Abstract
Streptococcus gallolyticus subsp. gallolyticus Sgg (formerly known as S. bovis type I) is the main causative agent of septicemia and infective endocarditis (IE) in elderly and immunocompromised persons. It belongs to the few opportunistic bacteria, which have been strongly associated to colorectal cancer (CRC). A literature survey covering a period of 40 years (1970–2010) revealed that 65% of patients diagnosed with an invasive Sgg infection had a concomitant colorectal neoplasia. Sgg is associated mainly with early adenomas and may thus constitute an early marker for CRC screening. Sgg has been described as a normal inhabitant of the rumen of herbivores and in the digestive tract of birds. It is more rarely detected in human intestinal tract (2.5–15%). Recent molecular analyses indicate possible zoonotic transmission of Sgg. Thanks to the development of a genetic toolbox and to comparative genomics, a number of factors that are important for Sgg pathogenicity have been identified. This review will highlight the role of Sgg pili in host colonization and how their phase-variable expression contributes to mitigate the host immune responses and finally their use as serological diagnostic tool. We will then present experimental data addressing the core question whether Sgg is a cause or consequence of CRC. We will discuss a few recent studies examining the etiological versus non-etiological participation of Sgg in colorectal cancer with the underlying mechanisms.
Keywords: S. gallolyticus, colorectal cancer, infective endocarditis, gut colonization, pili
Introduction
Streptococcus gallolyticus belongs to the Group D streptococci, a large group of phenotypically diverse bacteria known as the S. bovis/S. equinus complex (SBSEC), which consist of safe-graded bacteria used in food-fermentation, commensal bacteria of the gut and opportunistic pathogens in both humans and animals. About 15 years ago, a revised classification of this streptococcal group has been proposed (Poyart et al., 2002; Schlegel et al., 2003), but the new taxonomy is still not completely adopted by the scientific community, especially the clinicians, resulting in confusing names in the literature. The currently admitted classification based on multilocus sequence typing (MLST) data defines seven subspecies: Streptococcus gallolyticus subsp. gallolyticus (Sgg), S. gallolyticus subsp. macedonicus (Sgm), S. gallolyticus subsp. pasteurianus (Sgp), Streptococcus infantarius subsp. infantarius (Sii), Streptococcus lutetiensis, Streptococcus alactolyticus and Streptococcus equinus (Jans et al., 2015). Sgg is an opportunistic pathogen causing septicemia and endocarditis in elderly persons. Intriguingly, several clinical studies have demonstrated a strong association between invasive infections with Sgg and colon neoplasia in humans (Boleij et al., 2011). Colorectal cancer (CRC) is firstly a genetic disease that develop over several years involving a series of genetic changes (i.e., somatic mutations and epigenetic modifications) known as the adenoma-carcinoma sequence. Emerging studies have closely linked CRC development with gut microbiota changes (Braten et al., 2017; Flemer et al., 2017; Gao et al., 2017; Lucas et al., 2017). Earlier studies indicated a strong correlation between CRC and intestinal colonization by single bacterial species, such as colibactin-producing Escherichia coli, toxin-producing Bacteroides fragilis, Fusobacterium spp. and Streptococcus gallolyticus subspecies gallolyticus (Sgg). Sgg is commonly found in the flora of herbivores’ rumen, and therefore transmission from animal to human is highly suspected. In this review, we will discuss the specific traits that make Sgg a successful opportunistic pathogen in humans. We will then ask the million-dollar question whether Sgg is a cause or consequence of CRC. Recent evidence supporting the oncogenic role of Sgg but also evidences supporting the beneficial role of tumor microenvironment for Sgg outgrowth will be discussed.
S. gallolyticus subsp. gallolyticus (Sgg) Is an Opportunistic Pathogen
Sgg is a normal inhabitant of the gastrointestinal tract of different mammalian herbivores and birds. This bacterium was first isolated from Koala feces most probably because Sgg is able to degrade tannins, which are highly polar polyphenolic molecules, present in high quantity in eucalyptus leaves (Osawa, 1990). S. gallolyticus owes its name to its capacity to decarboxylate gallate, an organic acid derived from tannins hydrolysis (Osawa and Sasaki, 2004). S. gallolyticus can also be found outside the animal host as a saprophyte (Chamkha et al., 2002; Braten et al., 2017; Flemer et al., 2017; Gao et al., 2017; Lucas et al., 2017). Sgg was also detected in the human intestinal tract, but remains a low-abundance species (2.5–15%). However, a recent study conducted in Germany using sensitive PCR technique to detect Sgg indicated a higher carriage rate estimated at 62.5% in the stools of 99 healthy volunteers (Dumke et al., 2017). Rural residency and animal contact were shown to increase the detection rate of Sgg in humans further supporting the zoonotic potential of this bacterium. Indeed, the first complete genome of Sgg has provided several insights into the adaptation of Sgg to the rumen of herbivores and its capacity to cause endocarditis (Rusniok et al., 2010). In particular, it revealed the existence of many genes involved in plant carbohydrates degradation, two genes encoding tannases and a gene encoding a bile salt hydrolase conferring the bacterium the ability to survive in the gut. Metabolic pathways analysis indicates that Sgg should be able to synthesize all 20 amino-acids and most vitamins, thus displaying few nutritional requirements (Rusniok et al., 2010). Similar to other pathogenic streptococci, Sgg encodes an extracellular capsule exhibiting a high degree of similarity to S. pneumoniae serotype 23F. It was proposed that the polysaccharide capsule protects Sgg from the host innate immune responses, blocking for example pro-inflammatory IL-8 response in epithelial cells (Boleij et al., 2011). Surface proteins of pathogenic bacteria are often involved in the colonization of host tissues. Three pilus loci were revealed and further studied. To analyze the role of these pili, a genetic toolbox was developed enabling inactivation or overexpression of specific genes in Sgg UCN34 (Danne et al., 2013).
Colonization of Host Tissues by S. gallolyticus subsp. gallolyticus
Like their counterpart in Gram-negative bacteria, Gram-positive pili have often been associated with bacterial attachment and colonization of the host tissues (Danne and Dramsi, 2012). In Sgg, pili were first revealed by ultrastructural studies from pigeons virulent strains (Vanrobaeys et al., 1999). Molecular evidence was provided a decade later in strain TX20005 when a genome-wide analysis revealed the presence of three putative pilus operons (Sillanpaa et al., 2009). The first complete genome of another Sgg clinical isolate UCN34 confirmed the presence of three pilus operons, namely pil1, pil2 and pil3 (Rusniok et al., 2010). Both pil1 and pil3 are highly conserved loci among sequenced Sgg strains, whereas pil2 appears more variable (Sillanpaa et al., 2009; Rusniok et al., 2010). The first virulence factor characterized in Sgg was the Pil1 pilus. This pilus was shown to mediate Sgg binding to collagen types I and IV and in the bacterial attachment to the heart valves, thereby initiating endocarditis development (Danne et al., 2011). The Pil1-associated adhesin was shown to bind to various types of collagen with different affinities (Sillanpaa et al., 2009). More recently, the Pil1 adhesin was shown to bind to blood factor XII with a very high affinity, leading to activation of human contact system, which in turn results in prolongation of the coagulation time (Isenring et al., 2017). Manipulation of the host coagulation system by Sgg is proposed to contribute to virulence. Interestingly, Sgg isolates causing septicemia in pigeons are not able to bind to collagen type I (Vanrobaeys et al., 2000). While collagen type I is the major structural component of human heart, collagen type IV is found in the basal lamina layer underneath epithelial tissue. It is worth pointing out that colonic tumors display higher levels of collagen IV compared to normal tissues (Skovbjerg et al., 2009), which may explain a higher colonization of tumor sites by Sgg.
Next, it was shown that the Pil3 pilus was involved in Sgg binding to colonic mucus and thus promotes colonization of murine distal colon (Skovbjerg et al., 2009; Martins et al., 2015, 2016). By immunofluorescence on intestinal tissues following mice infection, Sgg was mainly found entrapped in the mucus layer (Skovbjerg et al., 2009; Martins et al., 2015, 2016). The Pil3A adhesin was shown to bind to MUC2 and MUC5AC mucins (Skovbjerg et al., 2009; Martins et al., 2015, 2016). MUC2 mucin is a major constituent of the adhesin mucus, whereas MUC5AC is not detected under normal circumstances. Importantly, aberrant and mislocalized expression of MUC5AC mucin in adenomas and carcinomas has been reported (Bartman et al., 1999; Sylvester et al., 2001; Bryant and Stow, 2004), as well as modification of mucins glycosylation patterns during colonic carcinogenesis (Devine and McKenzie, 1992; Itzkowitz et al., 1992; Mann et al., 1997; Jenab et al., 2001; Mesquita et al., 2003).
Pil1 and Pil3 pilus are expressed heterogeneously in Sgg UCN34 population (Danne et al., 2014). In UCN34, two distinct sub-populations were found: two-third of low-piliated bacteria (PilLow), and one third of high-piliated bacteria (Pilhigh). The molecular mechanism involved in this regulation has been identified as a combination of phase variation and transcriptional attenuation (Danne et al., 2014). Genetic evidence demonstrated that this heterogeneous expression is dependent on changes in the pil1 promoter region which includes a leader peptide composed of a variable number of GCAGA repeats followed by a transcription terminator. Addition or deletion of a single repeat by slip-strand mispairing during replication modifies the length of the regulatory leader peptide to be translated. The synthesis of a longer leader peptide controls the switch of pilus transcription through a destabilization of a stem-loop transcription terminator upstream of pil1 genes (Danne et al., 2014). Hyper-piliated bacteria were found more prone to phagocytosis by human macrophages and had a lower rate of survival in human blood compared to bacteria expressing low levels of pili (Danne et al., 2014). Thus, it was proposed that stochastic expression of the pilus in Sgg is an advantageous bacterial feature insuring an optimal tissue colonization and dissemination while evading the host immune responses.
Pilus components of pathogenic streptococci were shown to be promising vaccine candidates because they can induce protective immunity in mouse models (Margarit et al., 2009; Soriani and Telford, 2010). Since pili are often highly immunogenic surface appendages, detection of specific anti-pilins IgG may constitute an ideal serological diagnostic tool that could help in discriminating patients with early adenomas from healthy people. A small proof of concept study combining four pilus proteins demonstrated some potential for this approach (Boleij et al., 2012b). Using a larger cohort (576 CRC cases and 576 controls matched by sex, age and providence), it was shown that only 14% of CRC patients displayed Sgg-specific IgG antibodies (Butt et al., 2016). Detection of Sgg presence by measuring mucosal IgA antibodies may increase the sensitivity of this test.
Colorectal Cancer and Microbiota
Colorectal cancer (CRC) is one of the most commonly diagnosed tumors with a high mortality rate (Ferlay et al., 2015). The global burden of CRC is expected to increase by 60% to more than 2.2 million new cases and 1.1 million deaths by 2030 (Arnold et al., 2017). The majority of CRC cases are detected in Western countries with an incidence increasing every year, which correlates with population aging.
CRC development is a complex multi-factorial process occurring over many years as the result of an accumulation of genetic and epigenetic alterations in proto-oncogenes, tumor suppressor genes, and/or DNA repair genes, leading to transformation of normal colonic epithelium into glandular structures called adenocarcinomas (Fleming et al., 2012). The underlying causes of CRC are complex and heterogeneous. Both genetic and environmental factors can influence the initial steps and/or progression of CRC, which complicates the study of the disease etiology.
Depending on the origin of mutations, CRC can be classified as sporadic (70%) or inherited (30%). One key feature of both sporadic and familial CRC tumors is their high degree of genomic instability, arising from distinct molecular mechanisms defining different tumor molecular subtypes: (1) chromosomal instability (CIN), (2) microsatellite instability (MIS) resulting in hyper mutated tumors, (3) epigenetic instability with alteration in CpG island methylation (Muller et al., 2016). Despite this important molecular heterogeneity, defined signaling pathways are consistently altered in CRC tumors, including Wnt, TGF-beta, PI3K, RTK-RAS and P53 signaling (The Cancer Genome Atlas Network, 2012). Wnt pathway, which is crucial for intestinal epithelium homeostasis, is indeed constitutively activated in more than 90% of CRC tumors, and loss of function mutations in its negative regulator APC are found in 80% of non-hypermutated and 50% of hyper-mutated CRC tumors (The Cancer Genome Atlas Network, 2012).
In addition to genetic alterations, the tumor microenvironment plays a critical role in CRC development and important contributing factors are linked to nutrition, inflammation, epigenetics modifications and gut microbiota. The gut microbiota is currently considered as an organ which plays a crucial role in regulating host intestinal homeostasis through its capacity to modulate several biological processes including barrier, immunity and metabolic functions. A combination of external factors can influence microbial composition, including host genetics, diet, lifestyle, and environmental factors. These perturbations in the microbiota shift influence the balance between healthy and carcinogenesis. Alterations of the colon microbiota is recognized as an important player in the initiation and progression of CRC (Schwabe and Jobin, 2013 #337; Gagniere et al., 2016 #364; Braten et al., 2017; Flemer et al., 2017; Gao et al., 2017; Lucas et al., 2017). In addition, microbiota can modulate cancer therapy by its extensive metabolic capacity and profound immunomodulatory effects (reviewed by Pope et al., 2017 #462).
A critical question regarding CRC associated bacteria is whether they represent a consequence of altered host mucosal tissues or if the bacteria by themselves can have an oncogenic or pro-tumoral effect (Sears and Garrett, 2014; Raskov et al., 2017). Using microbiota transfer experiments in various CRC animal models, several studies have clearly shown that cancer-associated microbiota plays a role in cancer progression (Couturier-Maillard et al., 2013). Depletion of the intestinal bacterial microbiota in mice using antibiotics reduces the risk of colon cancer (Schwabe and Jobin, 2013 #337). Several molecular mechanisms have been proposed to explain bacteria-induced pro-tumoral effects: (i) induction of chronic inflammation; (ii) bacterial transformation of host metabolites into carcinogens; (iii) expression of specific bacterial factors such as toxins endowed with oncogenic properties, (iv) barrier failure (Schwabe and Jobin, 2013 #337; Gagniere et al., 2016 #364). A few bacterial species have been identified as playing a role in colorectal carcinogenesis such as Fusobacterium nucleatum (Fn), enterotoxigenic Bacteroides fragilis (ETBF), colibactin- and genotoxin-producing Escherichia coli, Enterococcus faecalis, Clostridium septicum (Gagniere et al., 2016 #364).
Both Fn and ETBF were shown to alter the Wnt/β-catenin signaling pathway. F. nucleatum was shown to adhere to and invade colonic cells through its unique surface adhesin FadA (Rubinstein et al., 2013). FadA binds to the host cell receptor E-cadherin promoting attachment and invasion of epithelial cells by Fn. FadA binding to E-cadherin leads to activation of β-catenin signaling (Bryant and Stow, 2004), resulting in increased cell proliferation (Rubinstein et al., 2013). ETBF are prevalent in the colon mucosa of CRC patients and able to modulate the mucosal immune responses and to induce epithelial cell changes (Sears et al., 2014; Boleij et al., 2015; Purcell et al., 2017). It was shown that ETBF secrete a zinc-dependent metalloprotease toxin called BFT which cleaves E-cadherin, thus causing nuclear translocation of β-catenin, increased c-Myc expression and cell proliferation (Wu et al., 2003). Of note, Helicobacter pylori, which is the sole bacterium clearly responsible for gastric cancer development, activates the β-catenin pathway resulting in increased cell proliferation (Parsonnet et al., 1991; Franco et al., 2005). H. pylori strain specific CagA protein is translocated into the host cell cytoplasm by a type IV secretion pilus where it interacts with tyrosine kinase c-Met receptor and its co-receptor CD44, leading to β-catenin activation and cellular proliferation (Suzuki et al., 2009; Bertaux-Skeirik et al., 2015). Besides CagA, several other mechanisms leading to β-catenin activation have been described in H. pylori altering the expression of Wnt ligands (Kirikoshi et al., 2001), activating Wnt receptors (Gnad et al., 2010), suppressing GSK3β (Sokolova et al., 2008; Nakayama et al., 2009), interfering with β-catenin/TCF4 complex by down-regulating the gastric tumor suppressor Runx3 (Liu et al., 2012), and interacting with E-cadherin to disrupt the E-cadherin/β-catenin complex (Murata-Kamiya et al., 2007) highlighting the importance of this signaling pathway in cancer development.
Epidemiological Association Between CRC and S. gallolyticus subsp. gallolyticus
Sgg is an important cause of endocarditis, an inflammation of the inner layer of the heart (the endocardium) (Hoen et al., 2002). A relationship between Sgg-induced endocarditis and CRC was established for the first time by McCoy and Mason (1951). Later on, several epidemiological studies confirmed this association ranging from 47 to 85% between Sgg and CRC, depending on the techniques used for Sgg detection (Klein et al., 1977; Waisberg et al., 2002; Kok et al., 2007; Gupta et al., 2010; Corredoira et al., 2015; Chand et al., 2016). Most of these studies were performed on a selected cohort of patients with a history of Sgg bacteremia/endocarditis. A recent molecular analysis of tumoral and adjacent normal tissues from unselected CRC patients by quantitative PCR using Sgg-specific primers showed that about 74% of tumor tissue and 47% of adjacent normal tissues were positive to Sgg (Kumar et al., 2017). In striking contrast, another study on unselected CRC patients showed a much lower prevalence using quantitative real-time PCR. Only 6 out of 190 patients included (3.2%) were positive for Sgg (Andres-Franch et al., 2017). Nevertheless, the six positive cases were all from tumor tissue samples, while none of the normal mucosa samples presented Sgg DNA (Andres-Franch et al., 2017). These contradictory results either result from differences in the sampled population, but more likely from differences in the methodology used to detect Sgg (site of detection, number of samples, sample processing, conservation, enrichment of Sgg in specific medium, primers, qPCR techniques).
A recent comparison of colorectal neoplasms associated to Clostridium septicum (Cs) to Sgg showed several differences in clinical presentation, underlying conditions, prognosis and long-term follow-up (Corredoira et al., 2017). Sgg positive cases were associated with advanced (52.3% vs. 5.2% for Cs) and non-advanced adenomas (28.1% vs. 0% for Cs) and the tumor was located mostly in the distal colon (65.6%) but also in the cecum/ascending colon (23.4%) and transverse colon (10.9%). In contrast, Cs positive cases were mostly associated with advanced neoplasia/invasive carcinoma (94.7% vs. 19.5% for Sgg) that were mostly located in the cecum/ascending colon (73.7%). These differences suggest that each bacterium (here Cs or Sgg) affects differently tumor development and that Sgg may play a role at the very early steps of CRC development. This hypothesis is in line with earlier observation that the majority of patients with Sgg positive endocarditis had asymptomatic colorectal tumors that were occasionally benign adenomas (Klein et al., 1977). Since the publication of the first complete genome of Sgg strain UCN34 (Rusniok et al., 2010), molecular studies were undertaken to determine if Sgg is a cause or a consequence of CRC. It is also possible for both scenarios to co-exist (Figure 1).
FIGURE 1.
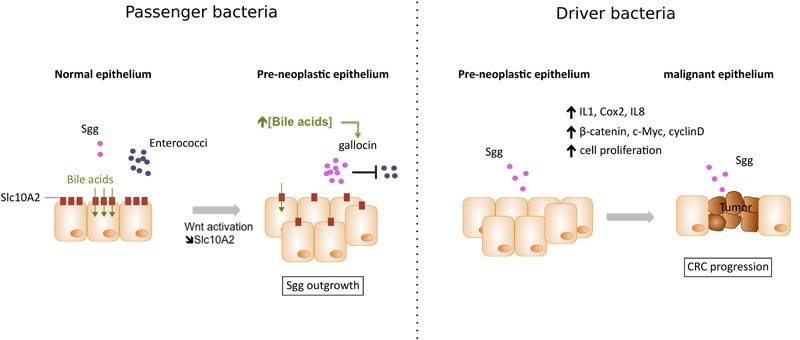
Two working models explaining Sgg association with colorectal cancer (CRC). (1) Sgg as a passenger bacterium: In pre-neoplastic epithelium, activation of the Wnt pathway leads to the downregulation of bile acids transporter Slc10A2 resulting in accumulation of bile acids- which in turn activates a specific “bacteriocin” enabling Sgg to kill related commensals (e.g., Enterococci). This local microbial imbalance can contribute to the development of CRC. (2) Sgg as a driver bacterium: High colonization of Sgg in pre-malignant epithelium can induce specific inflammatory responses (IL-1, COX-2, and IL-8) and increased cell proliferation associated with upregulation of β-catenin levels and its oncogenic downstream targets (c-Myc and cyclin D), thus accelerating transformation from pre-malignant to malignant epithelium.
Sgg as a Promoter of Colorectal Cancer
In favor of an etiological role of Sgg in CRC, the first experimental evidence that S. bovis could accelerate cancer development was reported in AOM-treated rats using S. bovis strain NCTC 8133 through increased inflammatory pathways (Ellmerich et al., 2000). However, following revision of S. bovis classification the strain NCTC 8133 was shown to belong to S. bovis type II/I now renamed S. infantarius (Biarc et al., 2004). In 2010, a molecular study demonstrating a significantly higher detection of Sgg in human neoplastic tissues versus normal adjacent tissue from the same patient was published (Abdulamir et al., 2010). It showed that bacterial colonization was accompanied by mRNA increase of genes encoding inflammatory molecules such as IL-1, COX-2, and IL-8; The authors proposed that this specific inflammatory response may drive the development of CRC (Abdulamir et al., 2010). A very recent study demonstrated that Sgg strain TX20005 promotes colorectal tumor development through increase of epithelial cell proliferation (Kumar et al., 2017). Using in vitro cell lines, Kumar et al. (2017) first showed that Sgg TX20005 increased cell proliferation in the following human colon cancer cell lines: HCT116, HT29, LoVo, but not in other colonic cell lines such as SW480, SW1116, or normal colonic cells CCD 841 CoN, FHC. Increased proliferation in responsive cell lines was associated with upregulation of β-catenin levels and its oncogenic downstream targets (c-Myc, cyclin D). Furthermore, using murine models, Kumar et al. (2017) showed that HCT116 cells cultured in vitro with Sgg and then injected into nude mice developed larger tumors as compared to control mice injected with HCTT16 co-cultured with non-pathogenic Lactococcus lactis MG1363. Secondly, in AOM-induced murine model of CRC, mice orally treated with Sgg displayed higher number of tumors, higher level of dysplasia, increased cell proliferation and β-catenin level in colon crypts as compared to control mice treated with L. lactis bacteria (Kumar et al., 2017). Interestingly, a preeminent early mutation in CRC is in the tumor suppressor gene APC found in 80% of human sporadic colon cancers and also responsible for the familial adenomatous polyposis syndrome, one of the main forms of hereditary colon cancer. Apc loss leads to the constitutive activation of Wnt/β-catenin pathway, which in turn induces cell proliferation and rapid loss of epithelial differentiation (reviewed in Zhan et al., 2017). Future studies will certainly aim at unraveling the molecular bases of Sgg-induced up-regulation of β-catenin in responsive cells.
Sgg Benefits From the Tumor Microenvironment
Consistent with intestinal dysbiosis reports in CRC, metabolomic studies revealed that CRC microenvironment is strongly altered when compared to normal mucosal environment (Hirayama et al., 2009). Key features indicate a drastic decrease in glucose and pyruvate levels and an increase in lactate (low pH), amino acids, lipids, and fatty acids. Growth of Sgg in spent media of human malignant colonic cells (Caco-2 and HCT116) was investigated and compared to other intestinal bacteria. It was shown that particular metabolites derived from increased glycolysis in tumor cells, such as F6P, 3PG or alanine, benefit to Sgg for its own multiplication (Boleij et al., 2012a). In line with these findings, S. bovis was found to be one of the most efficient bacteria to utilize glucose in an experimental human in vitro gut fermentation model (Egert et al., 2007).
In addition, it is tempting to hypothesize that interactions mediated by both Pil3 and Pil1 with colonic mucins expressed in tumors such as MUC5AC and collagen type IV, respectively, can further increase Sgg preferential colonization of dysplastic tissues within the colon.
Finally, we recently showed that Sgg strain UCN34 is able to produce a specific bacteriocin, named gallocin, which contributes to enhance bacterial colon colonization in tumor bearing mice (Aymeric et al., 2017). It was shown that gallocin is able to inhibit the growth of closely related Enterococci commensals, thus creating an appropriate colonization niche for Sgg. Gallocin activity is strongly potentiated in the presence of secondary bile acids such as deoxycholic and lithocholic acids, which are known risk factors for CRC. By comparing Apc+/- mice and their WT counterparts the authors showed that the presence of intestinal polyps per se, was sufficient to enhance Sgg UCN34 colonization in a gallocin-dependent manner. Indeed, following a one shot inoculation, Sgg UCN34 persisted for 3 months in adenoma-bearing host, whereas it was progressively excluded from the gut of healthy mice. This colonization advantage was lost with the gallocin-deficient mutant. The authors further unraveled a new link between Wnt pathway activation, an early step in CRC development, and increased luminal concentration of secondary bile acids by showing that Wnt activation resulted in decreased expression of the apical bile acids transporter Slc10A2 and reduced luminal bile acids reabsorption. Apc mutation, increased carcinogenic secondary bile acids and SGG colonization may thus be part of a vicious pro-tumoral triangle.
Conclusion
For more than 50 years, clinical studies have strongly linked the presence of Streptococcus bovis biotype I, renamed Streptococcus gallolyticus subsp. gallolyticus (Sgg), to CRC. The first direct demonstration of the etiological role of Sgg isolate TX20005 in promoting CRC development was provided very recently (Kumar et al., 2017). But Sgg has also been shown to behave as a passenger bacterium benefiting from tumor metabolites (Boleij et al., 2012a) and able to secrete a specific “bacteriocin” that can kill closely related gut commensals (Aymeric et al., 2017) thus enabling a better colonization of murine colon in CRC-context. We thus conclude that Sgg is both a passenger and a cancer promoting bacterium (Figure 1). But in order to become a driver bacterium, Sgg first needs to colonize the colon and it does so only if pre-malignant conditions exist. So Sgg is not the principal cause of CRC but an auxiliary factor accelerating the development of CRC. Ultimately, the strong association of Sgg with CRC constitutes a solid argument to recommend a systematic colonoscopy for assessment of occult neoplasia in patients suffering of Sgg infections. Development of new molecular tools for the sensitive and specific detection of specific CRC-associated bacteria should help in the early detection of subclinical colonic lesions but may also add a weapon in the oncologists’ arsenal as demonstrated recently (Bullman et al., 2017; Yu et al., 2017). A future area of investigation will be to study the relationship between Sgg and host immunity, another important player in CRC development.
Author Contributions
MM, EP-K, LA, and SD designed and wrote this review.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The handling Editor declared a past co-authorship with one of the authors SD.
Acknowledgments
We would like to thank Bruno Périchon, Camille Danne, Chloé Cassaro, Alexis Proutière, Laurence du Merle, and Patrick Trieu-Cuot for critical reading of the manuscript. We thank the Institut National du Cancer (INCA) for their financial support to this work, project PLBIO16-025 (to SD).
References
- Abdulamir A. S., Hafidh R. R., Bakar F. A. (2010). Molecular detection, quantification, and isolation of Streptococcus gallolyticus bacteria colonizing colorectal tumors: inflammation-driven potential of carcinogenesis via IL-1, COX-2, and IL-8. Mol. Cancer 9:249. 10.1186/1476-4598-9-249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres-Franch M., Galiana A., Sanchez-Hellin V., Ochoa E., Hernandez-Illan E., Lopez-Garcia P., et al. (2017). Streptococcus gallolyticus infection in colorectal cancer and association with biological and clinical factors. PLoS One 12:e0174305. 10.1371/journal.pone.0174305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold M., Sierra M. S., Laversanne M., Soerjomataram I., Jemal A., Bray F. (2017). Global patterns and trends in colorectal cancer incidence and mortality. Gut 66 683–691. 10.1136/gutjnl-2015-310912 [DOI] [PubMed] [Google Scholar]
- Aymeric L., Donnadieu F., Mulet C., du Merle L., Nigro G., Saffarian A., et al. (2017). Colorectal cancer specific conditions promote Streptococcus gallolyticus gut colonization. Proc. Natl. Acad. Sci. U.S.A. 115 E283–E291. 10.1073/pnas.1715112115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartman A. E., Sanderson S. J., Ewing S. L., Niehans G. A., Wiehr C. L., Evans M. K., et al. (1999). Aberrant expression of MUC5AC and MUC6 gastric mucin genes in colorectal polyps. Int. J. Cancer 80 210–218. [DOI] [PubMed] [Google Scholar]
- Bertaux-Skeirik N., Feng R., Schumacher M. A., Li J., Mahe M. M., Engevik A. C., et al. (2015). CD44 plays a functional role in Helicobacter pylori-induced epithelial cell proliferation. PLoS Pathog. 11:e1004663. 10.1371/journal.ppat.1004663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biarc J., Nguyen I. S., Pini A., Gosse F., Richert S., Thierse D., et al. (2004). Carcinogenic properties of proteins with pro-inflammatory activity from Streptococcus infantarius (formerly S. bovis). Carcinogenesis 25 1477–1484. 10.1093/carcin/bgh091 [DOI] [PubMed] [Google Scholar]
- Boleij A., Dutilh B. E., Kortman G. A., Roelofs R., Laarakkers C. M., Engelke U. F., et al. (2012a). Bacterial responses to a simulated colon tumor microenvironment. Mol. Cell. Proteomics 11 851–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boleij A., Hechenbleikner E. M., Goodwin A. C., Badani R., Stein E. M., Lazarev M. G., et al. (2015). The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin. Infect. Dis. 60 208–215. 10.1093/cid/ciu787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boleij A., Roelofs R., Danne C., Bellais S., Dramsi S., Kato I., et al. (2012b). Selective antibody response to Streptococcus gallolyticus pilus proteins in colorectal cancer patients. Cancer Prev. Res. 5 260–265. 10.1158/1940-6207.CAPR-11-0321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boleij A., Van Gelder M. M., Swinkels D. W., Tjalsma H. (2011). Clinical importance of Streptococcus gallolyticus infection among colorectal cancer patients: systematic review and meta-analysis. Clin. Infect. Dis. 53 870–878. 10.1093/cid/cir609 [DOI] [PubMed] [Google Scholar]
- Braten L. S., Sodring M., Paulsen J. E., Snipen L. G., Rudi K. (2017). Cecal microbiota association with tumor load in a colorectal cancer mouse model. Microb. Ecol. Health Dis. 28:1352433. 10.1080/16512235.2017.1352433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant D. M., Stow J. L. (2004). The ins and outs of E-cadherin trafficking. Trends Cell Biol. 14 427–434. 10.1016/j.tcb.2004.07.007 [DOI] [PubMed] [Google Scholar]
- Bullman S., Pedamallu C. S., Sicinska E., Clancy T. E., Zhang X., Cai D., et al. (2017). Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 358 1443–1448. 10.1126/science.aal5240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt J., Romero-Hernandez B., Perez-Gomez B., Willhauck-Fleckenstein M., Holzinger D., Martin V., et al. (2016). Association of Streptococcus gallolyticus subspecies gallolyticus with colorectal cancer: serological evidence. Int. J. Cancer 138 1670–1679. 10.1002/ijc.29914 [DOI] [PubMed] [Google Scholar]
- Chamkha M., Patel B. K., Traore A., Garcia J. L., Labat M. (2002). Isolation from a shea cake digester of a tannin-degrading Streptococcus gallolyticus strain that decarboxylates protocatechuic and hydroxycinnamic acids, and emendation of the species. Int. J. Syst. Evol. Microbiol. 52 939–944. [DOI] [PubMed] [Google Scholar]
- Chand G., Shamban L., Forman A., Sinha P. (2016). The association of Streptococcus gallolyticus subspecies pasteurianus bacteremia with the detection of premalignant and malignant colonic lesions. Case Rep. Gastrointest. Med. 2016:7815843. 10.1155/2016/7815843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corredoira J., Garcia-Pais M. J., Coira A., Rabunal R., Garcia-Garrote F., Pita J., et al. (2015). Differences between endocarditis caused by Streptococcus bovis and Enterococcus spp. and their association with colorectal cancer. Eur. J. Clin. Microbiol. Infect. Dis. 34 1657–1665. 10.1007/s10096-015-2402-1 [DOI] [PubMed] [Google Scholar]
- Corredoira J., Grau I., Garcia-Rodriguez J. F., Garcia-Pais M. J., Rabunal R., Ardanuy C., et al. (2017). Colorectal neoplasm in cases of Clostridium septicum and Streptococcus gallolyticus subsp. gallolyticus bacteraemia. Eur. J. Intern. Med. 41 68–73. 10.1016/j.ejim.2017.02.009 [DOI] [PubMed] [Google Scholar]
- Couturier-Maillard A., Secher T., Rehman A., Normand S., De Arcangelis A., Haesler R., et al. (2013). NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J. Clin. Invest. 123 700–711. 10.1172/JCI62236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danne C., Dramsi S. (2012). Pili of gram-positive bacteria: roles in host colonization. Res. Microbiol. 163 645–658. 10.1016/j.resmic.2012.10.012 [DOI] [PubMed] [Google Scholar]
- Danne C., Dubrac S., Trieu-Cuot P., Dramsi S. (2014). Single cell stochastic regulation of pilus phase variation by an attenuation-like mechanism. PLoS Pathog. 10:e1003860. 10.1371/journal.ppat.1003860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danne C., Entenza J. M., Mallet A., Briandet R., Debarbouille M., Nato F., et al. (2011). Molecular characterization of a Streptococcus gallolyticus genomic island encoding a pilus involved in endocarditis. J. Infect. Dis. 204 1960–1970. 10.1093/infdis/jir666 [DOI] [PubMed] [Google Scholar]
- Danne C., Guerillot R., Glaser P., Trieu-Cuot P., Dramsi S. (2013). Construction of isogenic mutants in Streptococcus gallolyticus based on the development of new mobilizable vectors. Res. Microbiol. 164 973–978. 10.1016/j.resmic.2013.09.002 [DOI] [PubMed] [Google Scholar]
- Devine P. L., McKenzie I. F. (1992). Mucins: structure, function, and associations with malignancy. Bioessays 14 619–625. 10.1002/bies.950140909 [DOI] [PubMed] [Google Scholar]
- Dumke J., Vollmer T., Akkermann O., Knabbe C., Dreier J. (2017). Case-control study: determination of potential risk factors for the colonization of healthy volunteers with Streptococcus gallolyticus subsp. gallolyticus. PLoS One 12:e0176515. 10.1371/journal.pone.0176515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egert M., De Graaf A. A., Maathuis A., De Waard P., Plugge C. M., Smidt H., et al. (2007). Identification of glucose-fermenting bacteria present in an in vitro model of the human intestine by RNA-stable isotope probing. FEMS Microbiol. Ecol. 60 126–135. 10.1111/j.1574-6941.2007.00281.x [DOI] [PubMed] [Google Scholar]
- Ellmerich S., Scholler M., Duranton B., Gosse F., Galluser M., Klein J. P., et al. (2000). Promotion of intestinal carcinogenesis by Streptococcus bovis. Carcinogenesis 21 753–756. 10.1093/carcin/21.4.753 [DOI] [PubMed] [Google Scholar]
- Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., et al. (2015). Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 136 E359–E386. 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- Flemer B., Lynch D. B., Brown J. M., Jeffery I. B., Ryan F. J., Claesson M. J., et al. (2017). Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut 66 633–643. 10.1136/gutjnl-2015-309595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming M., Ravula S., Tatishchev S. F., Wang H. L. (2012). Colorectal carcinoma: pathologic aspects. J. Gastrointest. Oncol. 3 153–173. 10.3978/j.issn.2078-6891.2012.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco A. T., Israel D. A., Washington M. K., Krishna U., Fox J. G., Rogers A. B., et al. (2005). Activation of beta-catenin by carcinogenic Helicobacter pylori. Proc. Natl. Acad. Sci. U.S.A. 102 10646–10651. 10.1073/pnas.0504927102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagniere J., Raisch J., Veziant J., Barnich N., Bonnet R., Buc E., et al. (2016). Gut microbiota imbalance and colorectal cancer. World J. Gastroenterol. 22 501–518. 10.3748/wjg.v22.i2.501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R., Gao Z., Huang L., Qin H. (2017). Gut microbiota and colorectal cancer. Eur. J. Clin. Microbiol. Infect. Dis. 36 757–769. 10.1007/s10096-016-2881-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnad T., Feoktistova M., Leverkus M., Lendeckel U., Naumann M. (2010). Helicobacter pylori-induced activation of beta-catenin involves low density lipoprotein receptor-related protein 6 and Dishevelled. Mol. Cancer 9:31. 10.1186/1476-4598-9-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A., Madani R., Mukhtar H. (2010). Streptococcus bovis endocarditis, a silent sign for colonic tumour. Colorectal Dis. 12 164–171. 10.1111/j.1463-1318.2009.01814.x [DOI] [PubMed] [Google Scholar]
- Hirayama A., Kami K., Sugimoto M., Sugawara M., Toki N., Onozuka H., et al. (2009). Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Res. 69 4918–4925. 10.1158/0008-5472.CAN-08-4806 [DOI] [PubMed] [Google Scholar]
- Hoen B., Alla F., Selton-Suty C., Beguinot I., Bouvet A., Briancon S., et al. (2002). Changing profile of infective endocarditis: results of a 1-year survey in France. JAMA 288 75–81. 10.1001/jama.288.1.75 [DOI] [PubMed] [Google Scholar]
- Isenring J., Kohler J., Nakata M., Frank M., Jans C., Renault P., et al. (2017). Streptococcus gallolyticus subsp. gallolyticus endocarditis isolate interferes with coagulation and activates the contact system. Virulence 9 248–261. 10.1080/21505594.2017.1393600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzkowitz S. H., Bloom E. J., Lau T. S., Kim Y. S. (1992). Mucin associated Tn and sialosyl-Tn antigen expression in colorectal polyps. Gut 33 518–523. 10.1136/gut.33.4.518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jans C., Meile L., Lacroix C., Stevens M. J. (2015). Genomics, evolution, and molecular epidemiology of the Streptococcus bovis/Streptococcus equinus complex (SBSEC). Infect. Genet. Evol. 33 419–436. 10.1016/j.meegid.2014.09.017 [DOI] [PubMed] [Google Scholar]
- Jenab M., Chen J., Thompson L. U. (2001). Sialomucin production in aberrant crypt foci relates to degree of dysplasia and rate of cell proliferation. Cancer Lett. 165 19–25. 10.1016/S0304-3835(00)00706-0 [DOI] [PubMed] [Google Scholar]
- Kirikoshi H., Sekihara H., Katoh M. (2001). Up-regulation of WNT10A by tumor necrosis factor alpha and Helicobacter pylori in gastric cancer. Int. J. Oncol. 19 533–536. [PubMed] [Google Scholar]
- Klein R. S., Recco R. A., Catalano M. T., Edberg S. C., Casey J. I., Steigbigel N. H. (1977). Association of Streptococcus bovis with carcinoma of the colon. N. Engl. J. Med. 297 800–802. 10.1056/NEJM197710132971503 [DOI] [PubMed] [Google Scholar]
- Kok H., Jureen R., Soon C. Y., Tey B. H. (2007). Colon cancer presenting as Streptococcus gallolyticus infective endocarditis. Singapore Med. J. 48 e43–e45. [PubMed] [Google Scholar]
- Kumar R., Herold J. L., Schady D., Davis J., Kopetz S., Martinez-Moczygemba M., et al. (2017). Streptococcus gallolyticus subsp. gallolyticus promotes colorectal tumor development. PLoS Pathog. 13:e1006440. 10.1371/journal.ppat.1006440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Xu X., Chen L., Li W., Sun Y., Zeng J., et al. (2012). Helicobacter pylori CagA inhibits the expression of Runx3 via Src/MEK/ERK and p38 MAPK pathways in gastric epithelial cell. J. Cell. Biochem. 113 1080–1086. 10.1002/jcb.23440 [DOI] [PubMed] [Google Scholar]
- Lucas C., Barnich N., Nguyen H. T. T. (2017). Microbiota, inflammation and colorectal cancer. Int. J. Mol. Sci. 18:1310. 10.3390/ijms18061310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann B., Klussmann E., Vandamme-Feldhaus V., Iwersen M., Hanski M. L., Riecken E. O., et al. (1997). Low O-acetylation of sialyl-Le(x) contributes to its overexpression in colon carcinoma metastases. Int. J. Cancer 72 258–264. [DOI] [PubMed] [Google Scholar]
- Margarit I., Rinaudo C. D., Galeotti C. L., Maione D., Ghezzo C., Buttazzoni E., et al. (2009). Preventing bacterial infections with pilus-based vaccines: the group B Streptococcus paradigm. J. Infect. Dis. 199 108–115. 10.1086/595564 [DOI] [PubMed] [Google Scholar]
- Martins M., Aymeric L., Du Merle L., Danne C., Robbe-Masselot C., Trieu-Cuot P., et al. (2015). Streptococcus gallolyticus Pil3 pilus is required for adhesion to colonic mucus and for colonization of mouse distal colon. J. Infect. Dis. 212 1646–1655. 10.1093/infdis/jiv307 [DOI] [PubMed] [Google Scholar]
- Martins M., Porrini C., Du Merle L., Danne C., Robbe-Masselot C., Trieu-Cuot P., et al. (2016). The Pil3 pilus of Streptococcus gallolyticus binds to intestinal mucins and to fibrinogen. Gut Microbes 7 526–532. 10.1080/19490976.2016.1239677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy W. C., Mason J. M., III (1951). Enterococcal endocarditis associated with carcinoma of the sigmoid; report of a case. J. Med. Assoc. State Ala. 21 162–166. [PubMed] [Google Scholar]
- Mesquita P., Peixoto A. J., Seruca R., Hanski C., Almeida R., Silva F., et al. (2003). Role of site-specific promoter hypomethylation in aberrant MUC2 mucin expression in mucinous gastric carcinomas. Cancer Lett. 189 129–136. 10.1016/S0304-3835(02)00549-9 [DOI] [PubMed] [Google Scholar]
- Muller M. F., Ibrahim A. E., Arends M. J. (2016). Molecular pathological classification of colorectal cancer. Virchows Arch. 469 125–134. 10.1007/s00428-016-1956-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata-Kamiya N., Kurashima Y., Teishikata Y., Yamahashi Y., Saito Y., Higashi H., et al. (2007). Helicobacter pylori CagA interacts with E-cadherin and deregulates the beta-catenin signal that promotes intestinal transdifferentiation in gastric epithelial cells. Oncogene 26 4617–4626. 10.1038/sj.onc.1210251 [DOI] [PubMed] [Google Scholar]
- Nakayama M., Hisatsune J., Yamasaki E., Isomoto H., Kurazono H., Hatakeyama M., et al. (2009). Helicobacter pylori VacA-induced inhibition of GSK3 through the PI3K/Akt signaling pathway. J. Biol. Chem. 284 1612–1619. 10.1074/jbc.M806981200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa R. (1990). Formation of a clear zone on tannin-treated brain heart infusion agar by a Streptococcus sp. isolated from feces of koalas. Appl. Environ. Microbiol. 56 829–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa R., Sasaki E. (2004). Novel observations of genotypic and metabolic characteristics of three subspecies of Streptococcus gallolyticus. J. Clin. Microbiol. 42 4912–4913. 10.1128/JCM.42.10.4912-4913.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsonnet J., Friedman G. D., Vandersteen D. P., Chang Y., Vogelman J. H., Orentreich N., et al. (1991). Helicobacter pylori infection and the risk of gastric carcinoma. N. Engl. J. Med. 325 1127–1131. 10.1056/NEJM199110173251603 [DOI] [PubMed] [Google Scholar]
- Pope J. L., Tomkovich S., Yang Y., Jobin C. (2017). Microbiota as a mediator of cancer progression and therapy. Transl. Res. 179 139–154. 10.1016/j.trsl.2016.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyart C., Quesne G., Trieu-Cuot P. (2002). Taxonomic dissection of the Streptococcus bovis group by analysis of manganese-dependent superoxide dismutase gene (sodA) sequences: reclassification of ‘Streptococcus infantarius subsp. coli’ as Streptococcus lutetiensis sp. nov. and of Streptococcus bovis biotype 11.2 as Streptococcus pasteurianus sp. nov. Int. J. Syst. Evol. Microbiol. 52 1247–1255. [DOI] [PubMed] [Google Scholar]
- Purcell R. V., Pearson J., Aitchison A., Dixon L., Frizelle F. A., Keenan J. I. (2017). Colonization with enterotoxigenic Bacteroides fragilis is associated with early-stage colorectal neoplasia. PLoS One 12:e0171602. 10.1371/journal.pone.0171602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskov H., Burcharth J., Pommergaard H. C. (2017). Linking gut microbiota to colorectal cancer. J. Cancer 8 3378–3395. 10.7150/jca.20497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein M. R., Wang X., Liu W., Hao Y., Cai G., Han Y. W. (2013). Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe 14 195–206. 10.1016/j.chom.2013.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusniok C., Couve E., Da Cunha V., El Gana R., Zidane N., Bouchier C., et al. (2010). Genome sequence of Streptococcus gallolyticus: insights into its adaptation to the bovine rumen and its ability to cause endocarditis. J. Bacteriol. 192 2266–2276. 10.1128/JB.01659-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel L., Grimont F., Ageron E., Grimont P. A., Bouvet A. (2003). Reappraisal of the taxonomy of the Streptococcus bovis/Streptococcus equinus complex and related species: description of Streptococcus gallolyticus subsp. gallolyticus subsp. nov., S. gallolyticus subsp. macedonicus subsp. nov. and S. gallolyticus subsp. pasteurianus subsp. nov. Int. J. Syst. Evol. Microbiol. 53 631–645. 10.1099/ijs.0.02361-0 [DOI] [PubMed] [Google Scholar]
- Schwabe R. F., Jobin C. (2013). The microbiome and cancer. Nat. Rev. Cancer 13 800–812. 10.1038/nrc3610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears C. L., Garrett W. S. (2014). Microbes, microbiota, and colon cancer. Cell Host Microbe 15 317–328. 10.1016/j.chom.2014.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears C. L., Geis A. L., Housseau F. (2014). Bacteroides fragilis subverts mucosal biology: from symbiont to colon carcinogenesis. J. Clin. Invest. 124 4166–4172. 10.1172/JCI72334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillanpaa J., Nallapareddy S. R., Qin X., Singh K. V., Muzny D. M., Kovar C. L., et al. (2009). A collagen-binding adhesin, Acb, and ten other putative MSCRAMM and pilus family proteins of Streptococcus gallolyticus subsp. gallolyticus (Streptococcus bovis Group, biotype I). J. Bacteriol. 191 6643–6653. 10.1128/JB.00909-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skovbjerg H., Anthonsen D., Lothe I. M., Tveit K. M., Kure E. H., Vogel L. K. (2009). Collagen mRNA levels changes during colorectal cancer carcinogenesis. BMC Cancer 9:136. 10.1186/1471-2407-9-136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolova O., Bozko P. M., Naumann M. (2008). Helicobacter pylori suppresses glycogen synthase kinase 3beta to promote beta-catenin activity. J. Biol. Chem. 283 29367–29374. 10.1074/jbc.M801818200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriani M., Telford J. L. (2010). Relevance of pili in pathogenic streptococci pathogenesis and vaccine development. Future Microbiol. 5 735–747. 10.2217/fmb.10.37 [DOI] [PubMed] [Google Scholar]
- Suzuki M., Mimuro H., Kiga K., Fukumatsu M., Ishijima N., Morikawa H., et al. (2009). Helicobacter pylori CagA phosphorylation-independent function in epithelial proliferation and inflammation. Cell Host Microbe 5 23–34. 10.1016/j.chom.2008.11.010 [DOI] [PubMed] [Google Scholar]
- Sylvester P. A., Myerscough N., Warren B. F., Carlstedt I., Corfield A. P., Durdey P., et al. (2001). Differential expression of the chromosome 11 mucin genes in colorectal cancer. J. Pathol. 195 327–335. 10.1002/path.951 [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Network (2012). Comprehensive molecular characterization of human colon and rectal cancer. Nature 487 330–337. 10.1038/nature11252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanrobaeys M., Haesebrouck F., Ducatelle R., De Herdt P. (2000). Adhesion of Streptococcus gallolyticus strains to extracellular matrix proteins. Vet. Microbiol. 74 273–280. 10.1016/S0378-1135(00)00180-2 [DOI] [PubMed] [Google Scholar]
- Vanrobaeys M., De Herdt P., Charlier G., Ducatelle R., Haesebrouck F. (1999). Ultrastructure of surface components of Streptococcus gallolyticus (S. bovis) strains of differing virulence isolated from pigeons. Microbiology 145 335–342. 10.1099/13500872-145-2-335 [DOI] [PubMed] [Google Scholar]
- Waisberg J., Matheus Cde O., Pimenta J. (2002). Infectious endocarditis from Streptococcus bovis associated with colonic carcinoma: case report and literature review. Arq. Gastroenterol. 39 177–180. 10.1590/S0004-28032002000300008 [DOI] [PubMed] [Google Scholar]
- Wu S., Morin P. J., Maouyo D., Sears C. L. (2003). Bacteroides fragilis enterotoxin induces c-Myc expression and cellular proliferation. Gastroenterology 124 392–400. 10.1053/gast.2003.50047 [DOI] [PubMed] [Google Scholar]
- Yu T., Guo F., Yu Y., Sun T., Ma D., Han J., et al. (2017). Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 170 548.e16–563.e16. 10.1016/j.cell.2017.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan T., Rindtorff N., Boutros M. (2017). Wnt signaling in cancer. Oncogene 36 1461–1473. 10.1038/onc.2016.304 [DOI] [PMC free article] [PubMed] [Google Scholar]


