOI] [Google Scholar]
- Juliano BO, Perez CM. Result of a collaborative test on the measurement of grain elongation of milled rice during cooking. J Cereal Sci. 1984;2:281–292. doi: 10.1016/S0733-5210(84)80016-8. [DOI] [Google Scholar]
- Kalode MB, Bentur JS, Prasada Rao U. Rice cultures resistant to rice gall midge (GM) biotypes 1 and four under artificial infestation in the greenhouse. Int Rice Res Newsl. 1993;10(2):17–18. [Google Scholar]
- Kamolsukyunyong W, Sukhaket W, Ruanjaichon V, Toojinda T, Vanavichit A. Single-feature polymorphism mapping of isogenic rice lines identify the influence of terpene synthase on brown planthopper feeding preferences. Rice. 2013;6:18. doi: 10.1186/1939-8433-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanjoo V (2012) Development of chromosome segment substitution lines related to drought tolerance in rice (Oryza sativa L.). Dissertation, Agricultural Biotechnology, Graduate School, Kasetsart University, p108
- Kanjoo V, Jearakongman K, Punyawaew K, Siangliw JL, Siangliw M, Vanavichit A, Toojinda T. Co-location of quantitative trait loci for drought and salinity tolerance in rice. Thai J Genet. 2011;4(2):126–138. [Google Scholar]
- Kano-Nakata M, Gowda VRP, Henry A, Serraj R, Inukai Y, Fujita D, Kobayashi N, Suralta RR, Yamauchi A. Functional roles of the plasticity of root system development in biomass production and water uptake under rainfed lowland conditions. Field Crop Res. 2013;144:288–296. doi: 10.1016/j.fcr.2013.01.024. [DOI] [Google Scholar]
- Korinsak S (2009) Marker-assisted pyramiding bacterial blight resistance genes (xa5, Xa21, xa33(t), Xa34(t) and qBB11) in rice. Dissertation, Kasetsart University
- Korinsak S, Siangliw M, Kotcharerk J, Jairin J, Siangliw JL, Jongdee B, Pantuwan G, Sidthiwong N, Toojinda T. Improvement of the submergence tolerance and the brown planthopper resistance of the Thai jasmine rice cultivar KDML105 by pyramiding Sub1 and Qbph12. Field Crop Res. 2016;188:105–112. doi: 10.1016/j.fcr.2015.10.025. [DOI] [Google Scholar]
- Korinsak SP (2010) Identification of blast resistance QTLs in two rice RIL populations and marker assisted selection for pyramiding of four QTLs in RD6 rice variety. Dissertation, Kasetsart University
- Kuaprasert B, Silprasit K, Horata N, Khunrae P, Wongpanya R, Boonyalai N, Vanavichit A, Choowongkomon K. Purification, crystallization and preliminary X-ray analysis of recombinant betaine aldehyde dehydrogenase 2 (OsBADH2), a protein involved in jasmine aroma, from Thai fragrant rice (Oryza sativa L.) Acta Cryst. 2011;67:1221–1223. doi: 10.1107/S1744309111030971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumboonreang N (2011) Effect of light duration on panicle initiation and advantages of off-season growing KDML105 rice using short day period. Dissertation, Suranaree University of Technology
- Lakshminarayana A, Khush GS. New genes for resistance to the brown planthopper in rice. Crop Sci. 1977;17:96–100. doi: 10.2135/cropsci1977.0011183X001700010028x. [DOI] [Google Scholar]
- Lanceras J, Huang ZL, Naivikul O, Vanavichit A, Ruanjaichon V, Tragoonrung S. Mapping of genes for cooking and eating qualities in Thai jasmine rice (KDML105) DNA Res. 2000;7:93–101. doi: 10.1093/dnares/7.2.93. [DOI] [PubMed] [Google Scholar]
- Lanceras J, Pantuwan G, Jongdee B, Toojinda T. Quantitative trait loci associated with drought tolerance at reproductive stage in rice. Plant Physiol. 2004;135:384–399. doi: 10.1104/pp.103.035527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LGCgroup (2015) KASP genotyping technology. https://www.lgcgroup.com/products/kasp-genotyping-chemistry/#.Wrhk3dR94_4
- Lin HX, Zhu MZ, Yano M, Gao JP, Liang ZW, Su WA, Hu XH, Ren ZH, Chao DY. QTLs for Na+ and K+ uptake of the shoots and roots controlling rice salt tolerance. Theor Appl Genet. 2004;108:253–260. doi: 10.1007/s00122-003-1421-y. [DOI] [PubMed] [Google Scholar]
- Ling Y, Weilin Z. Genetic and biochemical mechanisms of rice resistance to planthopper. Plant Cell Rep. 2016;35:1559–1572. doi: 10.1007/s00299-016-1962-6. [DOI] [PubMed] [Google Scholar]
- Liu Y, Wu H, Chen H, Liu Y, He J, Kang H, Sun Z, Pan G, Wang Q, Hu J, Zhou F, Zhou K, Zheng X, Ren Y, Chen L, Wang Y, Zhao Z, Lin Q, Wu F, Zhang X, Guo X, Cheng X, Jiang L, Wu C, Wang H, Wan J. A gene cluster encoding lectin receptor kinases confers broad-spectrum and durable insect resistance in rice. Nat Biotechnol. 2014;33:301–305. doi: 10.1038/nbt.3069. [DOI] [PubMed] [Google Scholar]
- Lorieux M, Petrov M, Huang N, Guiderdoni E, Ghesquière A. Aroma in rice: genetic analysis of a quantitative trait. Theor Appl Genet. 1996;93:1145–1151. doi: 10.1007/BF00230138. [DOI] [PubMed] [Google Scholar]
- McCall ER, Jurgens JF, Hoffpauir CL, Pons WA, Stark SM, Cucullu AF, Heinzelman DC, Cirino VO, Murray MD. Composition of rice: influence of variety and environment on physical and chemical composition. J Agric Food Chem. 1953;1:988–992. doi: 10.1021/jf60016a003. [DOI] [Google Scholar]
- Nguyen TT, Klueva N, Chamareck V, Aarti A, Magpantay G, Millena AC, Pathan MS, Nguyen HT. Saturation mapping of QTL regions and identification of putative candidate genes for drought tolerance in rice. Mol Gen Genomics. 2004;272:35–46. doi: 10.1007/s00438-004-1025-5. [DOI] [PubMed] [Google Scholar]
- Noenplab A, Vanavichit A, Toojinda T, Sirithunya P, Tragoonrung S, Sriprakhon S, Wongsaprom C. QTL mapping for leaf and neck blast resistance in Khao Dawk Mali 105 and Jao horn Nin recombinant inbred lines. ScienceAsia. 2006;32(2):133–142. doi: 10.2306/scienceasia1513-1874.2006.32.133. [DOI] [Google Scholar]
- Nounjan N, Siangliw JL, Toojinda T, Chadchawan S, Theerakulpisut P. Salt-responsive mechanisms in chromosome segment substitution lines of rice (Oryza sativa L. cv. KDML105) Plant Physiol Biochem. 2016;103:96–105. doi: 10.1016/j.plaphy.2016.02.038. [DOI] [PubMed] [Google Scholar]
- O’Toole JC, Bland WL. Genotypic variation in crop plant root systems. Adv Agron. 1987;41:91–145. doi: 10.1016/S0065-2113(08)60803-2. [DOI] [Google Scholar]
- Pitija K, Kamolsukyumyong W, Vanavichit A, Sookwong P, Mahatheeranont S. Monoterpenoid allelochemicals in resistance rice varieties against brown planthoppers, Nilaparvatalugens (Stål) JOAAT. 2014;1(2):82–88. doi: 10.12720/joaat.1.2.82-88. [DOI] [Google Scholar]
- Pitiphunpong S, Suwannaporn P. Physicochemical properties of KDML105 rice cultivar from different cultivated locations in Thailand. J Sci Food Agric. 2009;89:2186–2190. doi: 10.1002/jsfa.3701. [DOI] [Google Scholar]
- Prahalada GD, Shivakumar N, Lohithaswa HC, Sidde Gowda DK, Ramkumar G, Kim S-R, et al. Identification and fine mapping of a new gene, BPH31 conferring resistance to brown planthopper biotype 4 of India to improve rice, Oryza sativa L. Rice. 2017;10(1):41. doi: 10.1186/s12284-017-0178-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punyawaew K, Suriya-arunroj D, Siangliw M, Thida M, Lanceras-Siangliw J, Fukai S, Toojinda T. Thai jasmine rice cultivar KDML105 carrying Saltol QTL exhibiting salinity tolerance at seedling stage. Mol Breed. 2016;36:150. doi: 10.1007/s11032-016-0574-8. [DOI] [Google Scholar]
- Rao PSP, Kandalkar HG. Identification of a new Asian rice gall midge (GM) population in Bhandara district Maharashtra, India, and highly resistant genotypes. Int Rice Res Notes. 1992;17:9–10. [Google Scholar]
- Ren J, Gao F, Wu X, Zeng L, Lv J, Su X, Luo H, Ren G. BPH32, a novel gene encoding an unknown SCR domain-containing protein, confers resistance against the brown planthopper in rice. Sci Rep. 2016;6:37645. doi: 10.1038/srep37645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren ZH, Gao JP, Li LG, Cai XL, Huang W, Chao DY, Zhu MZ, Wang ZY, Luan S, Lin HX. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet. 2005;37:1141–1146. doi: 10.1038/ng1643. [DOI] [PubMed] [Google Scholar]
- Rice Knowledge Bank, IRRI. Salinity. http://www.knowledgebank.irri.org/decision-tools/rice-doctor/rice-doctor-fact-sheets/item/salinity
- Rice Today (2006) Improving the sacred. January–March, 2006. https://www.scribd.com/document/98480901/RT-Vol-5-No-1-All-in-the-genes
- Robin S, Pathan MS, Courtois B, Lafitte R, Carandang S, Lanceras JL, Amante M, Nguyen HT, Li Z. Mapping osmotic adjustment in an advanced back-cross inbred population of rice. Theor Appl Genet. 2003;107:1288–1296. doi: 10.1007/s00122-003-1360-7. [DOI] [PubMed] [Google Scholar]
- Ruengphayak S, Chaichumpoo E, Phromphan S, Kamolsukyunyong W, Sukhaket W, Phuvanartnarubal E, Korinsak S, Korinsak S, Vanavichit A. Pseudo-backcrossing design for rapidly pyramiding multiple traits into a preferential rice variety. Rice. 2015;8:7. doi: 10.1186/s12284-014-0035-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Yang Y, Chen S, Xu M. Discovery of a new fragrance allele and the development of functional markers for the breeding of fragrant rice varieties. Mol Breed. 2008;22:185–192. doi: 10.1007/s11032-008-9165-7. [DOI] [Google Scholar]
- Siangliw JL, Jongdee B, Pantuwan G, Toojinda T. Developing KDML105 backcross introgression lines using marker-assisted selection for QTL associated with drought tolerance in rice. ScienceAsia. 2007;33:207–214. doi: 10.2306/scienceasia1513-1874.2007.33.207. [DOI] [Google Scholar]
- Siangliw M, Phatanathara A, Toojinda T, Theerakulpisut P, Fukai S, Vanavichit A. The 4th international Rice congress (IRC2014), Bangkok, Thailand October 27 – November 1, 2014. 2014. QTL and candidate gene identification for salt tolerance and Na/K ratio at seedling stage under modified soil and hydroponic conditions in rice (Oryza Sativa L.) [Google Scholar]
- Siangliw M, Toojinda T, Tragoonrung S, Vanavichit A. Thai jasmine rice carrying QTLch9 is submergence tolerance. Ann Bot. 2003;91:255–261. doi: 10.1093/aob/mcf123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Singh RK, Sharma TR, Singh A, Singh NK. Fine mapping of aroma QTLs in basmati rice (Oryza sativa L) on chromosomes 3, 4 and 8. J Plant Biochem Biot. 2007;16:75–82. doi: 10.1007/BF03321978. [DOI] [Google Scholar]
- Sreewongchai T (2008) Identification of Magnaporthe grisea avirulence genes specific to rice blast resistance genes. Dissertation, Kasetsart University
- Sreewongchai T, Toojinda T, Thanintorn N, Kosawang C, Vanavichit A, Tharreau D, Sirithunya P. Development of elite indica rice lines with wide spectrum of resistance to Thai blast isolates by pyramiding multiple resistance QTLs. Plant Breed. 2010;129:176–180. doi: 10.1111/j.1439-0523.2009.01669.x. [DOI] [Google Scholar]
- Sun L, Su C, Wang C, Zhai H, Wan J. Mapping of a major resistance gene to the brown planthopper in the rice cultivar Rathu Heenati. Breed Sci. 2005;55:391–396. doi: 10.1270/jsbbs.55.391. [DOI] [Google Scholar]
- Sun SH, Gao FY, Lu XJ, Wu XJ, Wang XD, Ren GJ, Luo H. Genetic analysis and gene fine mapping of aroma in rice (Oryza sativa L, Cyperales, Poaceae) Genet Mol Biol. 2008;31:532–538. doi: 10.1590/S1415-47572008000300021. [DOI] [Google Scholar]
- Supapoj N, Boonyawit C, Jongdee B, Varavat O, Chamarerk V, Phengrat J (2009) RD33 (Hawm Ubon 80) rice variety. Thai Rice Research J 3(2):20-38
- Tamura Y, Hattori M, Yoshioka H, Yoshioka M, Takahashi A, Wu J, Sentoku N, Yasui H. Map-based cloning and characterization of a brown planthopper resistance gene BPH26 from Oryza sativa L. ssp. indica cultivar ADR52. Sci Rep. 2014;4:5872. doi: 10.1038/srep05872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng B, Zeng R, Wang Y, Liu Z, Zhang Z, Zhu H, et al. Detection of allelic variation at the Wx locus with single-segment substitution lines in rice (Oryza sativa L.) Mol Breed. 2012;30:583–595. doi: 10.1007/s11032-011-9647-x. [DOI] [Google Scholar]
- Thomson MJ, de Ocampo M, Egdane J, Rahman MA, Sajise AG, Adorada DL, Tumimbang-Raiz E, Blumwald E, Seraj ZI, Singh RK, Gregorio GB, Ismail AM. Characterizing the Saltol quantitative trait locus for salinity tolerance in rice. Rice. 2010;3:148–160. doi: 10.1007/s12284-010-9053-8. [DOI] [Google Scholar]
- Toojinda T, Siangliw JL, Punyawaew K, Kanjoo V. Research rep. National Center for genetic engineering and biotechnology. Pathumthani, Thailand: NSTDA; 2011. Development of single QTL near-isogenic lines (NILs) of KDML105 for drought tolerance. [Google Scholar]
- Toojinda T, Tragoonrung S, Vanavichit A, Siangliw JL, Pa-In N, Jantaboon J, Siangliw M, Fukai S. Molecular breeding for rainfed lowland rice in the Mekhong region. Plant Prod Sci. 2005;8:330–333. doi: 10.1626/pps.8.330. [DOI] [Google Scholar]
- Tragoonrung S, Sheng JQ, Vanavichit A (1996) Tagging an aromatic gene in lowland rice using bulk segregant analysis. In: IRRI (eds) Rice Genetics III. Proc 3rd Int. Rice Genet. Symp., Manila, Oct 16–20, 1995, pp 613–618
- Tripathy JN, Zhang J, Robin S, Nguyen TT, Nguyen HT. QTLs for cell-membrane stability mapped in rice (Oryza sativa L.) under drought stress. Theor Appl Genet. 2000;100:1197–1202. doi: 10.1007/s001220051424. [DOI] [Google Scholar]
- Uawisetwathana U, Graham S, Kamolsukyunyong W, Sukhaket W, Klanchui A, Toojinda T, Vanavichit A, Karoonuthaisiri N, Elliott C. Quantitative 1H NMR metabolome profiling of Thai jasmine rice (Oryza sativa) reveals primary metabolic response during brown planthopper infestation. Metabolomics. 2015;11:1640–1655. doi: 10.1007/s11306-015-0817-4. [DOI] [Google Scholar]
- Umemoto T, Yano M, Satoh H, Shomura A, Nakamura Y. Mapping of a gene responsible for the difference in amylopectin structure between Japonica-type and indica-type rice varieties. Theor Appl Genet. 2002;104:1–8. doi: 10.1007/s001220200000. [DOI] [PubMed] [Google Scholar]
- USDA (2013) Bibliography on salt tolerance. Fibres, grains and special crops. Riverside, CA: Brown GE, Jr. Salinity Lab. US Department Agriculture, Agriculture Research Service
- Vanavichit A, Kamolsukyurnyong W, Wanchana S, Wongpornchai S, Ruengphayak S, Toojinda T, Tragoonrung S. Proceedings the 1st international conference on Rice for the future, Kasetsart University, Bangkok, Thailand, 31 august – 3 September 2004. 2004. Discovering genes for rice grain aroma. [Google Scholar]
- Vanavichit A, Yoshihashi T. Molecular aspects of fragrance and aroma in rice. Adv Bot Res. 2010;56:49–73. doi: 10.1016/B978-0-12-381518-7.00002-9. [DOI] [Google Scholar]
- Vanavichit A, Yoshihashi T, Wanchana S, Areekit S, Saengsraku D, Kamolsukyunyong W J Lanceras J, Toojinda T, Tragoonrung S (2005) Positional cloning of Os2AP, the aromatic gene controlling the biosynthetic switch of 2-acetyl-1-pyrroline and gamma-aminobutyric acid (GABA). In: 5th International Rice Genetics Symposium. Manila: IRRI; p. 44.
- Varinruk B. Strengthening APEC cooperation on food security and climate change workshop, Hilton Hanoi opera hotel, Hanoi, Veit Nam 19–21 April 2017. 2017. Thailand rice production and rice R&D on climate change. [Google Scholar]
- Wang Y, Cao L, Zhang Y, Cao C, Liu F, Huang F, Qiu Y, Li R, Lou X. Map-based cloning and characterization of BPH29, a B3 domain-containing recessive gene conferring brown planthopper resistance in rice. J Exp Bot. 2015;66:6035–6045. doi: 10.1093/jxb/erv318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongpanya R, Boonyalai N, Thammachuchourat N, Horata N, Arikit S, Myint KM, Vanavichit A, Choowongkomon K. Biochemical and enzymatic study of rice BADH wild-type and mutants: an insight into fragrance in rice. Protein J. 2011;30:529–538. doi: 10.1007/s10930-011-9358-5. [DOI] [PubMed] [Google Scholar]
- Wongsaprom C, Sirithunya P, Vanavichit A, Pantuwan G, Jongdee B, Sidhiwong N, Lanceras-Siangliw J, Toojinda T. Two introgressed quantitative trait loci confer broad-spectrum resistance to blast disease in the genetic background of the cultivar RD6 Thai glutinous jasmine rice. Field Crops Res. 2010;119:245–251. doi: 10.1016/j.fcr.2010.07.013. [DOI] [Google Scholar]
- Yoshihashi T, Nguyen TT, Kabaki N. Area dependency of 2-acetyl-1-pyrroline content in an aromatic rice variety, Khao Dawk Mali 105. JARQ. 2004;38:105–109. doi: 10.6090/jarq.38.105. [DOI] [Google Scholar]
- Zhang J, Zheng HG, Aarti A, Pantuwan G, Nguyen TT, Tripathy JN, Sarial AK, Robin S, Babu RC, Nguyen BD, Sarkarung S, Blum A, Nguyen HT. Locating genomic regions associated with components of drought resistance in rice: comparative mapping within and across species. Theor Appl Genet. 2001;103:19–29. doi: 10.1007/s001220000534. [DOI] [Google Scholar]
- Zhao Y, Huang J, Wang Z, Jing S, Wang Y, Ouyang Y, Cai B, Xin X-F, Liu X, Zhang C, Pan Y, Ma R, Li Q, Jiang W, Zeng Y, Shangguan X, Wang H, Du B, Zhu LXX, Feng Y-Q, He SY, Chen R, Zhang Q, He G. Allelic diversity in an NLR gene BPH9 enables rice to combat planthopper variation. Proc Natl Acad Sci U S A. 2016;113:12850–12855. doi: 10.1073/pnas.1614862113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Evaluation of abiotic and biotic stress traits and recording of important agronomic traits method. (DOCX 22 kb)
Data Availability Statement
Not applicable


