Abstract
We used differential screening to isolate ripening-associated cDNAs from a Shiraz grape (Vitis vinifera L.) berry cDNA library. A rapid increase in the mRNA levels of a number of cDNAs not present in unripe fruit occurred in grape berries at the onset of ripening. The putative translation products of some of these clones had homologs in other species that are involved in cell wall structure. These included four proline-rich proteins, a small protein that is similar to the non-catalytic, N-terminal domain of some pectin methylesterases, and two other glutamate-rich proteins. The remainder of the clones encoded putative stress response proteins. These included two thaumatin-like proteins, a metallothionein, a transcription factor, a cytochrome P450 enzyme, and proteins induced by water, sugar, and/or cold stress in other species. Many of the homologs of the grape cDNAs thought to be involved in cell wall structure or stress-related responses also accumulate in a developmental manner in other plants. This may indicate that the grape mRNAs accumulate in response to stresses such as the storage of high concentrations of sugars and rapid cell expansion, or they may accumulate as part of the ripening developmental program.
Grape (Vitis vinifera L.) berries undergo considerable physical and biochemical changes as they develop, particularly during the ripening process. Grapes are considered to be a non-climacteric fruit, and berry development can be divided into three phases on the basis of berry growth. After fruit set, there is an initial phase of cell division (Harris et al., 1968) and cell expansion that results in rapid growth of the berry. This is followed by a lag phase during which berries do not increase in size. Following the lag phase, there is a second phase of berry growth during which ripening occurs (Coombe, 1992). The commencement of ripening is known as “véraison” by viticulturists. Ripening is characterized by a number of changes, including the degradation of chlorophyll, an increase in berry deformability, a rapid increase in the level of hexoses in the berry vacuole, an increase in berry volume, the catabolism of organic acids, the development of skin color (in red grapes), and the formation of compounds important for flavor and aroma. During this first phase of berry expansion, indole-3-acetic acid levels are elevated (Cawthon and Morris, 1982). As the auxin levels are low after véraison and cell division has ended, the second phase of expansion would appear to have a different biochemical basis. Not only does considerable cell expansion occur in this second phase of berry size increase, but this is also the period when berry softening occurs.
A relationship between fruit ripening and changes in mRNA levels has been demonstrated in grape berries by Boss et al. (1996), who showed that the accumulation of transcripts of genes in the flavonoid synthesis pathway was related to anthocyanin production in the berry skin during ripening. In many other fruit, the considerable changes that occur during ripening are also largely the result of changes in gene transcript levels. For example, much is known about the changes in mRNA levels that occur during the ethylene-driven ripening of climacteric fruit, particularly tomato. Screening of cDNA libraries from ripening wild-type and mutant tomato fruit has enabled the identification of a large number of cDNAs associated with the ripening process (Gray et al., 1992; Picton et al., 1993). This has lead to an enhanced understanding of tomato ripening and has allowed the development of transgenic plants with altered ripening characteristics (Gray et al., 1994). Only recently have researchers begun using similar techniques to investigate the molecular biology of ripening in non-climacteric fruit such as strawberry (Medina-Escobar et al., 1997; Manning, 1998) and pepper (Proust et al., 1996). The isolation of ripening-enhanced cDNAs from strawberry (Manning, 1998) and black currant (Woodhead et al., 1998) by differential screening has demonstrated that this technique will be useful in the study of ripening in non-climacteric fruit.
We used the differential screening technique to isolate “Grip” (grape ripening-induced) cDNAs from a ripening grape berry cDNA library. A number of differentially expressed clones were isolated and their sequences and expression patterns in grape tissues analyzed, and their possible function during ripening is discussed.
MATERIALS AND METHODS
Tissue Collection, Measurement of Ripening Parameters, and Isolation of RNA
Berries of grape (Vitis vinifera L. cv Shiraz) were sampled at 2-week intervals, beginning at flowering, during the 1995/1996 growing season from 20-year-old vines grown at a commercial vineyard in Willunga, South Australia. Deformability measurements of a number of berries were taken every 2 weeks using a Harpenden skinfold calliper gauge (British Indicators, Burgess Hill, West Sussex, UK) as described by Coombe and Bishop (1980). A randomly chosen sample was weighed and measured for total soluble solids (degrees Brix) with a refractometer (model 10430, Reichert, Vienna). Berries were picked, deseeded (except for the 2 weeks postflowering [wpf] sample), and the flesh (retaining the skins) and seeds were immediately frozen in liquid nitrogen and stored at −80°C until required. For the separation of skin from flesh, the skin was removed from frozen berries that had been partially thawed, and skin and flesh samples were extracted for RNA as described below. Leaf (three stages), root, and flower tissue were similarly frozen and stored.
Total RNA was extracted from the various tissues by the perchlorate method as described by Davies and Robinson (1996). This RNA was used in northern-blot analysis. For cDNA library production, mRNA was purified from total RNA using a mRNA isolation kit (PolyATract, Promega, Madison, WI) according to the manufacturer's instructions.
Preparation of Shiraz Berry cDNA Library
cDNA was prepared using a cDNA synthesis system (SuperScript Choice, Life Technologies/Gibco-BRL, Cleveland) according to the manufacturer's instructions using 5 μg of mRNA from 10 wpf, deseeded Shiraz berries. EcoRI adaptors were ligated to the cDNA, and the resultant fragments were cloned into predigested Lambda ZAP II/EcoRI/CIAP vector and packaged (Gigapack II Gold, Stratagene, La Jolla, CA). Titering, amplification, and screening of the library (with the appropriate probes, see below) were carried out as described in the manufacturer's instruction manual except for the hybridization conditions.
Differential Screening of cDNA Library
Duplicate lifts using Hybond N membrane (Amersham-Pharmacia Biotech, Uppsala) were hybridized with either the pre-véraison probe (made from 6-wpf cDNA, see below) or the post-véraison probe (made from 10-wpf cDNA). The filters were hybridized and washed as described by Davies and Robinson (1996). For the preparation of 6- and 10-wpf probes, 2 μg of mRNA from the 6- and 10-wpf samples was used to produce first-strand cDNA that was then A-tailed using terminal transferase and amplified by PCR using an oligo-dT primer (Chevalier et al., 1995). The mix of fragments thus generated was then labeled by random primer labeling (GIGAprime DNA labeling kit, Bresatec, Adelaide, South Australia), and unincorporated label was removed using a Sephadex G-50 column. Plaques showing hybridization with the 10-wpf but not the 6-wpf probe were isolated as a single species by secondary and (where required) tertiary screening. These single clones were then rescued into pBluescript plasmid vector as described by the manufacturer's instructions (Stratagene). Purified plasmid DNA of the rescued clones was digested with EcoRI endonuclease to determine the insert size. Selected clones were then sequenced by automated sequencing using T3 and T7 oligonucleotide primers and, where appropriate, specifically designed internal primers.
To reduce problems caused by the abundance of Grip 3 and 4 sequences, which predominated the early screening attempts, these sequences were selectively removed from the 10-wpf probe as follows. First-strand cDNA was prepared as described above. After the terminal transferase reaction, the fragments were ethanol precipitated, washed, dried, and taken up in 100 μL of water. A synthetic oligonucleotide with a 5′ biotin group attached was designed to a sequence common to both the Grip 3 and 4 sequences (CAAGGCTCCACCACCCATCC). Twenty microliters of 60 pm/μL primer was mixed with 100 μL of washed streptavidin paramagnetic particles (MagnaSphere, Promega) and 80 μL of 10× SSC and incubated at 25°C for 15 min. The beads were then washed three times with 5× SSC and resuspended in 60 μL of 10× SSC. One-hundred microliters of the first-strand cDNA was heated at 95°C for 10 min, snap-cooled on ice, and mixed with 30 μL of the probe/bead suspension. SSC was added to a final concentration of 3.5×. This mixture was incubated with gentle agitation at 38°C for 30 min. The beads were then captured and the cDNA was precipitated from the supernatant by ethanol precipitation, washed, dried, taken up in water, and then amplified and labeled as described above.
Sequence Analysis
Initially BLASTX and BLASTP programs (Genetics Computer Group, Madison, WI) were used to identify the sequences from other plant species and yeasts most closely related to the differentially screened grape clones. The degree of similarity between two sequences was calculated using the GAP program, and the theoretical pI was calculated using the ISOELECTRIC program (both from Genetics Computer Group). Sequence-property-based searches of protein databases were done using the PROPSEARCH program of Hobohn and Sander (1995).
Northern Analysis and Probe Preparation
Northern blotting, hybridization, and washing were done as described by Davies and Robinson (1996). 32P-Labeled probes to the various genes were prepared by random primer labeling (GIGAprime DNA labeling kit, Bresatec, Adelaide, South Australia) of the appropriate excised fragments.
Cloning of a Putative Cytochrome P450
A cDNA fragment encoding part of a putative cytochrome P450 enzyme was generated by chance by PCR using degenerate oligonucleotide primers designed to amplify plant glucanases (A. Jacobs, personal communication). This clone was used as a probe to isolate a full-length clone (gfh2) from the 10-wpf Shiraz cDNA library in Lambda Zap II. This cDNA clone was used to probe northern blots.
Cloning of the Thaumatin-Like VvTL2 cDNA
A partial clone of VvTL2 (homologous to the thaumatin-like clone from Sultanina [Loulakakis, 1997]) was cloned serendipitously by PCR from Shiraz 10-wpf cDNA (see above for the preparation of cDNA). The primers used were WHP5F: 5′-TGATTCAGGTAGCGGCAGT-3′ and B25 (Frohman et al., 1988). A 670-bp subclone was isolated and used to probe northern blots.
RESULTS
Differential Screening of a Ripening Shiraz Berry Library
The initial differential screening was conducted using a cDNA library made from poly(A+) RNA from Shiraz berries harvested 10 wpf (2 weeks after véraison). Duplicate lifts of this library were screened using either cDNA made from 6-wpf (2 weeks before véraison) berry RNA as a probe or cDNA made from RNA extracted from 10-wpf berry tissue. Autoradiographs of these filters showed many plaques that hybridized strongly with the 10-wpf, but not with the 6-wpf, probe (data not shown). The pattern of hybridization indicated that there is a change in the accumulation of transcripts of a number of genes during ripening. A number of these plaques were isolated as single species and rescued into pBluescript. DNA endonuclease digestion and DNA sequencing showed that the majority of these clones (11 of 19) comprised two very closely related sequences, Grip 3 and 4 (Table I). Although these cDNAs were apparently ripening related and therefore of interest to us, their dominance made screening for other ripening-related clones more difficult. To circumvent this problem, a biotinylated synthetic oligonucleotide designed to the Grip 3 and 4 sequences was used to subtract these sequences from the 10-wpf probe. Subsequent differential screening demonstrated that this approach was successful in reducing the number of Grip 3 and 4 clones isolated. Selected clones were sequenced and unique sequences were compared with the database sequences to identify homologs in other species and thereby determine possible functions.
Table I.
Ripening-induced cDNA clones encoding putative cell wall proteins in cv Shiraz grape
| Grip Clone No. (Accession No.) | Nearest Match (Accession no.) [Percent identity]a | Proposed Function | Features of Grape Clone | Mrb |
|---|---|---|---|---|
| Grip 3 (AJ237981) | Sunflower hydroxyproline-rich protein (M76546) [52%] | Cell wall structure | Pro/Lys-rich (37%/11%) repeats of PPVEKPP similar to Pro-rich repeats in nodulin genes | 22,539 |
| Grip 4 (AJ237982) | Similar to Grip 3 except for one amino acid substitution and a 13-amino acid insertion | 23,954 | ||
| Grip 13 (AJ237983) | Yellow lupin early nodulin precursor (X55371) [35%] | Pro-rich repeats (28% Pro, 15% Lys) | 20,275 | |
| Grip 15 (AJ237984) | Maackia amurensis early nodulin (AF039708) [53%] | Pro-rich repeats (36% Pro, 18% Lys) | 30,867 | |
| Grip 28 (AJ237985) | Protein from alfalfa nodules (Y11553) [56%] | Unknown | Matches N terminus of some pectin methylesterases | 21,128 |
| Grip 31 (AJ237986) | Alder protein (AG13) in nodule pericycle (Y08435) [57%] | Cell wall structure | 41% charged amino acids (29% Glu), pI 3.7, 54% amino acid identity with Grip 68 | 18,416 |
| Grip 68 (AJ237987) | Rubber tree latex allergen HEV B5 (U42640) [48%] | 41% charged amino acids (34% Glu), pI 3.7, 54% amino acid identity with Grip 31 | 15,921 |
The corresponding nearest matches were determined by comparing the deduced amino acid sequences against the database sequences. The other columns detail the proposed function of the homologs and particular features of the grape clones.
The percent identity, at the amino acid level, between the grape sequence and its nearest yeast/plant sequence match.
Mr calculated from protein sequence deduced from the cDNA sequence.
Transcript Accumulation Patterns of Ripening-Associated Genes
The cDNA clones described above were used to probe northern blots prepared with RNA from a number of tissues, i.e. roots, leaves at three stages of development, seeds, flowers, and a developmental series of berry samples. In the Shiraz samples used for RNA extraction the first signs of ripening, as judged by an increase in berry deformablity and hexose accumulation, occurred between 8 and 10 wpf (Davies et al., 1997). A number of cDNAs whose corresponding mRNAs accumulated during berry development were identified. Some of these had predominantly berry- and ripening-specific patterns of accumulation. The steady-state mRNA levels of others increased during ripening but were also present in other tissues.
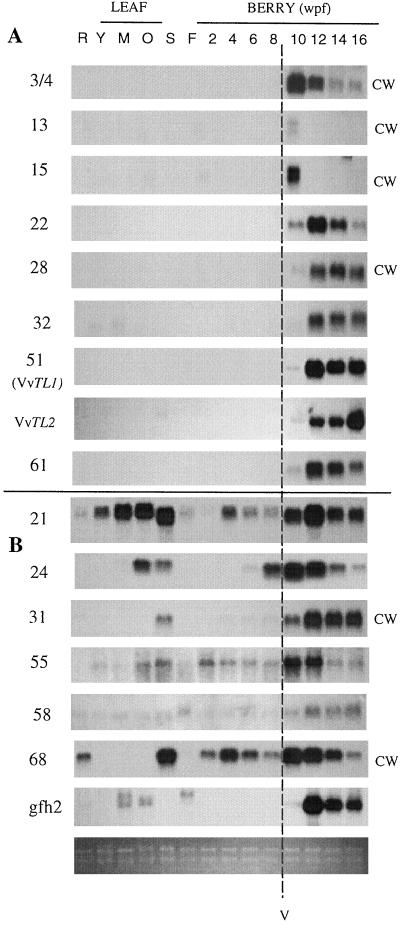
The clones that exhibited predominantly berry- and ripening-specific mRNA accumulation patterns included Grip 3, 4, 13, 15, 22, 28, 32, 51 (VvTL1), VvTL2, and Grip 61 (Fig. 1A). These clones can be further divided into two groups on the basis of the patterns of mRNA accumulation. The transcript levels for Grip 3, 4, 13, and 15 appear to be maximal early in ripening, because they are highest in the 10-wpf sample and then begin to decrease significantly. In contrast, the remaining Grip sequences shown in Figure 1A (Grip 22, 28, 32, 51, 61, and VvTL2) increase in transcript level later during ripening (12 wpf).
Figure 1.
Northern-blot analysis of the expression of Grip clones (and the gfh2 and VvTL2 clones) in various grape tissues. RNA from root (R), young leaf (Y), mid leaf (M), old leaf (O), seed from berries 4 wpf (S), flower (F), and a series of samples taken from developing berries at two weekly intervals (commencing at 2 wpf) were probed with the cDNAs as indicated. The dashed line indicates véraison. A, Genes exhibiting berry- and ripening-specific expression. B, Genes with up-regulated expression during ripening but also expressed in other tissues. CW, Clones encoding putative cell wall proteins. The bottom panel is a photograph of an ethidium-bromide-stained gel to show the intactness and relative loadings of the RNA samples used in the northern analysis.
The transcripts of the remainder of the Grip clones isolated (Fig. 1B) do not accumulate in a fruit- or ripening-specific manner; however, the levels of transcript are significantly increased during ripening. There is a diverse range of expression patterns displayed. For example, the Grip 21 message is present in all tissues at varying levels, while the Grip 24 sequence is only readily detectable in berries, seeds, and older leaves.
Properties of the Putative Ripening-Related cDNAs
General features of the grape clones, including their nearest match with other sequences in the databases, are given in Tables I and II. The values for percentage identity between the grape sequences and their homologs in other species vary considerably, so caution must be used in interpreting matches at the lower end of the range.
Table II.
Ripening-induced clones encoding putative stress response proteins in cv Shiraz grape
| Grip Clone No. (Accession No.) | Nearest Match (Accession No.) [Percent Identity]a | Proposed Function | Features of Grape Sequence | Mrb |
|---|---|---|---|---|
| Grip 21 (AJ237988) | Fission yeast YA15 protein (Z67757) [32%] | Maize homolog induced by sugar starvation | 33,371 | |
| Grip 22 (AJ237989) | No sig. matches | Unknown | pI 4.7, 12% Gly, 6% Cys, 6% Asn | 22,885 |
| Grip 24 (AJ237990) | Type 3 metallothionein from ripening banana (U49044) [75%] | Metal binding, control of free radicals | Conserved Cys, similar proteins found in other ripening fruit, up-regulated by cold in apple | 6,777 |
| Grip 32 (AJ237991) | SRC1 protein from Glycine max (AB000129) [64%] | Unknown, homologs water stress/cold regulated | Has putative nuclear targeting signal near C terminus | 13,554 |
| Grip 51 (VvTL1) (AJ237999) | Thaumatin-like protein from grape (AF003007) [98%] | PR protein, response to stress/pathogens | Thaumatin-like protein because has no C-terminal putative targeting signal | 24,051 |
| VvTL2 (AJ237998) | “Osmotin-like” protein from grape (Y10992) [94%] | NAc | ||
| Grip 55 (AJ237992) | Sunflower DC3 promoter binding factor (AF001453) [47%] | Transcription factor involved in control of ABA-responsive genes | Basic Leu-zipper-type transcription factor | 47,879 |
| Grip 58 (AJ237993) | Arabidopsis EST (AA712680) [68% over 280 amino acids] | pI 4.6, rich in small amino acids (Ala, Val, Leu, Ile, Gly) | 28,812 | |
| Grip 61 (AJ237994) | Strawberry ripening-induced protein (AJ001449) [44%] | Homologs involved in wounding response, storage of secondary metabolites | Member of a diverse group of low-Mr proteins related to latex proteins and pollen allergens, pI 5.0 | 17,076 |
| gfh2 (AJ237995) | 7-Ethoxycoumarin o-deethylase from Jerusalem artichoke (Y09920) [47%] | Detoxification, xenobiotic induced | Member of cytochrome P450 family | 56,182 |
The corresponding nearest matches were determined by comparing the deduced amino acid sequences against the database sequences. The other columns detail the proposed function of the homologs and particular features of the grape clones.
The percent identity, at the amino acid level, between the grape sequence and its nearest yeast/plant sequence match.
Mr calculated from protein sequence deduced from cDNA sequence.
The Mr for this sequence was not available as the clone was only partial length.
Putative Cell Wall Proteins
Grip 3 and 4
The cDNA clones that dominated the initial screen were two very closely related sequences, Grip 3 and 4. These two clones have identical deduced protein sequences except for a 13-amino acid insertion in Grip 4 and a single amino acid substitution. The best match with the deduced amino acid sequences of Grip 3 and 4 with database sequences is with a Hyp-rich, extensin-like protein from sunflower (Table I). However, closer examination suggests that the Grip 3 and 4 protein structure is more akin to certain legume proteins, many of which are nodulins. This is because the high-level match between Grip 3 and 4 and the extensins is due to the high levels of Pro in these sequences and does not account for the structure of the repeated motifs. Extensins are characterized by the Ser (Hyp)4-repeat or closely related sequences (Sommer-Knudsen et al., 1998). The Grip 3 and 4 sequences contain a different repeated motif, P-P-V/E-Y/E-K-P-P, that is repeated five times in the Grip 4 sequence. This is similar to the extended motif proposed for Pro/Hyp-rich glycoproteins (P/HRGPs) (Sommer-Knudsen et al., 1998) of K-P-P-Xaa-Yaa-K-P-P, where Xaa can be V, H, T, or A, and Yaa can be Y, T, E, or P. Some nodulin proteins expressed early in root nodule development, e.g. the nodulin precursor PRP4 from Medicago truncatula (accession no. L23504; Wilson et al., 1994), have an identical repeat to the Grip 3 and 4 sequences, i.e. P-P-V-E-K-P-P repeated numerous times throughout their length, suggesting that the Grip 3 and 4 sequences have a closer relationship with these proteins than with the extensins.
The high level of transcript in ripening berries was demonstrated by the high percentage of clones that were identified as Grip 3 and 4 in the initial differential screen and the high degree of hybridization detected by northern analysis (a 15-min exposure at room temperature with Kodak XOMAT film was sufficient to give readily detectable bands).
Grip 13 and 15
The Grip 13 and 15 cDNAs encode other putative members of the family of Pro-rich cell wall structural proteins. The pentapeptide PEHKP is repeated five times in Grip 13 and 11 times in Grip 15. This sequence is also found in more extensive motifs such as E-K-Xaa-Yaa-P-Zaa-H-K-P, where Xaa can be P or Q, Yaa can be P, L, or V, and Zaa can be Q or E, and is repeated six times in Grip 13 and eight times in Grip 15. This motif is not like that characteristic of extensins (there are none of the S-P-P-P-P motifs), but is more like the consensus motif described above by Sommer-Knudsen et al. (1998) for the P/HRGPs.
The Grip 13 cDNA sequence is 74% identical to the Grip 15 sequence at the nucleotide level (as determined by the GAP program) and only 57% identical to the Grip 4 sequence at the nucleotide level. We would therefore not expect significant cross-hybridization between these Pro-rich sequences at the stringencies used for hybridization and washing of the northern filters.
Like the Grip 3 and 4 putative proteins, the Grip 13 and 15 putative proteins may be involved in strengthening the cell walls by cross-linking other proteins and cell wall polysaccharides.
Grip 28
There are four plant gene sequences whose translation products have a Mr of approximately 20,000 and which are closely related to the putative translation product of the Grip 28 sequence. These include sequences from alfalfa (accession no. Y11553), Arabidopsis (accession no. AC002311), carrot (accession no. X52395), and Pinus radiata (accession no. AF049066). Interestingly, the putative protein from alfalfa (the nearest match to the grape sequence at 56% identity) is expressed in root nodules. A possible role for the Grip 28 protein in cell wall structure/metabolism is indicated by its match with the N-terminal portion of some pectin methylesterases (data not shown).
Grip 31 and 68
The nearest match for Grip 31 with other plant sequences in the databases is with ag13 from alder (Table I), a protein induced in nitrogen-fixing nodules induced by the actinomycetous bacteria Frankia (Guan et al., 1997). This gene is expressed in the pericycle in the young part of nodules and in the senescent cells that arise in the older parts of nodules. The nearest match to Grip 68 is with the rubber tree latex allergen HEV B5 (Table I). These sequences are members of a small group of Glu-rich plant proteins related by sequence similarity and include sequences from kiwifruit (accession no. L27810), black currant (accession no. AJ007576), buckwheat (accession no. D87983), Lotus japonicus (accession no. AF000402), and potato (accession no. Z11679). Transcripts of two of the homologs accumulate in ripening fruit. The mRNA from KIWI501 cDNA is present in kiwifruit for a brief period just after the onset of ripening and again briefly as the fruit approach full size (Ledger and Gardner, 1994). Transcripts of the cDNA from black currant are present during berry ripening, with maximum levels accumulating in fully ripe fruit (Woodhead et al., 1998). It has been speculated that these proteins may perform a structural role in cell walls due to their high Pro content and hydrophilic nature (Guan et al., 1997).
Putative Stress-Induced Proteins
Grip 21
The Grip 21 cDNA clone encodes a putative protein whose nearest match with other database sequences is with a theoretical protein from yeast (Table II). The nearest match with a plant sequence (27% identity at the amino acid level) is with a protein sequence deduced from a maize cDNA (accession no. X82617) whose expression is induced in root tips during Glc starvation (Chevalier et al., 1995). The maize clone does not seem to be full length, as it does not have a Met residue at the N terminus. The level of amino acid sequence identity between these two sequences is low, so this match must be viewed with caution.
Grip 22
The Grip 22 sequence was cloned frequently during differential screening, suggesting that it is an abundant message in ripening grape berries. Database searches using BLASTP did not reveal any close matches to this sequence. However, thaumatin-like proteins were highly represented in the results obtained with the sequence-property-based PROPSEARCH program, which ignores the order of the amino acids in the primary sequence in favor of parameters such as the amino acid composition, Mr, and charge. It is therefore possible that the Grip 22 sequence represents a divergent member of this gene family.
Grip 24
The deduced protein sequence encoded by the small Grip 24 cDNA showed that it is closely related to plant metallothionein-like proteins. These are low-Mr, Cys-rich proteins found in a wide variety of organisms including animals, plants, and fungi, which are thought to be involved in the sequestration of metal ions (Robinson et al., 1993). The banana and grape sequences form part of a group of metallothionein-like sequences that have been defined as type 3 on the basis of the pattern of Cys residues (Reid and Ross, 1997). Many members of this group are associated with fruit ripening, e.g. in apple (Reid and Ross, 1997), banana (Clendennen and May, 1997), kiwifruit (Ledger and Gardner, 1994), papaya (Lam and Abu Bakar, 1996), black currant (Woodhead et al., 1998), and cherry (Wiersma et al., 1998). Metallothionein-like proteins are also involved in the response to stresses such as treatment with metal ions and heat shock (Hsieh et al., 1995), Glc starvation (Chevalier et al., 1995), high levels of Suc (Chatthai et al., 1997), Suc starvation (Hsieh et al., 1995), low temperature (Reid and Ross, 1997), wounding, and viral infection (Choi et al., 1996).
Various functions, including roles in metal metabolism and detoxification, activated oxygen detoxification, and control of cellular redox potential, have been proposed but there is little evidence in support of these suggestions. Metal ion binding capability has been demonstrated for the plant metallothionein-like proteins from wheat (Lane et al., 1987) and pea (Evans et al., 1992). The presence of the Grip 24 mRNA in fully expanded leaves is analogous to other reports of metallothionein-like gene expression in senescing leaves (Buchanan-Wollaston, 1994), so it is possible that its function in grape is part of a senescence event in which any of the functions mentioned above may be important.
Grip 32
The protein deduced from the Grip 32 sequence is related to a group of small plant proteins of unknown function induced by stimuli such as low temperature and water stress. Near the C terminus, there is a Lys-rich region that is highly similar to the bipartite nuclear targeting signal present in proteins from other organisms (Monroy et al., 1993). This is followed by a Ser-rich region that is highly conserved among these proteins. The Grip 32 sequence also contains a motif, K-G-E-G-Q/Y-G, that is repeated five times.
Of the Grip 32 homologs, those from alfalfa (Monroy et al., 1993), citrus (Cai et al., 1995), rice (Takahashi et al., 1994), and soybean (Takahashi and Shimosaka, 1997) have all been shown to be induced by low-temperature treatment and may be involved in the adaptation of plants to low-temperature stress (Monroy et al., 1993; Takahashi et al., 1994). While the accumulation of transcripts of some of these genes is also inducible by other stresses such as high temperature, drought, wounding, and viral infection (Takahashi and Shimosaka, 1997), the homologs from rice (Takahashi et al., 1994) and soybean (Takahashi and Shimosaka, 1997) are not inducible by abscisic acid (ABA). The presence of a putative nuclear targeting signal indicates that the Grip 32 putative protein may be located in the nucleus. As suggested by Monroy et al. (1993) for the alfalfa homolog Cas15, the Grip 32 putative protein may function to control gene expression or may perform a role in stabilizing the nuclear structure.
Thaumatin-Like Proteins
Grip 51 (VvTL1) and VvTL2
The Grip 51 cDNA encodes a putative protein that is virtually identical to the VvTL1 thaumatin-like protein from Muscat Gordo Blanco previously described by Tattersall et al. (1997). A second thaumatin-like cDNA, VvTL2 (closely related to the sequence from Sultanina reported by Loulakakis, 1997), was also cloned and used as a probe for northern-blot analysis. Both Grip 51 and VvTL2 were expressed in post-véraison berries (Tattersall et al., 1997; Fig. 1A), and were induced in leaves and pre-véraison berries by ethephon treatment and powdery mildew infection (Jacobs et al., 1999).
Both the Grip 51 (VvTL1) and VvTL2 sequences appear to encode thaumatin-like proteins that are located to the extracellular space because they have acid pIs, putative N-terminal signal peptides, and lack the C-terminal extension thought to be involved in vacuolar targeting. Thaumatin-like proteins or their mRNAs have also been recorded in other ripening fruit, including banana (Clendennen and May, 1997; Tattersall et al., 1997), kiwifruit (Tattersall et al., 1997), and cherry (Fils-Lycaon et al., 1996). The function of the grape thaumatin-like proteins is unknown, but an involvement in disease resistance has been suggested (Tattersall et al., 1997; Salzman et al., 1998; Jacobs et al., 1999).
Grip 55
The putative protein derived from the Grip 55 cDNA sequence appears to be a member of the basic Leu zipper family of transcription factors. The most closely related database sequence, a Dc3 promoter-binding factor from sunflower (Table II), has been shown to interact with the ABA-responsive and embryo-specification elements of the carrot gene Dc3 (Kim et al., 1997). The precise N terminus of the Grip 55 sequence is not identifiable because there is more than one in-frame start codon in reasonable context. The Dc3 gene is a member of the lea (late embryogenesis abundant) gene family, which may act to protect the embryo during desiccation. Because of the close sequence similarity between the putative grape Grip 55 and DPBF-1 proteins, the grape protein may play a role in controlling ABA-/water-stress-inducible gene expression during ripening in grape berries.
Grip 58
Grip 58 does not have any fully sequenced homologs in other plants, but database searches revealed matches with a number of Arabidopsis expressed sequence tags. The best of these matches at the nucleotide level was with accession number AA712680. When all possible open reading frames of this expressed sequence tag were translated and compared with the grape sequence, one of the putative translation products was closely related to the Grip 58 sequence (Table II).
Grip 61
The putative grape protein encoded by Grip 61 is a member of a group of low-Mr proteins that includes the poppy major latex protein, proteins from Arabidopsis (accession no. X91914) and tobacco (Neale et al., 1990), and ripening-related proteins found in fruit such as strawberry (accession no. AJ001449), bell pepper (Pozueta-Romero et al., 1995), and melon (Aggelis et al., 1997). In bell pepper the Sn1 protein and its message are present in ripening fruit and are inducible in green fruit by wounding. In melon the Grip 61 homolog Mel 7 is also present in ripening fruit and is inducible in unripe fruit by ethylene treatment (Aggelis et al., 1997). The suggestion was made in both these papers that the expression of these genes may be related to disease resistance. The major latex proteins from poppy and the Sn1 protein from bell pepper have been localized in vesicles (Griffing and Nessler, 1989; Pozueta-Romero et al., 1995). Interestingly, small vesicles with a similar appearance are also found in some exocarp cells of ripening grape berries (Hardie et al., 1996).
Gfh2
The gfh2 cDNA encodes a putative protein that matches closely with a plant enzyme that has been shown to have 7-ethoxycoumarin o-deethylase activity in yeast (Table II). This enzyme is a member of the cytochrome P450 monooxygenase family, a large family with diverse functions including roles in the phenylpropanoid pathway. The gfh2 cDNA sequence was not cloned as part of the differential screening experiment, but has been included in this paper because its corresponding mRNA exhibits a differential pattern of accumulation in developing berries. Transcripts of gfh2 accumulated mainly in berries after véraison (Fig. 1B). There was a band in the flower RNA sample that hybridized with the gfh2 probe but corresponded to a larger transcript, possibly due to cross-hybridization with a related sequence. Due to the large numbers of different cytochrome P450s present in plants, it is possible that there may be some cross-hybridizing RNA species.
DISCUSSION
It is apparent from the differential screening analysis presented in this work that there is a dramatic change in the mRNA population in grape berries as they enter ripening. The transcript levels of a range of genes increase at véraison, including, in addition to the cDNAs described in this paper, genes encoding alcohol dehydrogenase, chitinase, thaumatin-like proteins, and anthocyanin synthesis pathway enzymes (Boss et al., 1996; Robinson et al., 1997; Sarni-Manchado et al., 1997; Tattersall et al., 1997). There are also genes whose transcript levels decrease at around the time of véraison (e.g. the genes encoding the grape putative vacuolar invertases [Davies and Robinson, 1996]). These changes in mRNAs levels are likely to be involved in the physical and metabolic changes that occur during ripening. Similar differences in steady-state mRNA levels have been observed between pre- and post-ripening states in other fruit. An example of this in nonclimacteric fruit is provided by strawberries, in which ripening commences when the auxin concentration decreases below a threshold level (Given et al., 1988). In strawberry, auxins reduce the levels of some mRNAs and proteins and increase the levels of others (Veluthambi and Poovaiah, 1984; Reddy and Poovaiah, 1990; Manning, 1994; Manning, 1998).
The different patterns of mRNA accumulation observed by northern analysis (Fig. 1) suggest that these genes are under a range of regulatory controls. Many of the sequences isolated by the differential screening were present at high levels in ripening fruit (as shown by northern-blot analysis and the frequency of cloning during library screening). The ripening-associated P/HPRGs Grip 3 and 4 sequences, for example, are major components of the profile of mRNAs present during berry ripening. Transcript levels for some cDNAs were much lower (as assayed by northern analysis) and were isolated only once during the differential screening procedure (e.g. Grip 55 and 58). This suggests that although the methodology used was more likely to isolate more populous ripening-associated species, it did not exclude the isolation of less-prevalent cDNAs. Further studies using techniques such as differential display may identify ripening-induced cDNAs present at lower levels.
The putative Grip proteins described in this paper were divided into two general groups based on their proposed functions in the berry. One group consists of proteins that may be involved in cell wall structure and includes members of the P/HRGP family, a diverse group of proteins, many of which do not have proven functions but are thought to be involved in providing additional support to the polysaccharide network in the cell wall by the formation of intermolecular cross-links (Sommer-Knudsen et al., 1998). The expression of genes encoding these proteins in other species is variously induced by wounding, by pathogen attack, during normal developmental processes, and during root nodule formation. The proposal that the putative Pro-rich cell wall proteins described in this paper are expressed to high levels after véraison gives further credence to the observations of Nunan et al. (1998). These authors have shown that the protein content of berry mesocarp cell walls increased from approximately 8% by weight early in development to nearly 12% after véraison. Amino acid analysis showed that the levels of Hyp increased dramatically over the same period. A similar increase in the Hyp content of the Shiraz berries used in this study was found using a colorimetric assay (data not shown).
The P/HRGPs produced in the berry at véraison could have two not necessarily exclusive functions. One would be to provide additional strength to the cell walls during rapid expansion of the cells; the other would be to restrict the invasion of pathogens. The berries from which the 10-wpf cDNA library was constructed did not have any visible signs of pathogenic attack, so a direct response to infection seems unlikely. Also, the expression of Grip 3, 4, 13, and 15 in berries only after ripening had commenced suggests that their expression is under developmental control rather than being a response to environmental factors or pathogenic infection. It is possible that the expression of these putative cell wall genes in ripening berries is a developmentally regulated prophylactic measure against pathogenic attack. The existence of such a program in grapes is indicated by the ripening-associated induction of expression of pathogenesis-related genes and the expression of the corresponding proteins in uninfected berries after véraison (Robinson et al., 1997; Tattersall et al., 1997; Jacobs et al., 1999).
The considerable increase in cell volume after véraison (Coombe, 1992) requires the berry cell walls to expand rapidly. Pericarp cell walls do not appear to thicken appreciably during ripening (Hardie et al., 1996) and there are no major changes to the composition of the cell wall polysaccharides during ripening (Nunan et al., 1998). Thus, the expression of the putative P/HPGPs during ripening may be a developmentally controlled part of the normal ripening process designed to hold the cell walls together as they undergo the second phase of rapid expansion and softening.
The remaining Grip putative proteins appear to be non-cell wall proteins likely to be involved in stress response. This is suggested by the proposed functions of their homologs in other plants and by the stimuli that enhance the expression of these homologous genes. Stress due to changes in osmotic potential may occur during low-temperature treatments, pathogen infection, drought, salinity (Bray, 1997), and, as argued below, due to the storage of large amounts of osmotically active substances in storage tissues such as fruit. During berry ripening, cells expand rapidly as they accumulate large amounts of the hexoses, Glc, Fru, and water. This results in considerable changes in osmotic pressure and water potential. Fruit such as grapes, which store particularly high levels of sugars (approximately 20% [w/w] hexoses when fully ripe [Lott and Barrett, 1967]), may be especially vulnerable to the reduction in water activity caused by the high vacuolar levels of sugars. Part of the adjustment to the rapid increase in vacuolar sugar levels may be the synthesis of proteins involved in stress management. A number of cDNAs cloned in this study have homologs implicated in the response to water deficit in other plants.
Some of the putative Grip proteins also have homologs that are expressed during morphogensis in other plants. Neale et al. (1990) concluded that some genes normally associated with the response to pathogens (including a chitinase, a β-1,3-glucanse, and an extensin) are involved in normal developmental processes in healthy tobacco plants. Metallothionein-like proteins are expressed in a developmental manner during such processes as leaf senescence (Buchanan-Wollaston, 1994), the ethylene-promoted abscission of leaflets (Coupe et al., 1995), and embryogenesis (Chatthai et al., 1997).
The changes observed in the mRNA population at véraison may be influenced by changes in the levels of plant growth regulators. In grape berries indole acetic acid levels are highest early in fruit development and decrease to low levels during the period leading up to véraison (Cawthon and Morris, 1982). When a synthetic, auxin-like compound is applied to grape berries just prior to véraison, the transcript levels of genes normally expressed prior to véraison are maintained and the accumulation of transcripts for ripening-associated genes is suppressed (Davies et al., 1997). This coincides with a delay in the commencement of ripening and in the usual post-véraison increase in ABA levels. ABA is often associated with the control of expression of stress-related genes (Bray, 1997; Zhu et al., 1997), and many of the cDNAs isolated by differential screening have homologs implicated in the stress response.
In summary, the putative Grip proteins may function to protect tissues that are more at risk of pathogenic infection due to changes in cell wall properties during ripening (and possibly other morphogenic events). Equally important may be the role of some of the putative Grip proteins in protecting ripening berry tissues from the consequences of an important part of the ripening process itself, i.e. the storage of large amounts of hexoses (and water) in the cell vacuole.
ACKNOWLEDGMENTS
We thank John and Di Harvey for the care and provision of grapevine material, Melissa Pickering and Jude Osborne for technical assistance, and Dr. Paul Boss, Prof. Geoff Fincher, Dr. Maria Hrmova, Dr. Anna Koltunow, and Andrew Jacobs for helpful discussions.
LITERATURE CITED
- Aggelis A, John I, Karvouni Z, Grierson D. Characterization of two cDNA clones for mRNAs expressed during ripening of melon (Cucumis meloL.) fruits. Plant Mol Biol. 1997;33:313–322. doi: 10.1023/a:1005701730598. [DOI] [PubMed] [Google Scholar]
- Boss PK, Davies C, Robinson SP. Analysis of the expression of anthocyanin pathway genes in developing Vitis viniferaL. cv Shiraz grape berries and the implications for pathway regulation. Plant Physiol. 1996;111:1059–1066. doi: 10.1104/pp.111.4.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray EA. Plant responses to water deficit. Trends Plant Sci. 1997;2:48–54. [Google Scholar]
- Buchanan-Wollaston V. Isolation of cDNA clones for genes that are expressed during leaf senescence in Brassica napus. Plant Physiol. 1994;105:839–846. doi: 10.1104/pp.105.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Moore GA, Guy CL. An unusual group 2 LEA gene family in citrus responsive to low temperature. Plant Mol Biol. 1995;29:11–23. doi: 10.1007/BF00019115. [DOI] [PubMed] [Google Scholar]
- Cawthon DL, Morris JR. Relationship of seed number and maturity to berry development, fruit maturation, hormonal changes and uneven ripening of ‘Concord’ (Vitis labruscaL.) grapes. J Am Hortic Soc. 1982;107:1097–1104. [Google Scholar]
- Chatthai M, Kaukinen KH, Tranbarger TJ, Gupta PK, Misra S. The isolation of a novel metallothionein-related cDNA expressed in somatic and zygotic embryos of Douglas-fir: regulation by ABA, osmoticum, and metal ions. Plant Mol Biol. 1997;34:243–254. doi: 10.1023/a:1005839832096. [DOI] [PubMed] [Google Scholar]
- Chevalier C, Bourgeois E, Pradet A, Raymond P. Molecular cloning and characterization of six cDNAs expressed during glucose starvation in excised maize (Zea maysL.) root tips. Plant Mol Biol. 1995;28:473–485. doi: 10.1007/BF00020395. [DOI] [PubMed] [Google Scholar]
- Choi D, Kim HM, Yun HK, Park J-A, Kim WT, Bok SH. Molecular cloning of a metallothionein-like gene from Nicotiana glutinosaL. and its induction by wounding and tobacco mosaic virus infection. Plant Physiol. 1996;112:353–359. doi: 10.1104/pp.112.1.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clendennen SK, May GD. Differential gene expression in ripening banana fruit. Plant Physiol. 1997;115:463–469. doi: 10.1104/pp.115.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombe BG. Research on development and ripening of the grape berry. Am J Enol Vitic. 1992;43:101–110. [Google Scholar]
- Coombe BG, Bishop GR. Development of the grape berry: II. Changes in the diameter and deformability during veraison. Aust J Agr Res. 1980;31:499–509. [Google Scholar]
- Coupe SA, Taylor JE, Roberts JA. Characterisation of an mRNA encoding a metallothionein-like protein that accumulates during ethylene-promoted abscission of Sambucus nigraL. leaflets. Planta. 1995;197:442–447. doi: 10.1007/BF00196665. [DOI] [PubMed] [Google Scholar]
- Davies C, Boss PK, Robinson SP. Treatment of grape berries, a nonclimacteric fruit with a synthetic auxin, retards ripening and alters the expression of developmentally regulated genes. Plant Physiol. 1997;115:1155–1161. doi: 10.1104/pp.115.3.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies C, Robinson SP. Sugar accumulation in grape berries: cloning of two putative vacuolar invertase cDNAs and their expression in grapevine tissue. Plant Physiol. 1996;111:275–283. doi: 10.1104/pp.111.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans KM, Gatehouse JA, Lindsay WP, Shi J, Tommey AM, Robinson NJ. Expression of the pea metallothionein-like gene PsMTA in Escherichia coli and Arabidopsis thaliana and analysis of trace metal ion accumulation: implications for function PsMTA. Plant Mol Biol. 1992;20:1019–1028. doi: 10.1007/BF00028889. [DOI] [PubMed] [Google Scholar]
- Fils-Lycaon BR, Wiersma PA, Eastwell KC, Sautiere P. A cherry protein and its gene, abundantly expressed in ripening fruit, have been identified as thaumatin-like. Plant Physiol. 1996;111:269–273. doi: 10.1104/pp.111.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohman A, Dush MK, Martin GR. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Given NK, Venis MA, Grierson D. Hormonal regulation of ripening in the strawberry, a non-climacteric fruit. Planta. 1988;174:402–406. doi: 10.1007/BF00959527. [DOI] [PubMed] [Google Scholar]
- Gray J, Picton S, Shabbeer J, Schuch W, Grierson D. Molecular biology of fruit ripening and its manipulation with antisense genes. Plant Mol Biol. 1992;19:69–87. doi: 10.1007/BF00015607. [DOI] [PubMed] [Google Scholar]
- Gray JE, Picton S, Giovannoni JJ, Grierson D. The use of transgenic and naturally occurring mutants to understand and manipulate tomato fruit ripening. Plant Cell Environ. 1994;17:557–571. [Google Scholar]
- Griffing LR, Nessler CL. Immunolocalization of the major latex proteins in developing laticifers of opium poppy (Papaver somniferum) J Plant Physiol. 1989;134:357–363. [Google Scholar]
- Guan C, Akkermans ADL, van Kammen A, Bisseling T, Pawlowski K. ag13 is expressed in Alnus glutinosanodules in infected cells during endosymbiont degradation and in the nodule pericycle. Physiol Plant. 1997;99:601–606. [Google Scholar]
- Hardie WJ, O'Brien TP, Jaudzems VG. Morphology, anatomy and development of the pericarp after anthesis in grape, Vitis viniferaL. Aust J Grape Wine Res. 1996;2:97–142. [Google Scholar]
- Harris JM, Kriedemann PE, Possingham JV. Anatomical aspects of grape berry development. Vitis. 1968;7:106–119. [Google Scholar]
- Hobohn U, Sander C. A sequence property approach to searching protein databases. J Mol Biol. 1995;251:390–399. doi: 10.1006/jmbi.1995.0442. [DOI] [PubMed] [Google Scholar]
- Hsieh H-M, Liu W-K, Huang PC. A novel stress-inducible metallothionein-like gene from rice. Plant Mol Biol. 1995;28:381–389. doi: 10.1007/BF00020388. [DOI] [PubMed] [Google Scholar]
- Jacobs AK, Dry IB, Robinson SP. Induction of different pathogenesis-related cDNAs in grapevine infected with powdery mildew and treatment with ethephon. Plant Pathol. 1999;48:325–336. [Google Scholar]
- Kim SY, Chung H, Thomas TL. Isolation of a novel class of bZIP transcription factors that interact with ABA-responsive and embryo-specification elements in the Dc3 promoter using a modified yeast one-hybrid system. Plant J. 1997;11:1237–1251. doi: 10.1046/j.1365-313x.1997.11061237.x. [DOI] [PubMed] [Google Scholar]
- Lam PF, Abu Bakar UK. Nucleotide sequence of cDNA clone (accession no. Y08322) encoding a metallothionein-like protein from papaya fruit (PGR 96-120) Plant Physiol. 1996;112:1735. [Google Scholar]
- Lane B, Kajioka R, Kennedy T. The wheat germ Ec protein is a zinc-containing metallothionein. Biochem Cell Biol. 1987;49:71–83. [Google Scholar]
- Ledger SE, Gardner RC. Cloning and characterization of five cDNAs for genes differentially expressed during fruit development of kiwifruit (Actinidia deliciosa var deliciosa) Plant Mol Biol. 1994;25:877–886. doi: 10.1007/BF00028882. [DOI] [PubMed] [Google Scholar]
- Lott RV, Barrett HC. The dextrose, levulose, sucrose and acid content of the juice from 39 grape clones. Vitis. 1967;6:257–268. [Google Scholar]
- Loulakakis KA. Nucleotide sequence of a Vitis vinifera L. cDNA (accession no. Y10992) encoding for osmotin-like protein (PGR 97-064) Plant Physiol. 1997;113:1464. [Google Scholar]
- Manning K. Changes in gene expression during strawberry fruit ripening and their regulation by auxin. Planta. 1994;194:62–68. [Google Scholar]
- Manning K. Isolation of a set of ripening-related genes from strawberry: their identification and possible relationship to fruit quality traits. Planta. 1998;205:622–631. doi: 10.1007/s004250050365. [DOI] [PubMed] [Google Scholar]
- Medina-Escobar N, Cardenas J, Valpuesta V, Munoz-Blanco J, Caballero JL. Cloning and characterization of cDNAs from genes differentially expressed during the strawberry fruit ripening process by a MAST-PCR-SBDS method. Anal Biochem. 1997;248:288–296. doi: 10.1006/abio.1997.2110. [DOI] [PubMed] [Google Scholar]
- Monroy AF, Castonguay Y, Laberge S, Sarhan F, Venzina LP, Dhindsa RS. A new cold-induced alfalfa gene is associated with enhanced hardening at subzero temperature. Plant Physiol. 1993;102:873–879. doi: 10.1104/pp.102.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale AD, Wahleithner JA, Lund M, Bonnett HT, Kelly A, Meeks-Wagner DR, Peacock WJ, Dennis ES. Chitinase, β-1,3-glucanase, osmotin, and extensin are expressed in tobacco explants during flower formation. Plant Cell. 1990;2:672–684. doi: 10.1105/tpc.2.7.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunan KJ, Sims IM, Bacic A, Robinson SP, Fincher GB. Changes in cell wall composition during ripening of grape (Vitis viniferaL.) berries. Plant Physiol. 1998;118:783–792. doi: 10.1104/pp.118.3.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picton S, Gray J, Barton S, AbuBakar U, Lowe A, Grierson D. cDNA cloning and characterisation of novel ripening-related mRNAs with altered patterns of accumulation in the ripening inhibitor (rin) tomato ripening mutant. Plant Mol Biol. 1993;23:193–207. doi: 10.1007/BF00021431. [DOI] [PubMed] [Google Scholar]
- Pozueta-Romero J, Klein M, Houlne G, Schantz M-L. Characterization of a family of genes encoding a fruit-specific wound-stimulated protein of bell pepper (Capsicum annum): identification of a new family of transposable elements. Plant Mol Biol. 1995;28:1011–1025. doi: 10.1007/BF00032663. [DOI] [PubMed] [Google Scholar]
- Proust J, Houlne G, Schantz M-L, Schantz R. Characterization and gene expression of an annexin during fruit development in Capsicum annuum. FEBS Lett. 1996;383:208–212. doi: 10.1016/0014-5793(96)00252-9. [DOI] [PubMed] [Google Scholar]
- Reddy ASN, Poovaiah BW. Molecular cloning and sequencing of a cDNA for an auxin-repressed mRNA: correlation between fruit growth and the repression of the auxin regulated gene. Plant Mol Biol. 1990;14:127–136. doi: 10.1007/BF00018554. [DOI] [PubMed] [Google Scholar]
- Reid SJ, Ross GS. Up-regulation of two cDNA clones encoding metallothionein-like proteins in apple fruit during cool storage. Physiol Plant. 1997;100:183–189. [Google Scholar]
- Robinson NJ, Tommey AM, Kuske C, Jackson PJ. Plant metallothioneins. Biochem J. 1993;295:1–10. doi: 10.1042/bj2950001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson SP, Jacobs AJ, Dry IB. A class IV chitinase is highly expressed in grape berries during ripening. Plant Physiol. 1997;114:771–778. doi: 10.1104/pp.114.3.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman RA, Tikhonova I, Bordelon BP, Hasegawa PM, Bressan RA. Coordinate accumulation of antifungal proteins and hexoses constitutes a developmentally controlled defense response during fruit ripening in grape. Plant Physiol. 1998;117:465–472. doi: 10.1104/pp.117.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarni-Manchado P, Verries C, Tesniere C. Molecular characterisation and structural analysis of one alcohol dehydrogenase gene (GV-Adh1) expressed during ripening of grapevine (Vitis viniferaL.) berry. Plant Sci. 1997;125:177–187. [Google Scholar]
- Sommer-Knudsen J, Bacic A, Clarke AE. Hydroxyproline-rich plant glycoproteins. Phytochemistry. 1998;47:483–497. [Google Scholar]
- Takahashi R, Joshee N, Kitagawa Y. Induction of chilling resistance by water stress, and cDNA sequence analysis and expression of water stress-regulated genes in rice. Plant Mol Biol. 1994;26:339–352. doi: 10.1007/BF00039544. [DOI] [PubMed] [Google Scholar]
- Takahashi R, Shimosaka E. cDNA sequence analysis and expression of two cold-regulated genes in soybean. Plant Science. 1997;123:93–104. [Google Scholar]
- Tattersall DB, van Heeswijck R, Hoj PB. Identification and characterization of a fruit-specific, thaumatin-like protein that accumulates at very high levels in conjunction with the onset of sugar accumulation and berry softening in grapes. Plant Physiol. 1997;114:759–769. doi: 10.1104/pp.114.3.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veluthambi K, Poovaiah BW. Auxin-regulated polypeptide changes at different stages of strawberry fruit development. Plant Physiol. 1984;75:349–353. doi: 10.1104/pp.75.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiersma PA, Wu Z, Wilson SM. A fruit-related metallothionein-like cDNA clone from sweet cherry (accession no. AF028013) corresponds to fruit genes from diverse species (PGR 98-015) Plant Physiol. 1998;116:867. [Google Scholar]
- Wilson RC, Long F, Maruoka EM, Cooper JB. A new proline-rich early nodulin from Medicago truncatulais highly expressed in nodule meristem cells. Plant Cell. 1994;6:1265–1275. doi: 10.1105/tpc.6.9.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhead M, Taylor MA, Brennan R, McNicol RJ, Davies HV. Cloning and characterisation of the cDNA clones of five genes that are differentially expressed during ripening in the fruit of blackcurrant (Ribes nigramL.) J Plant Physiol. 1998;153:381–393. [Google Scholar]
- Zhu J-K, Hasegawa PM, Bressan RA. Molecular aspects of osmotic stress in plants. Crit Rev Plant Sci. 1997;16:253–277. [Google Scholar]