Abstract
The genome of a fungal strain Penicillium chrysogenum strain HKF42, which can grow on 20% sucrose has been annotated for 7595 protein coding sequences. On mining of CAZymes, we could annotate a β-fructofuranosidase gene responsible for fructo-oligosaccharides (FOS) synthesis which is a known prebiotic. The enzyme activity was demonstrated and validated with the generation of FOS as kestose and nystose.
Electronic supplementary material
The online version of this article (10.1007/s12088-017-0704-y) contains supplementary material, which is available to authorized users.
Keywords: Prebiotic, Fructo-oligosaccharides, Penicillium, β-Fructofuranosidase
Prebiotics which can impart nutritional as well as health benefits to human population have recently gained the attention of the people worldwide [1]. Fructo-oligosaccharides (FOS) sometimes also mentioned as oligofructose come under the category of prebiotic food segment which selectively stimulates the beneficial colon bacteria and boosts the health of the host [2, 3]. Owing to their low caloric value, FOS are the most appropriate ingredients which have been incorporated in dairy and bakery products [4]. Hydrolysis of inulin by inulinase and transformation of sucrose by fructosyltransferase are the two widely used methods for the production of FOS [5]. Production of FOS from sucrose is an economical process as compared to FOS obtained by inulin hydrolysis because of low-cost of sucrose [6]. FOS synthesis from sucrose is mediated by two major enzymes Sucrase (sucrose fructosyltransferase, FTase, EC 2.4.1.9) and invertase (β-fructofuranosidase fructohydrolase, FFase, EC 3.2.1.26) [1]. Various FOS synthesized from sucrose include 1-kestose (GF2), nystose (GF3), and 1F-β-fructofuranosylnystose (GF4) where 1–3 units of fructose are attached in β-(2-1) linkage to the sucrose [4]. The recent increase in market-share of FOS among the prebiotics has propelled the need for exploring novel enzymes and microorganisms having the capacity for high FOS yields along with an increase in the cost effectiveness of the process [7].
Several studies have reported Penicillium sp. to be a potential producer of FFase which is utilised for the FOS production [8–10]. In the present study, Penicillium chrysogenum strain HKF42 isolated from effluent treatment plant (ETP) was screened for FFase enzyme activity and subsequent synthesis of FOS [11]. For a better understanding of the genes coding for required enzymes, de novo whole genome sequencing approach was followed. Genomic DNA was isolated with FastDNA SPIN Kit (MP Biomedicals, USA) followed by preparation of paired-end and mate-pair sequencing libraries with mean sizes 629 and 690 bp, respectively. The libraries were sequenced (2 × 150 bp) on Illumina HiSeq 2500 platform. High quality reads generated were used for de novo assembly using Soapdenovo2 assembler [12] resulting in a draft genome of 31.4 Mbp containing 160 contigs and 143 scaffolds (Table 1).
Table 1.
Genome features of Penicillium chrysogenum strain HKF42
| Features | Penicillium chrysogenum strain HKF42 genome |
|---|---|
| No. of reads | 61,456,596 |
| Pair end read length (bp) | 150 |
| Genome assembly size | 31,410,885 |
| Number of contigs | 160 |
| Number of scaffolds | 143 |
| Contigs N50 | 2,015,765 |
| Gaps between scaffolds | 0 |
| Scaffold N50 | 2,810,928 |
| No. of gene model | 11,251 |
| No. of exon per gene | 3.14 |
| Mean protein length | 475.45 |
| GC content (%) | 53.22 |
| No. of tRNA | 188 |
| NR annotated | 11,184 |
| SwissProt annotated | 7595 |
| KEGG annotated | 2154 |
| KOG annotated | 5482 |
| Pfam annotated | 8154 |
| CAZymes | 600 |
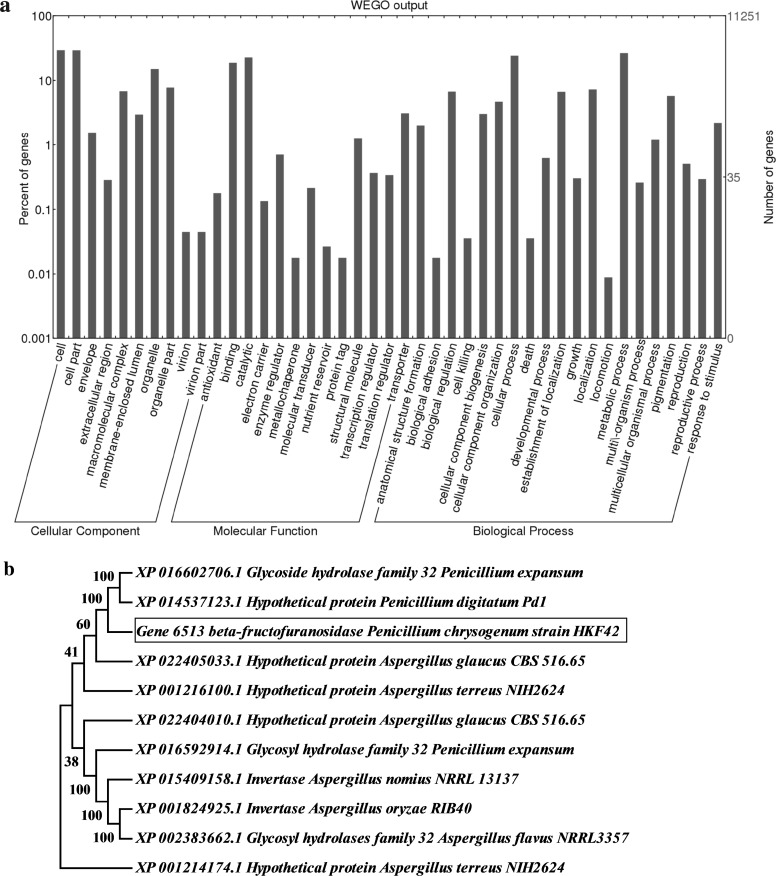
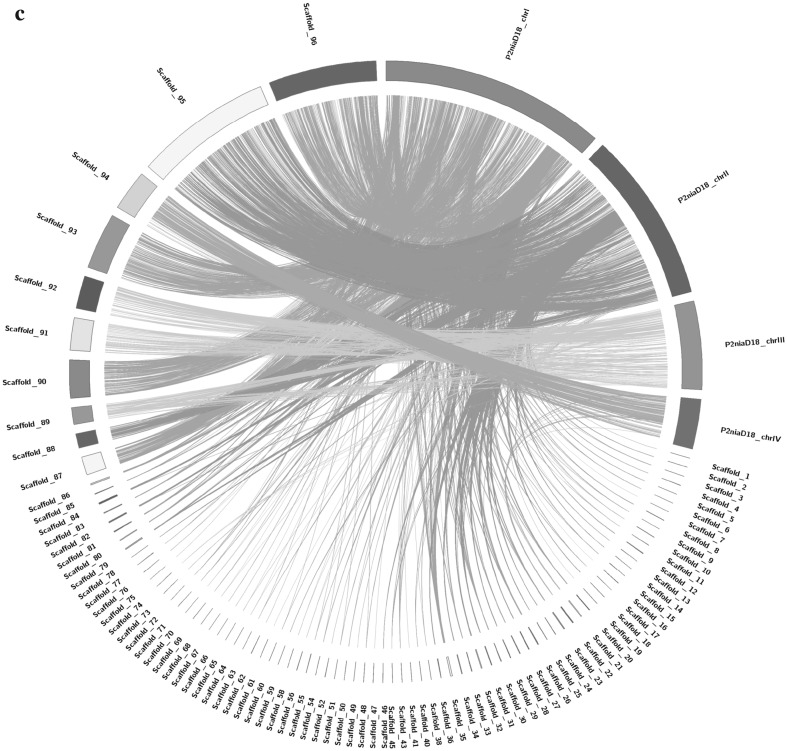
A total of 568,994 bp (1.81%) repeat sequences were masked using RepeatMasker and the RepBase library and afterwards, the repeat masked assembly was used for coding gene prediction. Coding sequences (CDS) in the genome were predicted by GeneMarkES [13] which resulted in a total of 11,251 protein-coding genes. Later, NCBI’s non-redundant (nr) database was used for similarity search of predicted CDS using the BLASTP algorithm. The total proteins were also searched for similarity against Swiss-Prot, Pfam and KOG databases using BLASTP, rpsblast and Hmmscan algorithms (via webMGA). Gene Ontology (GO) annotation obtained through nr database using blast2GO Pro and CDS associated with similar functions were assigned to the same GO functional groups. GO sequence analysis distributions revealed the presence of all three GO domains i.e. cellular components (3926), biological processes (3981) and molecular function (3348) (Fig. 1a). Proteins were mapped and orthologs were assigned to the biological pathways through KEGG automatic annotation server (KAAS) and compared with the KEGG database using BLASTP. For CAZymes analysis, CDS were annotated using dbCAN, the data generated in dbCAN was based on the family classification from CAZy database [14, 15]. Gene annotation disclosed the presence of an enzyme belonging to CAZymes family which was located on gene 6513 and corresponded to FFase with molecular weight of 72.8 kDa respectively. The nearest neighbour analysis of FFase gene 6513 carried out through BLASTP (version 2.7.1), amino acid sequence was searched for similarity against reference proteins (refseq_proteins) and 10 hits were chosen based on query coverage more than 80%. Sequences were aligned using ClustalW interface in MEGA 7 based on the neighbour joining method, and bootstrap values were based on 1000 replicates (Fig. 1b). Gene 6513 shares 78% similarity with Penicillium digitatum Pd1 protein, the nearest neighbor highlighted the diversity of the FFase enzyme in Penicillium and Aspergillus sp. We, further characterized the gene product using program MotifFinder, I-TASSER and COACH for binding and active site prediction as described in Table 2; Fig. S1 and S2 [16, 17]. Ancestral relationship of the isolate P. chrysogenum strain HKF42 with P. chrysogenum strain P2niaD18 was carried out using synteny analysis through BLASTN and CIRCOS which gave a visual overview of the alignment (Fig. 1c) [18].
Fig. 1.
a WEGO plot visualization of GO terms identified in P. chrysogenum strain HKF42. b Neighbor Joining tree calculated by ClustalW for β-fructofuranosidase gene from Penicillium sp. constructed by MEGA 7. Numbers displayed on the branches are the bootstrap support obtained through 1000 replications. c The synteny relationship between Penicillium chrysogenum strain P2niaD18 reference genome and P. chrysogenum strain HKF42 scaffolds. The right hand side of circle starting from P2niaD18_chrI to P2niaD18_chrIV represents the reference genome, and the P. chrysogenum strain HKF42 genome sample is displayed on the left side of circle represented as scaffolds. Different colors of lines connect the scaffolds of the P. chrysogenum strain HKF42 genome on the left, to the matched sections of the reference genome on the right
Table 2.
Characterization of active site motifs and domains identified in gene 6513 coding for β-fructofuranosidase (FFase)
| Domain | Pfam | Predicted binding site residues using different tools | Active site residues predicted by I-Tasser | ||||||
|---|---|---|---|---|---|---|---|---|---|
| NCBI-CDD | 214,757 | 185,737 | 185,718 | PF00251 | PF08244 | TM-SITE | S-SITE | COACH | |
| Motif | Glycosyl hydrolases family 32 | Glycosyl hydrolase family 32, beta-fructosidases | Glycosyl hydrolase families: GH43, GH62, GH32, GH68 | Glycosyl hydrolases family 32 N-terminal | Glycosyl hydrolases family 32 C-terminal | ||||
| Gene 6513 |
Position 46–526 | Position 52–397 | Position 62–395 | Position 46–340 | Position 468–667 | Ligands (fructose, sucrose, nystose) G55, D56, L74, T78, F114, D115, R186, D187, E264, T265, Y337 |
Ligands (glucose, kestose, sucrose) D56, L74, F114, D115, L137, P138, I139, H140, R186, E264, E289, S298, S300, Y337, W365, F371, G372 |
Ligands (fructose) G55, D56, L74, T78, F114, D115, R186, D187, E264, Y337, W365 |
D56 and E264 |
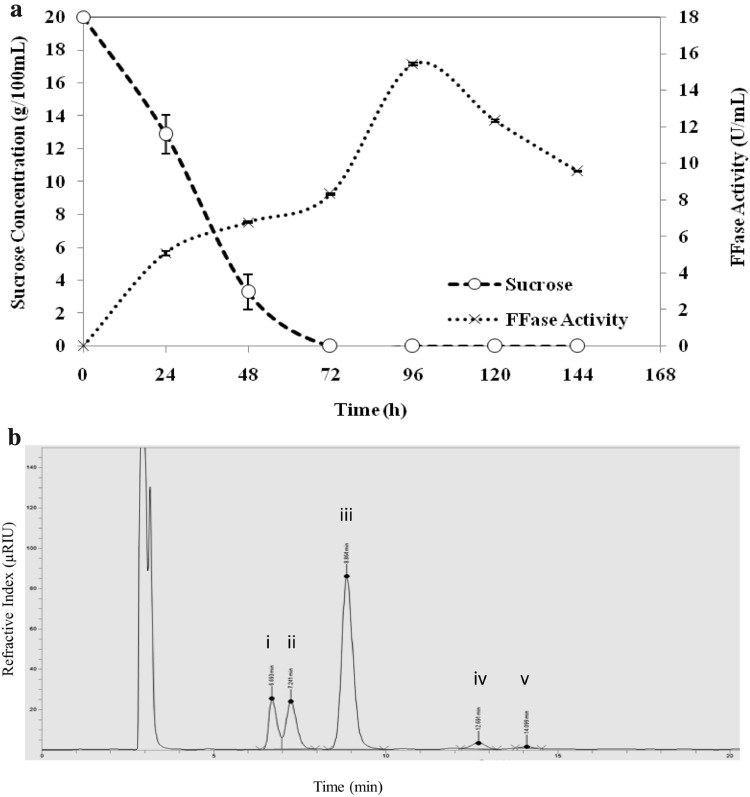
The genetic capacity of the isolate for FFase production and FOS synthesis was further validated by submerged fermentation studies. Inoculum development and submerged fermentation strategy was followed as per Prata et al. [19]. The samples were withdrawn at a regular interval of 24 h; centrifuged using a refrigerated centrifuge (4 °C, 10,000 RPM) and the supernatant without further purification was used as the source of FFase. Assay of FFase activity was carried out as per the method of Sangeetha et al. [20] wherein the quantitative analysis of the FOS was done using HPLC and retention time of products were compared with the FOS standards for identification. One unit of FFase activity was defined as the amount of enzyme required to produce 1 µmol of glucose per minute at 55 °C with 55% sucrose at pH 5.5 [8]. The consumption of sucrose by the fungal isolate and concomitant production of FFase is shown in Fig. 2a. A rapid reduction in sucrose concentration was observed with 85% sucrose being hydrolyzed in 48 h followed by complete hydrolysis in 72 h. FFase activity increased gradually to 8.3 U/mL in 72 h which doubled to 15.4 U/mL in 96 h resulting in complete hydrolysis of sucrose. HPLC analysis of the assay reaction mixture confirmed the hydrolysis of sucrose to glucose and fructose by FFase along with the formation of two types of FOS viz., kestose and nystose as indicated in Fig. 2b.
Fig. 2.
a Sucrose utilization and concomitant production of extracellular β-fructofuranosidase (FFase) from P. chrysogenum strain HKF42. b HPLC chromatogram of the short chain FOS synthesized by crude β-fructofuranosidase (FFase) from P. chrysogenum strain HKF42 (i) fructose; (ii) glucose; (iii) sucrose; (iv) kestose; (v) nystose
In conclusion, P. chrysogenum strain HKF42 can be considered a promising candidate for the production of extracellular β-fructofuranosidase and synthesis of short chain FOS and the genome data further can be utilized for mining of other prebiotic synthesizing enzymes.
This Whole Genome Shotgun project has been deposited in GenBank under the accession number MWKT00000000. The version described in this paper is MWKT01000000.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Authors would like to acknowledge to CSIR-NEERI for providing essential resources for the research work [KRC No. CSIR-NEERI/KRC/2017/SEP/EBGD/2]. Vaibhav Gujar is thankful to University Grants Commission (UGC), New Delhi for providing Junior and Senior Research Fellowship for carrying out this research.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s12088-017-0704-y) contains supplementary material, which is available to authorized users.
References
- 1.Vega R, Zuniga-Hansen ME. A new mechanism and kinetic model for the enzymatic synthesis of short-chain fructooligosaccharides from sucrose. Biochem Eng J. 2014;82:158–165. doi: 10.1016/j.bej.2013.11.012. [DOI] [Google Scholar]
- 2.Huang MP, Wu M, Xu QS, Mo DJ, Feng JX. Highly efficient synthesis of fructooligosaccharides by extracellular fructooligosaccharide-producing enzymes and immobilized cells of Aspergillus aculeatus M105 and purification and biochemical characterization of a fructosyltransferase from the fungus. J Agric Food Chem. 2016;64:6425–6432. doi: 10.1021/acs.jafc.6b02115. [DOI] [PubMed] [Google Scholar]
- 3.Lorenzoni ASG, Aydos LF, Klein MP, Rodrigues RC, Hertz PF. Fructooligosaccharides synthesis by highly stable immobilized β-fructofuranosidase from Aspergillus aculeatus. Carbohydr Polym. 2014;103:193–197. doi: 10.1016/j.carbpol.2013.12.038. [DOI] [PubMed] [Google Scholar]
- 4.Flores-Maltos DA, Mussatto SI, Contreras-Esquivel JC, Rodrigues-Herrera R, Teixeira JA, Aguilar CN. Biotechnological production and application of fructooligosaccharides. Crit Rev Biotechnol. 2014;36:259–267. doi: 10.3109/07388551.2014.953443. [DOI] [PubMed] [Google Scholar]
- 5.Marin-Navarro J, Talens-Perales D, Polaina J. One-pot production of fructooligosaccharides by a Saccharomyces cerevisiae strain expressing an engineered invertase. Appl Microbiol Biotechnol. 2015;99:2549–2555. doi: 10.1007/s00253-014-6312-4. [DOI] [PubMed] [Google Scholar]
- 6.Aguiar-oliveira E, Maugeri F. Effects of lyophilization on the catalytic properties of extracellular fructosyltransferase from Rhodotorula sp. LEB-V10. Int Res J Biotechnol. 2012;3:96–111. [Google Scholar]
- 7.Bali V, Panesar PS, Bera MB, Panesar R. Fructo-oligosaccharides: production, purification and potential applications. Crit Rev Food Sci Nutr. 2013;55:1475–1490. doi: 10.1080/10408398.2012.694084. [DOI] [PubMed] [Google Scholar]
- 8.Mussatto SI, Prata MB, Rodrigues LR, Teixeira JA. Production of fructooligosaccharides and β-fructofuranosidase by batch and repeated batch fermentation with immobilized cells of Penicillium expansum. Eur Food Res Technol. 2012;235:13–22. doi: 10.1007/s00217-012-1728-5. [DOI] [Google Scholar]
- 9.Nascimento AKC, Nobre C, Cavalcanti MTH, Teixeira JA, Porto ALF. Screening of fungi from the genus Penicillium for production of β-fructofuranosidase and enzymatic synthesis of fructooligosaccharides. J Mol Catal B Enzym. 2016;134:70–78. doi: 10.1016/j.molcatb.2016.09.005. [DOI] [Google Scholar]
- 10.Xu Q, Zheng X, Huang M, Wu N, Yan Y, Pan J, Yang Q, Duan CJ, Liu JL, Feng JX. Purification and biochemical characterization of a novel β-fructofuranosidase from Penicillium oxalicum with transfructosylating activity producing neokestose. Process Biochem. 2014;50:1237–1246. doi: 10.1016/j.procbio.2015.04.020. [DOI] [Google Scholar]
- 11.Deshmukh R, Mathew A, Purohit HJ. Characterization of antibacterial activity of bikaverin from Fusarium sp. HKF15. J Biosci Bioeng. 2014;117:443–448. doi: 10.1016/j.jbiosc.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 12.Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, Tang J. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borodovsky M, Lomsadze A. Eukaryotic gene prediction using GeneMark.hmm-E and GeneMark-ES. Curr Protoc Bioinforma. 2011;33:6494–6506. doi: 10.1002/0471250953.bi0406s35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu L, Taujale R, Liu F, Song J, Yin Q, Zhang Y, Guo J, Yin Y. Draft genome sequence of Talaromyces verruculosus (“Penicillium verruculosum”) strain TS63-9, a fungus with great potential for industrial production of polysaccharide-degrading enzymes. J Biotechnol. 2016;219:5–6. doi: 10.1016/j.jbiotec.2015.12.017. [DOI] [PubMed] [Google Scholar]
- 15.Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y. DbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012;40:445–451. doi: 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang J, Roy A, Zhang Y. Protein–ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics. 2013;29:2588–2595. doi: 10.1093/bioinformatics/btt447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang J, Zhang Y. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res. 2015;43:W174–W181. doi: 10.1093/nar/gkv342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Specht T, Dahlmann TA, Zadra I, Kürnsteiner H, Kück U. Complete sequencing and chromosome-scale genome assembly of the industrial progenitor strain P2niaD18 from the penicillin producer Penicillium chrysogenum. Genome Announc. 2014;2:4–5. doi: 10.1128/genomeA.00577-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prata MB, Mussatto SI, Rodrigues LR, Teixeira JA. Fructooligosaccharide production by Penicillium expansum. Biotechnol Lett. 2010;32:837–840. doi: 10.1007/s10529-010-0231-y. [DOI] [PubMed] [Google Scholar]
- 20.Sangeetha PT, Ramesh MN, Prapulla SG. Production of fructo-oligosaccharides by fructosyl transferase from Aspergillus oryzae CFR 202 and Aureobasidium pullulans CFR 77. Process Biochem. 2004;39:755–760. doi: 10.1016/S0032-9592(03)00186-9. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.