Abstract
Coronary heart disease (CHD) is a leading cause of morbidity in people over 65 years of age; >40% of all deaths are due to this condition. The association between increasing age and CHD is well documented; the accumulation of senescent cells in cardiac and vascular tissues may represent one factor underpinning this observation. We aimed to identify senescence-related expression changes in primary human senescent cardiomyocytes and endothelial cells and to relate transcript expression in peripheral blood leucocytes to prevalent and incident CHD in the InCHIANTI study of aging. We quantified splicing factor expression and splicing patterns of candidate transcripts in proliferative and senescent later passage endothelial cells and cardiomyocytes using qRTPCR. Senescence-associated isoforms also expressed in peripheral blood leucocytes were then examined for associations with CHD status in 134 pairs of age, sex and BMI-matched CHD cases and controls. Splicing factor expression was dysregulated in senescent cardiomyocytes, as previously reported for endothelial cells, as was the expression of alternatively expressed cardiac and vascular candidate genes in both cell types. We found nominal associations between the expression of VEGFA156b and FNI-EIIIIA isoforms in peripheral blood mRNA and CHD status. Dysregulated splicing factor expression is a key feature of senescent cardiomyocytes and endothelial cells. Altered splicing of key cardiac or endothelial genes may contribute to the risk of CHD in the human population.
Introduction
Cardiovascular disease is the major cause of mortality and morbidity in the developed world [1,2]. Age is one of the predominant risk factors for this group of disorders, and produces structural, molecular and functional changes in cardiac and vascular tissues [3–5]. The accumulation of senescent cells may be a contributor to cardiovascular pathology, since cells in the myocardium and vasculature are subject to cellular aging [6,7] and have known roles in cardiovascular dysfunction [8]. Senescent cells are one of the major drivers of aging phenotypes; selective removal of such cells from transgenic mice results in rejuvenation and amelioration of multiple aging phenotypes [9,10]. Perfusion of aged myocardium with neonatal cells was able to restore more youthful patterns of gene expression to human cardiomyocyte progenitors or senescent rat myocytes, and these molecular changes were also associated with significant improvements in cardiovascular outcomes [11].
Senescent cells display altered gene expression patterns compared with non-senescent cells [12]. In particular, senescent cells and older organisms display reductions in cellular plasticity and adaptive capacity [13,14], which is in part determined by patterns of alternative mRNA processing. Over 95% of genes express more than one gene product (isoform) under different physiological conditions [15]. Splice-site usage and patterns of alternative splicing are driven by a battery of splicing regulatory proteins termed as splicing factors [16,17]. Some studies have shown that genes encoding splicing factors are strongly dysregulated with aging in population studies and in senescent cells cultured in vitro, and are associated with lifespan in mammalian and invertebrate model systems [17–19]. Furthermore, small molecule restoration of splicing factor expression was recently demonstrated to be associated with rescue from multiple senescence phenotypes in aged fibroblasts [20].
In the present study, we aimed to examine senescence-related gene expression changes in human primary cardiomyocytes and endothelial cells, and to relate these changes to cardiovascular risk in a human population. As previously reported for endothelial cells and fibroblasts [17], we found that senescent cardiomyocytes demonstrate some dysregulation of splicing factor expression. We noted changes to the expression of genes with roles in cardiac or endothelial cell function in senescent cells. Both cell types demonstrated different alterations in the expression of senescence-associated secretory phenotype (SASP) proteins. Some deregulated isoforms expression in senescent cardiomyocytes or endothelial cells were also detected in mRNA extracted from human peripheral blood. The expression of the anti-angiogenic vascular endothelial growth factor 165b isoform (VEGFA165b) isoform and the alternatively expressed FN-1EIIIA isoform showed some evidence of a nominal association with prevalent and incident coronary heart disease (CHD) in participants from the InCHIANTI study of aging [21]. This work provides evidence that dysregulation of alternative splicing may alter the transcriptomic output of aged cardiomyocytes or endothelial cells, and that these changes may be associated with the development of cardiovascular diseases.
Methods
Culture of early passage and senescent cardiomyocytes and endothelial cells
Cultures of non-senescent and senescent endothelial cells and cardiomyocytes were used in the present study. Although the cultures were not from multiple isolates, each culture underwent senescence independently in three biological replicates to produce cultures that were not absolutely identical. Human aortic endothelial cells (HAoEC) and human cardiac myocytes (HCM) were seeded at a density of 6 × 103 cell/cm2 and were cultured in specific growth medium (C-22022 for HAoEC and C-22070 for HCM, PromoCell). Earlier passage proliferative cultures were at population doubling (PD) = 24 for endothelial cells and PD = 28 for cardiomyocytes. For the production of senescent cultures, cells were counted and equal numbers of cells seeded at each passage in continuous culture until the growth of the culture slowed to less than 0.5 PD/week. This occurred at PD = 65 and PD = 75 for endothelial cells and cardiomyocytes respectively. Cells were maintained at 37°C and 5% CO2 and used when not confluent to ensure that cessation of growth was not due to contact inhibition. Cell senescence was assessed in three biological replicates by the biochemical senescence marker senescence-associated β-galactosidase (SA β-Gal), tested in triplicate using a commercial kit (Sigma–Aldrich, U.K.) according to manufacturer’s instructions, with a minimum of 300 cells assessed per replicate. Senescence was also quantified in molecular terms by assessing the expression of the cyclin-dependent kinase inhibitor 2A (CDKN2A) gene (a known molecular marker of cell senescence) and by changes in cell morphology typical of senescence as in our previous work [17,20]. Total RNA (100 ng) was reverse transcribed in 20 μl reactions using the Superscript III VILO kit (Life Technologies, Foster City, U.S.A.). CDKN2A expression was measured by qRTPCR relative to three empirically determined endogenous control genes (GUSB, PPIA and GADPH) on the QuantStudio 12K Flex platform (Applied Biosystems, Foster City, U.S.A.). PCR reactions contained 2.5 μl Taq-Man Universal Mastermix (no AMPerase) (Applied Biosystems, Foster City, U.S.A.), 900 nM each primer, 250 nM probe and 0.5 μl cDNA in a total volume of 5 μl. PCR conditions were a single cycle at 95°C for 10 min followed by 40 cycles at 95°C for 15 s and 60°C for 1 min.
Quantification of secretion of SASP cytokines in early passage and senescent cardiomyocytes and endothelial cells
Cells were seeded at 6 × 104 cells per well in a six-well plate and cultured until 95% confluence. Cell supernatants were then harvested and stored at −80°C. Levels of SASP proteins GM-CSF, IFNγ, IL1β, IL2, IL6, IL8, IL10, IL-12p70, and TNFα were measured in cell supernatants using the K15007B MesoScale Discovery multiplex ELISA immunoassay (MSD, Rockville, U.S.A.) in six biological replicates. Proteins were quantified relative to a standard curve using a Sector Imager SI-6000 according to the manufacturer’s instructions. Statistical analysis was carried out with the computer-assisted Prism GraphPad Program (Prism version 5.00, GraphPad Software, San Diego, CA).
Quantification of splicing factor expression in early passage and senescent cardiomyocytes
An a priori list of splicing factor candidate genes were chosen on the basis that they were associated with human aging in populations and had been characterised in primary human fibroblasts and endothelial cells in our previous work [17,18]. The list of genes included the positive regulatory splicing factors SRSF1, SRSF2, SRSF3, SRSF6, SRSF7, PNSIR and TRA2B, the negative regulatory splicing inhibitors HNRNPA0, HNPNPA1, HNPNPA2B1, HNPNPD, HNPNPH3, HNPNPK, HNPNPM, HNPNPUL2 and the core spliceosomal factors AKAP17A, LSM2, LSM14, IMP3 and SF3B1. Gene expression was measured as described above, using a custom TaqMan Low Density Array (TLDA) format from Life Technologies (Foster City, U.S.A.) as previously described [17]. Transcript expression was assessed by the Comparative Ct approach, relative to the IDH3B, GUSB and PPIA endogenous control genes and normalised to their expression in RNA from early passage cells.
Quantification of candidate gene expression in early passage and senescent cardiomyocytes and endothelial cells
A panel of candidate genes were selected for analysis based on biological relevance, documented roles in cardiac or endothelial dysfunction and insight into the differential functionality of alternatively expressed isoforms. The identity of genes tested in each cell type and the rationale for their inclusion is given in Table 1. Probes specific to particular isoforms were designed to unique regions of the transcripts in question where possible (assay sequences are available upon request). Assays were validated by standard curve analysis of seven serial 1:2 dilutions of pooled cDNA from cardiomyocytes and endothelial cells. Reverse transcription and qRTPCR conditions are described above.
Table 1.
Genes and transcripts selected for expression analysis
| Gene name | Transcript accession | Reason for selection | |
|---|---|---|---|
| ENDOTHELIAL CELS | |||
| Total expression | |||
| CTNNB1 | Catenin β1 | NM 001098209 | Key component of Wnt signalling, implicated in several aspects of cardiovascular differentiation and disease |
| EDNRB | Endothelin receptor type B | NM 000115.3 | Involved in cardiac remodelling following myocardial infarction |
| ICAM-1 | Intracellular cell adhesion molecule 1 | NM 000201 | Key regulators of atherosclerotic plaque development |
| VCAM-1 | Vascular cell adhesion molecule 1 | NM 001078 | Key regulators of atherosclerotic plaque development |
| FN1 | Fibronectin | NM 001306129 | Extracelullar matrix protein with roles in atherosclerosis |
| Alternative isoforms | |||
| FN1(EIIIA) | Fibronectin EIIIA isoform | NM 212482 | Mouse isoform specific knockout exhibit cardiovascular defects |
| FN1(EIIIB) | Fibronectin EIIIB isoform | NM 001306130 | Mouse isoform specific knockout exhibit cardiovascular defects |
| ENG(L) | Endoglin long form | NM 001114753 | Negatively regulates angiogenesis and plays a role in heart development |
| ENG(S) | Endoglin short form | NM 001278138 | Positively regulates angiogenesis and plays a role in heart development role |
| ID1(1) | Inhibitor of DNA binding 1, HLH protein transcript 1 | NM 002165.3 | Involved in endothelial cell differentiation |
| ID1(2) | Inhibitor of DNA binding 1, HLH protein transcript 2 | NM 181353.2 | Involved in endothelial cell differentiation and associated with senescence |
| VEGFA121 | Vascular endothelial growth factor 121 isoform | NM 001171628 | Pro-angiogenic isoform of VEGFA |
| VEGFA165b | Vascular endothelial growth factor 165b isoform | NM 001171626 | Anti-angiogenic isoform of VEGFA |
| TJP1(A) | Tight junction protein 1 A isoform | NM 175610.3 | Controls angiogenesis and endothelial barrier formation to stabilise cardiomyocyte integrity |
| TJP1(B) | Tight junction protein 1 B isoform | NM 003257.4 | Controls angiogenesis and endothelial barrier formation to stabilise cardiomyocyte integrity |
|
| |||
| CARDIOMYOCYTES | |||
| Total expression | |||
| GATA4 | GATA-binding protein 4 | NM 001308093 | Cardiogenic transcription factor |
| Alternative isoforms | |||
| ANK1(L) | Ankyrin-B long isoform | NM 001127493.1 | Localisation and membrane stabilisation of ion transporters and ion channels in cardiomyocytes, involved in cardiac arrhythmias |
| ANK1(S) | Ankyrin-B short isoform | NM 001148.4 | Localisation and membrane stabilisation of ion transporters and ion channels in cardiomyocytes, involved in cardiac arrhythmias |
| GJA5(L) | Gap junction α-5 long isoform | NM 005266 | Associated with cardiac arrhythmias |
| GJA5(S) | Gap junction α-5 short isoform | NM 181703 | Associated with cardiac arrhythmias |
| TNNT2 | Cardiac muscle troponin T | NM 001276346 | Associated with cardiomyopathy |
| TNNT2(S) | Cardiac muscle troponin T short form | NM 001001431 | Associated with cardiomyopathy |
| NPPA | Natriuretic peptide A | NM 006172 | Low levels associated with atrial fibrillation |
| NPPA(AS) | Natriuretic peptide A – antisense1 | ENST00000446542.5 | Negative regulator of NPPA |
| NPPA(S) | Natriuretic peptide A short form | ENST00000376476.1 | Alternative isoform of NPPA involved in cardiac development |
| ATP2A2(A) | Sarco/endoplasmic reticulum Ca2+-ATPase A isoformy | NM 170665 | Involved in cardiac muscle contraction |
| ATP2A2(B) | Sarco/endoplasmic reticulum Ca2+-ATPase B isoform | NM 001681 | Involved in cardiac muscle contraction |
The gene acronym, gene name, transcript accession number(s) and known function of a panel of candidate genes selected for analysis are given above.
Associations between candidate gene expression and CHD in the InCHIANTI population
The InCHIANTI study of aging is a population study of aging [21]. Participants undertook detailed assessment of health and lifestyle parameters at baseline, and again at four subsequent follow-ups (FU2; 2004–2006, FU3; 2007–2009 and FU4; 2012–2014). We selected 268 participants for study for whom we had both an RNA sample at FU3 and clinical information at FU3 and FU4. Participants were categorised according to the presence of CHD as either: (i) no CHD (mean age: 75.9 years, S.D.: 13.4 years, n=134), (ii) CHD at least 3 years prior to FU3 (mean age: 78.8 years, S.D.: 7.4 years, n=45), (iii) CHD within 3 years before FU3 (mean age: 79.8 years, S.D.: 8.9 years, n=80) and (iv) no overt disease at the time of RNA sampling but CHD onset between FU3 and FU4 (mean age: 80.3 years, S.D.: 9.2 years, n=9). CHD was defined as the presence of myocardial infarction or angina. Each CHD case was paired with a control participant matched for age (5 year age bins), sex and BMI (four categories; underweight (<18.5), normal (18.5–25), overweight (25–30) and obese (30+). The mean age of controls was 75.9 years (S.D. = 13.4 years). All human studies have been carried out in accordance with the ethical standards laid out in the Declaration of Helsinki.
Blood was collected into PAXgene tubes (BD Biosciences) at FU3, and extracted using the PAXgene blood RNA kit (Qiagen, Paisley, U.K.). One hundred nanograms RNA reverse transcribed using the High-Capacity cDNA RT kit (Thermo Fisher, U.K.) and transcript expression levels were quantified as described above. Gene expression data were normalised where necessary prior to analysis, according to the most appropriate transformation metric indicated by the Statistics software (STATA-SE 14).
We used paired logistic regression models to investigate the association between the gene expression levels of the senescence-related alternative isoforms and both prevalent and incident CHD (STATA-SE 14, Statacorp LLC, U.K.). Models were adjusted for smoking (pack-years), study-site and cell counts (% neutrophils, monocytes, basophils, eosinophils). Associations with age were assessed by fully adjusted linear regression. Analyses were performed on the entire cohort, but also stratified by sex. We investigated the effect of multiple testing (Bonferroni correction), but also report suggestive associations with nominal P-value <0.05.
Results
Senescent cardiomyocytes and endothelial cells demonstrate different senescence characteristics
Senescence reached at PD = 75 and 65 for cardiomyocyte and endothelial cell cultures respectively. Earlier passage proliferative cardiomyocytes and endothelial cell PD times were 4 and 5 days compared with 10 and 12 days in the senescent cultures. Despite the similarities in cell kinetics, the senescent cell percentage for late passage cardiomyocyte cultures was consistently lower than seen in later passage endothelial cells. SA-β-Gal staining in the cultures rose from 12 to 52% for the endothelial cells but from 9 to 30% in the cardiomyocyte cultures (Figure 1A). Differences were also evident in molecular markers of senescence, where we noted a five-fold increase in the expression of CDKN2A in senescent endothelial cells but less than one-fold increase in late passage cardiomyocytes (Figure 1B). The SASP of late passage endothelial cells was characterised by significantly more of the signature SASP cytokine IL-6 at senescence, whereas levels of GM-CSF and IL-8 were reduced compared with early passage cells. In contrast, senescent cardiomyocytes unexpectedly displayed significantly reduced expression of many SASP components (Table 2).
Figure 1. Senescence parameters in cardiomyocytes and endothelial cells.
(A) Senescent cell load was assessed by SA β-Gal staining in early and late passage endothelial cells and cardiomyocytes. (B) Senescent cell load was assessed by mRNA expression of the senescence associated transcript of the CDKN2A gene. Statistical significance is indicated by stars, ***, P=0.0001. Error bars represent ± S.E.M. of six independent experiments.
Table 2.
Expression of SASP proteins in senescent endothelial cells and cardiomyocytes
| Young endothelial cells | Old endothelial cells | Young cardiomyocytes | Old cardiomyocytes | |
|---|---|---|---|---|
| GM-CSF | 10.55 (0.33) | 4.55 (0.15)*** | 3.76 (0.14) | 1.60 (0.08)*** |
| IFNγ | 3.51 (0.42) | 4.19 (0.34) | 1.44 (0.19) | 0.05 (0.01)*** |
| IL-12 | 0.51 (0.12) | 0.56 (0.13) | 0.12 (0.07) | 0.08 (0.02) |
| IL-1β | 0.39 (0.14) | 0.35 (0.03) | 0.41 (0.04) | 0.39 (0.05) |
| IL-2 | 2.35 (0.42) | 1.49 (0.15) | 0.44 (0.05) | 0.30 (0.06) |
| IL-6 | 710.30 (27.1) | 1238.58 (46.8)*** | 2567.7 (275.6) | 1682.5 (174.1)* |
| IL-8 | 17756.78 (937.3) | 3477.4 (355.3)*** | 334.3 (12.7) | 281.8 (12.1)** |
| IL-10 | 0.29 (0.04) | 0.31 (0.04) | 0.47 (0.08) | 0.10 (0.02)*** |
| TNFα | 0.44 (0.09) | 0.55 (0.14) | 0.15 (0.03) | 0.14 (0.05) |
The expression of SASP proteins in early passage cardiomyocytes and endothelial cells compared with those in late passage cells, as determined by multiplex ELISA is given above. The results are expressed as pg/ml and are the mean ± S.E.M. of six independent experiments. Statistical significance by two-way ANOVA is indicated by stars, with * = 0.05, ** = 0.001 and *** = <0.0001 compared with the young value.
Changes in splicing factor expression in senescent cardiomyocytes
We previously reported changes in splicing factor expression in senescent endothelial cells [17]. Here, we demonstrate that similar changes were present in senescent cardiomyocytes (Table 3), although these were less marked than those previously identified in endothelial cells; 7 of the 20 splicing factor transcripts analysed were senescence-associated in cardiomyocytes compared with 9 out of 20 transcripts in senescent endothelial cells [17]. Different splicing factors were affected in late passage cardiomyocytes compared with late passage endothelial cells [17], with only TRA2B showing changes in both cell types.
Table 3.
Disrupted splicing factor expression in senescent cardiomyocytes
| Early passage cardiomyocytes | Late passage cardiomyocytes | |
|---|---|---|
| AKAP17A | 1.011 (0.067) | 0.970 (0.039) |
| HNRNPA0 | 1.025 (0.101) | 0.917 (0.030) |
| HNRNPA1 | 1.021 (0.092) | 1.063 (0.037) |
| HNRNPA2B1 | 1.026 (0.105) | 1.254 (0.04) |
| HNRNPD | 1.035 (0.143) | 1.732*** (0.271) |
| HNRNPH3 | 1.033 (0.119) | 1.579* (0.170) |
| HNRNPK | 1.025 (0.108) | 0.745* (0.037) |
| HNRNPM | 1.015 (0.079) | 0.951 (0.028) |
| HNRNPUL2 | 1.041 (0.136) | 1.031 (0.064) |
| IMP3 | 1.016 (0.081) | 0.983 (0.035) |
| LSM14A | 1.025 (0.102) | 0.817 (0.025) |
| LSM2 | 1.009 (0.057) | 0.927 (0.163) |
| PNISR | 1.046 (0.143) | 1.197 (0.052) |
| SF3B1 | 0.996 (0.083) | 1.201* (0.028) |
| SRSF1 | 1.012 (0.071) | 1.054 (0.098) |
| SRSF2 | 1.006 (0.047) | 0.851* (0.022) |
| SRSF3 | 1.015 (0.079) | 1.181 (0.155) |
| SRSF6 | 1.014 (0.075) | 1.265 (0.242) |
| TRA2B | 1.049 (0.146) | 0.634* (0.086) |
| SRSF7 | 1.015 (0.083) | 0.668* (0.089) |
The expression profiles of transcripts encoding splicing regulatory factors in early and late passage cardiomyocytes are given below. Quantifications refer to the level of each splicing factor for each gene, normalised to the geometric mean of the level of each gene in early passage cells. The results are expressed as the mean of six independent experiments. Statistical significance is indicated by stars, with * = 0.05, ** = 0.001 and *** = <0.0001. Figures in parentheses refer to the S.E.M. Transcripts significantly altered in senescent cardiomyocytes are indicated in bold italic type.
Changes in patterns of alternative splicing are evident in both senescent cardiomyocytes and endothelial cells
We also assessed expression of a panel of a priori candidate genes, selected on the basis of key roles in cardiomyocyte or endothelial cell function or dysfunction (Table 1). We saw total expression changes for Catenin β1 (CTNNB1), intracellular cell adhesion molecule (ICAM) and vascular cell adhesion molecule 1 (VCAM) genes, and also changes for the fibronectin ((FN1), ENG, ID1, VEGFA and TJP1 isoforms in senescent endothelial cells. In senescent cardiomyocytes, the expression of GJA5, cardiac muscle troponin T (TNNT2) and ATP2A2 isoforms was altered (Table 4). In some cases, the dysregulated expression was limited to one alternatively expressed isoform (e.g. TNNT2, ID1), while in other cases multiple alternatively expressed isoform was affected (e.g. VEGFA, ENG, GJA5, ATP2A2).
Table 4.
Disrupted gene expression and patterns of alternative splicing in senescent cardiomyocytes and endothelial cells
| Early passage cells | Late passage cells | |
|---|---|---|
| ENDOTHELIAL CELLS | ||
| Total expression | ||
| CTNNB1 | 1.187 (0.074) | 2.134 (0.151)*** |
| ENDRB | 1.240 (0.132) | 1.364 (0.224) |
| FN1 | 1.191 (0.083) | 2.312 (0.212)*** |
| VCAM1 | 1.135 (0.089) | 6.063 (0.513)*** |
| ICAM1 | 1.248 (0.205) | 3.811 (0.384)*** |
| Alternative isoforms | ||
| FN1(EIIIA) | 1.186 (0.068) | 2.193 (0.180)*** |
| FN1(EIIIB) | 1.255 (0.155) | 2.335 (0.196)*** |
| ENG(L) | 1.183 (0.064) | 2.471 (0.271)*** |
| ENG(S) | 1.139 (0.065) | 1.556 (0.156)* |
| ID1(1) | 1.450 (0.133) | 1.911 (0.167)* |
| ID1(2) | 1.270 (0.162) | 1.044 (0.110) |
| VEGFA121 | 1.287 (0.199) | 2.760 (0.532)* |
| VEGFA165b | 1.172 (0.122) | 2.438 (0.369)** |
| TJP1(A) | 1.025 (0.102) | 0.921 (0.102) |
| TJP1(B) | 1.270 (0.176) | 1.309 (0.158) |
|
| ||
| CARDIOMYOCYTES | ||
| Total expression | ||
| GATA4 | 1.584 (0.291) | 1.332 (0.157) |
| Alternative isoforms | ||
| ANK1(L) | 1.277 (0.191) | 1.477 (0.102) |
| ANK1(S) | 1.463 (0.228) | 2.368 (0.393) |
| GJA5(L) | 1.614 (0.346) | 2.789 (0.414)* |
| GJA5(S) | 1.687 (0.398) | 4.324 (0.810)*** |
| TNNT2(S) | 1.464 (0.217) | 0.589 (0.074)*** |
| TNNT2 | 1.930 (0.500) | 1.923 (0.225) |
| NPPA | 1.474 (0.196) | 1.420 (0.324) |
| NPPA(AS) | 1.343 (0.158) | 1.541 (0.223) |
| NPPA(S) | 1.642 (0.348) | 1.450 (0.211) |
| ATP2A2(A) | 1.382 (0.064) | 2.013 (0.457)* |
| ATP2A2(B) | 1.375 (0.036) | 2.113 (0.158)** |
The expression profiles of transcripts encoding splicing regulatory factors in early and late passage cardiomyocytes and endothelial cells are given above. Quantifications refer to the level of each transcript for each gene, normalised to the geometric mean of the level of each gene in early passage cells. The results are expressed as the mean of six independent experiments. Statistical significance is indicated by stars, with * = 0.05, ** = 0.001 and *** = < 0.0001. Figures in parentheses refer to the S.E.M. Transcripts showing differential expression in late passage cells are indicated in bold italic type.
Expression of alternative isoforms of the FN1 and VEGFA genes, and changes in ICAM expression in peripheral blood may be correlated with CHD status
We first assessed whether genes demonstrating senescence-related expression changes in endothelial cells and cardiomyocytes were expressed in peripheral blood. Of the 11 transcripts that were significantly dysregulated in senescent endothelial cells, 10 (CTNNB1, endothelin receptor type B (EDNRB), FN1-EIIIA, FN1-EIIIB, endoglin long form (ENG(L)), ICAM, inhibitor of DNA binding 1, HLH protein transcript 1 (ID1(1)), VCAM, vascular endothelial growth factor 121 isoform (VEGFA121) and VEGFA165b) were also expressed in peripheral blood. Of the five transcripts showing dysregulated expression in senescent cardiomyocytes, only two (ATP2A2-A and ATP2A2-B) were detected in peripheral blood.
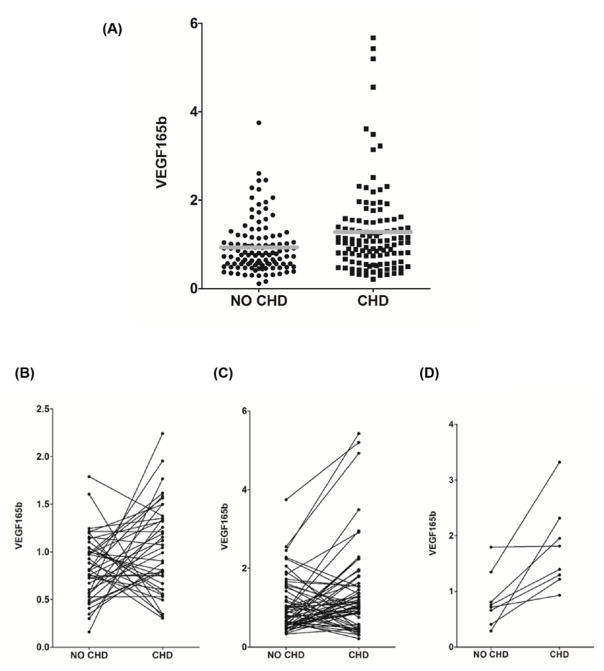
We next assessed whether senescence-related changes in the expression or processing of these isoforms could impact on cardiovascular phenotypes by exploring associations between their expression and prevalent or incident CHD (defined as angina or MI) in participants from the InCHIANTI study of aging using paired logistic regression analysis. In 125 paired cases and controls matched for age, sex and BMI, we noted associations between prevalent CHD and expression of the FN-1EIIIA isoform and the VEGFA165b isoform, although these associations did not make the multiple testing threshold (coefficients: 0.98 and −0.65, S.E.M.: 0.45 and 0.31, P=0.024 and 0.039 for VEGFA165b and FN1-EIIIA respectively; Table 5). Although both these associations were nominal, the positive association between CHD and expression of the VEGFA165b isoform was most persuasive (Figure 2A). Only ATP2A2(B) was associated with age in peripheral blood (β coefficient: −0.007, SE: 0.003, P=0.022).
Table 5.
Associations between the presence of CHD and expression of senescence-related endothelial and cardiomyocyte transcripts in peripheral blood
| Transcript | β coefficient | S.E.M. | 95% CI | P-value |
|---|---|---|---|---|
| Prevalent CHD at the time of assessment (125 cases compared with 125 controls) | ||||
| CTNNB1 | −0.07 | 0.26 | −0.59 to 0.44 | 0.786 |
| EDNRB | 0.26 | 0.16 | −0.04 to 0.56 | 0.092 |
| FN1(EIIIA) | −0.65 | 0.31 | −1.27 to −0.03 | 0.039 |
| FN1(EIIIB) | −0.16 | 0.29 | −0.72 to 0.40 | 0.580 |
| ICAM1 | −0.52 | 0.33 | −1.17 to 0.14 | 0.125 |
| IDV(1) | −0.09 | 0.20 | −0.48 to 0.30 | 0.655 |
| IDV(2) | −0.16 | 0.15 | −0.45 to 0.14 | 0.312 |
| ATP2A2(A) | −0.16 | 0.26 | −0.64 to 0.38 | 0.616 |
| ATP2A2(B) | −0.06 | 0.25 | −0.55 to 0.43 | 0.810 |
| VCAM1 | 0.09 | 0.10 | −0.12 to 0.29 | 0.394 |
| VEGFA121 | −0.05 | 0.23 | −0.50 to 0.40 | 0.832 |
| VEGFA165b | 0.98 | 0.44 | 0.13 to 1.83 | 0.024 |
| Incident CHD: Controls (n=134), Cases: category 1 (n=45), category 2 (n=80), category 3 (n=9) | ||||
| CTNNB1 | −0.08 | 0.24 | −0.55 to 0.40 | 0.758 |
| EDNRB | 0.25 | 0.15 | −0.04 to 0.54 | 0.090 |
| FN1(EIIIA) | −0.55 | 0.28 | −1.10 to −0.01 | 0.049 |
| FN1(EIIIB) | −0.21 | 0.28 | −0.76 to 0.35 | 0.465 |
| ICAM1 | −0.63 | 0.32 | −1.25 to −0.01 | 0.046 |
| IDV(1) | −0.06 | 0.19 | −0.44 to 0.32 | 0.770 |
| IDV(2) | −0.12 | 0.14 | −0.39 to 0.15 | 0.392 |
| ATP2A2(A) | −0.12 | 0.26 | −0.62 to 0.39 | 0.647 |
| ATP2A2(B) | −0.08 | 0.25 | −0.57 to 0.41 | 0.743 |
| VCAM1 | 0.15 | 0.10 | −0.04 to 0.34 | 0.131 |
| VEGFA121 | 0.03 | 0.23 | −0.41 to 0.41 | 0.892 |
| VEGFA165b | 0.91 | 0.41 | 0.11 to 1.70 | 0.025 |
| Incident CHD (men only): Controls (n=65), Cases: category 1 (n=21), category 2 (n=41), category 3 (n=3) | ||||
| CTNNB1 | −1.05 | 0.54 | −2.12 to 0.01 | 0.052 |
| EDNRB | 0.35 | 0.27 | −0.18 to 0.87 | 0.200 |
| FN1(EIIIA) | −3.91 | 5.20 | −14.11 to 6.28 | 0.452 |
| FN1(EIIIB) | 1.83 | 1.47 | −4.72 to 1.05 | 0.213 |
| ICAM1 | −1.07 | 0.49 | −2.03 to −0.12 | 0.028 |
| IDV(1) | −0.03 | 0.29 | −0.59 to 0.54 | 0.927 |
| IDV(2) | −0.44 | 0.23 | −0.90 to 0.02 | 0.062 |
| ATP2A2(A) | −0.18 | 0.39 | −0.94 to 0.58 | 0.648 |
| ATP2A2(B) | −0.12 | 0.40 | −0.90 to 0.65 | 0.757 |
| VCAM1 | 0.31 | 0.17 | −0.04 to 0.65 | 0.080 |
| VEGFA121 | −0.08 | 0.35 | −0.77 to 0.62 | 0.827 |
| VEGFA165b | 1.97 | 0.75 | 0.50 to 3.44 | 0.01 |
| Incident CHD (women only): Controls (n=69), Cases: category 1 (n=24), category 2 (n=39), category 3 (n=6) | ||||
| CTNNB1 | 0.53 | 0.36 | −0.18 to 1.24 | 0.142 |
| EDNRB | 0.23 | 0.24 | −0.23 to 0.70 | 0.325 |
| FN1(EIIIA) | −1.03 | 0.47 | −1.96 to −0.10 | 0.030 |
| FN1(EIIIB) | 0.20 | 0.37 | −0.51 to 0.92 | 0.578 |
| ICAM1 | −0.30 | 0.48 | −1.25 to 0.65 | 0.534 |
| IDV(1) | −0.12 | 0.30 | −0.71 to 0.48 | 0.701 |
| IDV(2) | 0.07 | 0.20 | −0.31 to 0.45 | 0.726 |
| ATP2A2 (A) | −0.17 | 0.38 | −0.92 to 0.57 | 0.648 |
| ATP2A2 (B) | −0.12 | 0.33 | −0.77 to 0.53 | 0.724 |
| VCAM1 | 0.10 | 0.14 | −0.18 to 0.38 | 0.489 |
| VEGFA121 | 0.09 | 0.31 | −0.53 to 0.70 | 0.777 |
| VEGFA165b | 0.19 | 0.55 | −0.89 to 1.26 | 0.735 |
Associations between transcript expression and CHD assessed by adjusted paired logistic regression analysis are given above. CHD is defined as the presence of angina or myocardial infarction. Cases are divided into category 1 (individuals with CHD diagnosed more than 3 years prior to the time of sampling), category 2 (individuals with CHD diagnosed less than 3 years prior to the time of sampling) and category 3 (individuals who did not have CHD at the time of sampling but who subsequently developed CHD more than 3 years after the time of sampling). Significant associations are given in bold italic type. Abbreviations: 95% CI, 95% confidence interval.
Figure 2. Association of peripheral blood mRNA levels of the anti-angiogenic VEGFA165b isoform and presence of CHD.
(A) The relationship between VEGFA165b expression and prevalent CHD. The expression levels of the VEGFA165b isoform of the VEGFA gene calculated relative to the level of three endogenous control genes and expressed relative to the median level in the sample cohort is given on the Y-axis. The presence or absence of CHD in the assayed participants at the time of assessment is given on the Y-axis. Each black circle represents levels in a single participant without CHD, each black square represents levels in a single participant with CHD. The mean level in participants with and without CHD are indicated by grey bars. (B–D) The relationship between the expression levels of the VEGFA165b isoform and incident CHD. The expression levels of the VEGFA165b isoform calculated relative to the level of three endogenous control genes and expressed relative to the median level in the sample cohort is given on the Y-axis in samples paired for age, sex and BMI. Each pair of linked circles represents a single case and paired control. Category 1: cases with CHD diagnosed more than 3 years prior to the time of sampling (B). Category 2: cases with CHD diagnosed less than 3 years prior to the time of sampling (C). Category 3: individuals who did not have CHD at the time of sampling but who subsequently developed CHD >3 years after the time of sampling (D).
We then assessed associations between transcript expression and incident CHD. Cases were designated as (i) CHD diagnosed more than 3 years from time of sampling (n=45), (ii) incident CHD diagnosed within 3 years from time of sampling (n=80) and (iii) future CHD (i.e. no CHD at time of sampling, but CHD incident up to 3 years following sampling) (n=9). Controls in each analysis were participants without CHD who did not develop CHD by the final follow-up (n=134). Again, we observed associations between FN1-EIIIA and VEGFA165b expression and CHD status. We also observed a nominal inverse correlation between CHD status and the expression of the ICAM gene (coefficient: −0.63, S.E.M.: 0.31, P=0.052) (Table 5). While again not surviving adjustment for multiple testing, the data were most persuasive for an association between VEGFA165b expression and CHD (Figure 2B–D), where the expression level of VEGFA165b was elevated in 7/8 of the samples from participants who had not yet developed CHD compared with their paired controls, although the numbers in this group (n=9) were too small for definitive conclusion (Figure 2D). We also noted sex differences, with VEGFA165b and ICAM associations being driven by the men (coefficients 1.96 and −1.07, S.E.M.: 0.75 and 0.49, P=0.009 and 0.028 respectively; Table 5) and the FN1-EIIIA effect being driven by women (coefficient: −1.02, S.E.M.: 0.47, P=0.030; Table 5).
Discussion
There is rising evidence that some aging phenotypes are driven by the accumulation of senescent cells in multiple tissues [9,10]. Recent data suggests that dysregulated alternative splicing may contribute to the development of senescence [17,18,20,22] and here, we report changes in splicing factor expression and alternative splicing in senescent endothelial cells and cardiomyocytes. Furthermore, the expression of the anti-angiogenic VEGFA165b isoform and the embryonic FN1-EIIIA isoform in peripheral blood demonstrate nominal associations with CHD in participants from the InCHIANTI study of aging.
The characteristics of senescent cells differ from tissue to tissue. Despite similarities in growth kinetics, late passage endothelial cells cultures show higher senescent cell loads, different expression of SASP proteins and more dysregulation of splicing factors and alternative splicing patterns than do late passage cardiomyocytes. Some of our observations may point to the fraction of growth arrested, but not senescent, cells in the cardiomyocyte cultures being higher, which may partially explain the attenuated senescence characteristics we observe. The endothelial cell SASP is also known to be attenuated in response to DNA damaging agents [23], and more generally, the SASP response in other cell models demonstrates differences in composition depending on the stimulus for senescence [24]. The differences between cell types we identify are perhaps unsurprising, since patterns of gene expression and alternative splicing are understandably highly tissue specific. It is also known that SASP differs from cell type to cell type. Our data suggest that the changes occurring in senescent endothelial cells may have more impact on the aging cardiovascular system than do changes in senescent cardiomyocytes. One caveat of our study is that although our tissue culture work was carried out in three biological replicates, we did not have data from independent isolates of cardiomyocytes or endothelial cells. This is because of the scarcity of well-characterised primary isolates. However, since each of our biological replicates has undergone independent replicative senescence in parallel they are not identical cultures as each will have accrued different mutations and epigenetic changes during the senescence process, although they do of course share a genetic background.
The portfolio of splicing factor changes in senescent endothelial cells and cardiomyocytes is interesting. In our previous work, we found the most dysregulated splicing factors to belong to the HNRP class of regulators, which typically inhibit splice-site usage. In contrast, only a single SR splice activator, SRSF7, demonstrated dysregulation at the level of mRNA transcript in senescent endothelial cells [17]. In contrast, senescent cardiomyocytes demonstrated dysregulation of both SR and HNRNP transcripts. This may indicate that a higher proportion of aberrant splice-site usage in endothelial cells than it does in cardiomyocytes, although it is difficult however to predict the precise consequences of these changes at the level of individual genes, since each individual splice site is controlled by a unique balance of activators and inhibitors [25].
Our data are consistent with splicing patterns in senescent cardiomyocytes and endothelial cells contributing to age-related cardiac and endothelial dysfunction. We observed dysregulated expression of both GJA5 isoforms, both ATP2A2 isoforms and the short isoform of TNNT2 in senescent cardiomyocytes. GJA5 encodes a gap junction protein, polymorphisms in this gene have been associated with atrial fibrillation [26,27]. The ATP2A2 gene encodes a sarcoplasmic/endoplasmic reticulum calcium transporter, which has previously been implicated in chronic heart failure [28]. The TNNT2 gene encodes cardiac troponin T2, mutations which have been associated with familial hypertrophic cardiomyopathy and stroke [29,30]. In senescent endothelial cells, we noted dysregulated expression of multiple genes, including FN1, VCAM, ICAM, ENG, ID1 and VEGFA. FN1 encodes fibronectin, an important component of the extracellular matrix, isoforms of which have been implicated in thrombosis [31]. VCAM and ICAM encode cellular adhesion molecules, and have been implicated in vascular inflammation, coronary flow and stroke [32–34]. ENG encodes endoglin, a molecule with roles in cardiac remodelling, which has been implicated in myocardial fibrosis [35]. ID1 encodes an inhibitor of DNA binding, with roles in cardiac cell growth, senescence and differentiation [36]. Finally, VEGFA is a key mediator of angiogenesis, which has been implicated in multiple pathologies including CHD [37].
We observed nominal (P<0.05) associations between the peripheral blood expression of specific FN1 and VEGFA isoforms and CHD in InCHIANTI study participants. Our data are most supportive of a nominal association of elevated VEGFA165b levels in people with CHD, driven by the men in the cohort. VEGFA is a secreted protein which undergoes extensive alternative splicing, and encodes isoforms with both angiogenic and anti-angiogenic potential through the differential use of an alternative splice site within exon 8 [38]. Increased expression of the consensus, angiogenesis promoting VEGFA isoforms following cardiac ischemia would serve to promote the formation of new blood vessels and allow reperfusion. The anti-angiogenic VEGFA165b isoform however, may act to inhibit neovascularisation and impede cardiac recovery. Accordingly, VEGFA165b has previously been associated with several cardiovascular phenotypes including coronary artery disease, hypertension, myocardial infarction and peripheral artery disease in humans [39,40]. Elevated levels of VEGFA165b have also been documented in ischemic heart tissue from patients who have undergone surgical reperfusion following myocardial infarction [40]. Although the number of patients in our cohort with incident CHD between follow-up 3 and 4 was too small (n=9) to be able to draw definitive conclusions, there is observationally a trend towards elevated VEGFA165b expression in participants who did not have overt CHD at the time of collection, but who went on to exhibit symptoms. Should this observation be confirmed in a larger study, this would raise the exciting possibility that the expression levels of this isoform in the peripheral circulation could represent a potential biomarker for future CHD. Although less convincing, we also noted a nominal association with the expression of the FN1-EIIIA isoform with CHD which was predominantly driven by effects in women. This isoform is predominantly embryonic, but has been suggested to play a role in cardiac wound healing responses after MI [41]. If this tenuous association is later confirmed in larger studies, it is possible that the lower levels of expression we note in CHD patients may reflect poorer capacity for repair.
In conclusion, our data are supportive of a model whereby senescent endothelial cells and cardiomyocytes in situ may express a different portfolio of alternatively expressed isoforms leading to gradual impairment of cardiac and endothelial function and increased probability of cardiovascular events. Increased expression of alternatively expressed transcripts such as the VEGFA165b isoform, may therefore lead to not only progressive cardiovascular remodelling, but also impaired neovascularisation following ischemic events in both the heart and the periphery. Strategies to specifically target splicing events such as those that lead of the production of VEGFA165b are now being explored; such as Translarna™, one such splicing therapeutic, was recently licenced for the treatment of Duchenne Muscular Dystrophy. Characterisation of senescence-related splicing events in aging heart and vascular tissues may therefore identify potential new therapeutic targets for future cardiovascular protection.
Clinical perspectives.
CHD is a leading cause of morbidity and mortality in the Western world. Like many other common chronic diseases, age is a major risk factor for the onset of CHD.
Cellular senescence is a major driving force for organismal aging. Selective removal of such cells is sufficient to ameliorate multiple aging phenotypes in animal models. Cell senescence is in turn partly driven by dysregulated mRNA processing.
We identified alterations to splicing regulation and alternative splicing in senescent cardiomyocytes and endothelial cells. The anti-angiogenic VEGFA156b isoform of the VEGFA gene was seen to be associated with both prevalent and incident CHD. These data suggest that changes to the regulation of key cardiovascular genes may contribute to age-associated risk of CHD.
Acknowledgments
Funding
This work was supported by the Dunhill Medical Trust [grant number R386/1114].
Abbreviations
- BMI
body mass index
- CDKN2A
cyclin-dependent kinase inhibitor 2A
- CHD
coronary heart disease
- HAoEC
human aortic endothelial cell
- HCM
human cardiac myocyte
- MI
myocardial infarction
- PD
population doubling
- SASP
senescence-associated secretory phenotype
- VEGFA165b
vascular endothelial growth factor 165b isoform
Footnotes
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
Author contribution
E.L. carried out the experiments, analysed the data and contributed to the manuscript. L.C.P. contributed to data analysis and reviewed the manuscript. B.P.L. contributed to data analysis and reviewed the manuscript. S.B. is the curator of the InCHIANTI study and reviewed the manuscript. D.M. contributed to discussion, data analysis and reviewed the manuscript. L.F. organised the sample cohort and contributed to the manuscript. L.W.H. designed and managed the study, interpreted the data and wrote the manuscript.
References
- 1.Kuller LH, Lopez OL, Mackey RH, Rosano C, Edmundowicz D, Becker JT, et al. Subclinical cardiovascular disease and death, dementia, and coronary heart disease in patients 80+ years. J Am Coll Cardiol. 2016;67:1013–1022. doi: 10.1016/j.jacc.2015.12.034. https://doi.org/10.1016/j.jacc.2015.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Newman AB, Naydeck BL, Ives DG, Boudreau RM, Sutton-Tyrrell K, O’Leary DH, et al. Coronary artery calcium, carotid artery wall thickness, and cardiovascular disease outcomes in adults 70 to 99 years old. Am J Cardiol. 2008;101:186–192. doi: 10.1016/j.amjcard.2007.07.075. https://doi.org/10.1016/j.amjcard.2007.07.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dai DF, Chen T, Johnson SC, Szeto H, Rabinovitch PS. Cardiac aging: from molecular mechanisms to significance in human health and disease. Antioxid Redox Signal. 2012;16:1492–1526. doi: 10.1089/ars.2011.4179. https://doi.org/10.1089/ars.2011.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niccoli T, Partridge L. Ageing as a risk factor for disease. Curr Biol. 2012;22:R741–R752. doi: 10.1016/j.cub.2012.07.024. https://doi.org/10.1016/j.cub.2012.07.024. [DOI] [PubMed] [Google Scholar]
- 5.Strait JB, Lakatta EG. Aging-associated cardiovascular changes and their relationship to heart failure. Heart Fail Clin. 2012;8:143–164. doi: 10.1016/j.hfc.2011.08.011. https://doi.org/10.1016/j.hfc.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morgan RG, Ives SJ, Lesniewski LA, Cawthon RM, Andtbacka RH, Noyes RD, et al. Age-related telomere uncapping is associated with cellular senescence and inflammation independent of telomere shortening in human arteries. Am J Physiol Heart Circ Physiol. 2013;305:H251–H258. doi: 10.1152/ajpheart.00197.2013. https://doi.org/10.1152/ajpheart.00197.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyer K, Hodwin B, Ramanujam D, Engelhardt S, Sarikas A. Essential role for premature senescence of myofibroblasts in myocardial fibrosis. J Am Coll Cardiol. 2016;67:2018–2028. doi: 10.1016/j.jacc.2016.02.047. https://doi.org/10.1016/j.jacc.2016.02.047. [DOI] [PubMed] [Google Scholar]
- 8.Matthews C, Gorenne I, Scott S, Figg N, Kirkpatrick P, Ritchie A, et al. Vascular smooth muscle cells undergo telomere-based senescence in human atherosclerosis: effects of telomerase and oxidative stress. Circ Res. 2006;99:156–164. doi: 10.1161/01.RES.0000233315.38086.bc. https://doi.org/10.1161/01.RES.0000233315.38086.bc. [DOI] [PubMed] [Google Scholar]
- 9.Baker DJ, Childs BG, Durik M, Wijers ME, Sieben CJ, Zhong J, et al. Naturally occurring p16(Ink4a)-positive cells shorten healthy lifespan. Nature. 2016;530:184–189. doi: 10.1038/nature16932. https://doi.org/10.1038/nature16932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baker DJ, Wijshake T, Tchkonia T, LeBrasseur NK, Childs BG, van de Sluis B, et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479:232–236. doi: 10.1038/nature10600. https://doi.org/10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grigorian-Shamagian L, Liu W, Fereydooni S, Middleton RC, Valle J, Cho JH, et al. Cardiac and systemic rejuvenation after cardiosphere-derived cell therapy in senescent rats. Eur Heart J. 2017;38:2957–2967. doi: 10.1093/eurheartj/ehx454. https://doi.org/10.1093/eurheartj/ehx454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Purcell M, Kruger A, Tainsky MA. Gene expression profiling of replicative and induced senescence. Cell Cycle. 2014;13:3927–3937. doi: 10.4161/15384101.2014.973327. https://doi.org/10.4161/15384101.2014.973327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Tollefsbol TO. Age-related epigenetic drift and phenotypic plasticity loss: implications in prevention of age-related human diseases. Epigenomics. 2016;8:1637–1651. doi: 10.2217/epi-2016-0078. https://doi.org/10.2217/epi-2016-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pomatto LCD, Davies KJA. The role of declining adaptive homeostasis in ageing. J Physiol. 2017 doi: 10.1113/JP275072. https://doi.org/10.1113/JP275072. [DOI] [PMC free article] [PubMed]
- 15.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. https://doi.org/10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 16.Fu XD, Ares M., Jr Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet. 2014;15:689–701. doi: 10.1038/nrg3778. https://doi.org/10.1038/nrg3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holly AC, Melzer D, Pilling LC, Fellows AC, Tanaka T, Ferrucci L, et al. Changes in splicing factor expression are associated with advancing age in man. Mech Ageing Dev. 2013;134:356–366. doi: 10.1016/j.mad.2013.05.006. https://doi.org/10.1016/j.mad.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harries LW, Hernandez D, Henley W, Wood AR, Holly AC, Bradley-Smith RM, et al. Human aging is characterized by focused changes in gene expression and deregulation of alternative splicing. Aging Cell. 2011;10:868–878. doi: 10.1111/j.1474-9726.2011.00726.x. https://doi.org/10.1111/j.1474-9726.2011.00726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee BP, Pilling LC, Emond F, Flurkey K, Harrison DE, Yuan R, et al. Changes in the expression of splicing factor transcripts and variations in alternative splicing are associated with lifespan in mice and humans. Aging Cell. 2016;15:903–913. doi: 10.1111/acel.12499. https://doi.org/10.1111/acel.12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Latorre E, Birar VC, Sheerin AN, Jeynes JCC, Hooper A, Dawe HR, et al. Small molecule modulation of splicing factor expression is associated with rescue from cellular senescence. BMC Cell Biol. 2017;18:31. doi: 10.1186/s12860-017-0147-7. https://doi.org/10.1186/s12860-017-0147-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferrucci L, Bandinelli S, Benvenuti E, Di Iorio A, Macchi C, Harris TB, et al. Subsystems contributing to the decline in ability to walk: bridging the gap between epidemiology and geriatric practice in the InCHIANTI study. J Am Geriatr Soc. 2000;48:1618–1625. doi: 10.1111/j.1532-5415.2000.tb03873.x. https://doi.org/10.1111/j.1532-5415.2000.tb03873.x. [DOI] [PubMed] [Google Scholar]
- 22.Deschenes M, Chabot B. The emerging role of alternative splicing in senescence and aging. Aging Cell. 2017;16:918–933. doi: 10.1111/acel.12646. https://doi.org/10.1111/acel.12646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bent EH, Gilbert LA, Hemann MT. A senescence secretory switch mediated by PI3K/AKT/mTOR activation controls chemoprotective endothelial secretory responses. Genes Dev. 2016;30:1811–1821. doi: 10.1101/gad.284851.116. https://doi.org/10.1101/gad.284851.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maciel-Baron LA, Morales-Rosales SL, Aquino-Cruz AA, Triana-Martinez F, Galvan-Arzate S, Luna-Lopez A, et al. Senescence associated secretory phenotype profile from primary lung mice fibroblasts depends on the senescence induction stimuli. Age (Dordr) 2016;38:26. doi: 10.1007/s11357-016-9886-1. https://doi.org/10.1007/s11357-016-9886-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith CW, Valcarcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem Sci. 2000;25:381–388. doi: 10.1016/s0968-0004(00)01604-2. https://doi.org/10.1016/S0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- 26.Hailati J, Yang YC, Zhang L, He PY, Baikeyi M, Muhuyati W, et al. Association between −44G/A and +71A/G polymorphisms in the connexin 40 gene and atrial fibrillation in Uyghur and Han populations in Xinjiang, China. Genet Mol Res. 2016:15. doi: 10.4238/gmr15048628. https://doi.org/10.4238/gmr15048628. [DOI] [PubMed]
- 27.Sun Y, Hills MD, Ye WG, Tong X, Bai D. Atrial fibrillation-linked germline GJA5/connexin40 mutants showed an increased hemichannel function. PLoS ONE. 2014;9:e95125. doi: 10.1371/journal.pone.0095125. https://doi.org/10.1371/journal.pone.0095125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lyon AR, Bannister ML, Collins T, Pearce E, Sepehripour AH, Dubb SS, et al. SERCA2a gene transfer decreases sarcoplasmic reticulum calcium leak and reduces ventricular arrhythmias in a model of chronic heart failure. Circ Arrhythm Electrophysiol. 2011;4:362–372. doi: 10.1161/CIRCEP.110.961615. https://doi.org/10.1161/CIRCEP.110.961615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mochmann HC, Scheitz JF, Petzold GC, Haeusler KG, Audebert HJ, Laufs U, et al. Coronary angiographic findings in acute ischemic stroke patients with elevated cardiac troponin: the Troponin Elevation in Acute Ischemic Stroke (TRELAS) study. Circulation. 2016;133:1264–1271. doi: 10.1161/CIRCULATIONAHA.115.018547. https://doi.org/10.1161/CIRCULATIONAHA.115.018547. [DOI] [PubMed] [Google Scholar]
- 30.Jachymova M, Muravska A, Palecek T, Kuchynka P, Rehakova H, Magage S, et al. Genetic variation screening of TNNT2 gene in a cohort of patients with hypertrophic and dilated cardiomyopathy. Physiol Res. 2012;61:169–175. doi: 10.33549/physiolres.932157. [DOI] [PubMed] [Google Scholar]
- 31.Chauhan AK, Kisucka J, Cozzi MR, Walsh MT, Moretti FA, Battiston M, et al. Prothrombotic effects of fibronectin isoforms containing the EDA domain. Arterioscler Thromb Vasc Biol. 2008;28:296–301. doi: 10.1161/ATVBAHA.107.149146. https://doi.org/10.1161/ATVBAHA.107.149146. [DOI] [PubMed] [Google Scholar]
- 32.Denys A, Clavel G, Lemeiter D, Schischmanoff O, Boissier MC, Semerano L. Aortic VCAM-1: an early marker of vascular inflammation in collagen-induced arthritis. J Cell Mol Med. 2016;20:855–863. doi: 10.1111/jcmm.12790. https://doi.org/10.1111/jcmm.12790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turhan H, Saydam GS, Erbay AR, Ayaz S, Yasar AS, Aksoy Y, et al. Increased plasma soluble adhesion molecules; ICAM-1, VCAM-1, and E-selectin levels in patients with slow coronary flow. Int J Cardiol. 2006;108:224–230. doi: 10.1016/j.ijcard.2005.05.008. https://doi.org/10.1016/j.ijcard.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 34.Zhang MJ, Zhang M, Yin YW, Li BH, Liu Y, Liao SQ, et al. Association between intercellular adhesion molecule-1 gene K469E polymorphism and the risk of stroke in a Chinese population: a meta-analysis. Int J Neurosci. 2015;125:175–185. doi: 10.3109/00207454.2014.919916. https://doi.org/10.3109/00207454.2014.919916. [DOI] [PubMed] [Google Scholar]
- 35.Shyu KG. The role of endoglin in myocardial fibrosis. Acta Cardiol Sin. 2017;33:461–467. doi: 10.6515/ACS20170221B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cunningham TJ, Yu MS, McKeithan WL, Spiering S, Carrette F, Huang CT, et al. Id genes are essential for early heart formation. Genes Dev. 2017 doi: 10.1101/gad.300400.117. https://doi.org/10.1101/gad.300400.117. [DOI] [PMC free article] [PubMed]
- 37.Wang Y, Huang Q, Liu J, Wang Y, Zheng G, Lin L, et al. Vascular endothelial growth factor A polymorphisms are associated with increased risk of coronary heart disease: a meta-analysis. Oncotarget. 2017;8:30539–30551. doi: 10.18632/oncotarget.15546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qiu Y, Hoareau-Aveilla C, Oltean S, Harper SJ, Bates DO. The anti-angiogenic isoforms of VEGF in health and disease. Biochem Soc Trans. 2009;37:1207–1213. doi: 10.1042/BST0371207. https://doi.org/10.1042/BST0371207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ganta VC, Choi M, Kutateladze A, Annex BH. VEGF165b modulates endothelial VEGFR1-STAT3 signaling pathway and angiogenesis in human and experimental peripheral arterial disease. Circ Res. 2017;120:282–295. doi: 10.1161/CIRCRESAHA.116.309516. https://doi.org/10.1161/CIRCRESAHA.116.309516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hueso L, Rios-Navarro C, Ruiz-Sauri A, Chorro FJ, Nunez J, Sanz MJ, et al. Dynamics and implications of circulating anti-angiogenic VEGF-A165b isoform in patients with ST-elevation myocardial infarction. Sci Rep. 2017;7:9962. doi: 10.1038/s41598-017-10505-9. https://doi.org/10.1038/s41598-017-10505-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ulrich MM, Janssen AM, Daemen MJ, Rappaport L, Samuel JL, Contard F, et al. Increased expression of fibronectin isoforms after myocardial infarction in rats. J Mol Cell Cardiol. 1997;29:2533–2543. doi: 10.1006/jmcc.1997.0486. https://doi.org/10.1006/jmcc.1997.0486. [DOI] [PubMed] [Google Scholar]