Abstract
Intravenous leiomyomatosis is an unusual smooth muscle neoplasm with quasi-malignant intravascular growth but a histologically banal appearance. Herein, we report expression and molecular cytogenetic analyses of a series of 12 intravenous leiomyomatosis cases to understand better the pathogenesis of intravenous leiomyomatosis. All cases were analyzed for expression of HMGA2, MDM2 and CDK4 proteins by immunohistochemistry based on our previous finding of der(14)t(12;14)(q14.3;q24) in intravenous leiomyomatosis. Seven of 12 (58%) intravenous leiomyomatosis cases expressed HMGA2, and none expressed MDM2 or CDK4. Co-localization of hybridization signals for probes from the HMGA2 locus (12q14.3) and from 14q24 by interphase fluorescence in situ hybridization (FISH) was detected in a mean of 89.2% of nuclei in HMGA2-positive cases by immunohistochemistry, but in only 12.4% of nuclei in negative cases, indicating an association of HMGA2 expression and this chromosomal rearrangement (p=8.24×10−10). Four HMGA2-positive cases had greater than two HMGA2 hybridization signals per cell. No cases showed loss of a hybridization signal by interphase FISH for the frequently deleted region of 7q22 in uterine leiomyomata. One intravenous leiomyomatosis case analyzed by array comparative genomic hybridization revealed complex copy number variations. Finally, expression profiling was performed on three intravenous leiomyomatosis cases. Interestingly, hierarchical cluster analysis of the expression profiles revealed segregation of the intravenous leiomyomatosis cases with leiomyosarcoma rather than with myometrium, uterine leiomyoma of the usual histological type, or plexiform leiomyoma. These findings suggest that intravenous leiomyomatosis cases share some molecular cytogenetic characteristics with uterine leiomyoma, and expression profiles similar to that of leiomyosarcoma cases, further supporting their intermediate, quasi-malignant behavior.
INTRODUCTION
Smooth muscle tumors arising from the uterus range from benign uterine leiomyomata to malignant leiomyosarcoma and include a variety of tumors with unusual growth patterns. uterine leiomyoma is the most common tumor of the female reproductive tract (1), and approximately 25–40% of uterine leiomyomata have non-random tumor-specific cytogenetic abnormalities (2, 3). In addition to “usual type” uterine leiomyoma, clinically benign histologic variants are recognized including atypical (a.k.a. bizarre, pleomorphic, or symplastic), plexiform, and cellular leiomyomata (4–6). In contrast to uterine leiomyoma, leiomyosarcoma is rare, has an aggressive clinical behavior, complex cytogenetic and genomic rearrangements, and is histologically distinguishable from uterine leiomyoma by the presence of coagulative tumor necrosis, severe nuclear or cytological atypia, and elevated mitotic activity (7–9). In addition to the histologic spectrum of smooth muscle tumors, tumors resembling uterine leiomyoma at both gross and microscopic levels but presenting in unusual locations with quasi-malignant behavior include intravenous leiomyomatosis, disseminated peritoneal leiomyomatosis, and benign metastasizing leiomyoma.
intravenous leiomyomatosis is a rare entity characterized by intravascular nodular masses of histologically benign smooth muscle cells growing in uterine and pelvic veins, and sometimes extending into the inferior vena cava and chambers of the right heart (10–13). Intravenous leiomyomatosis occurs most commonly in women in the fifth decade, characteristically presenting with abnormal uterine bleeding or pain due to concomitant presence of uterine leiomyoma. If the intravenous leiomyomatosis mass extends along the inferior vena cava, venous return to the right heart becomes obstructed, and patients can present with findings of hemodynamic compromise, such as dyspnea, syncope, congestive heart failure, or even sudden death (14). Clinical examination usually reveals an enlarged uterus or a pelvic mass. On pathologic examination, multiple myometrial masses are typically associated with worm-like plugs within parametrial vessels. Despite the presence of extensive intravascular involvement, patients with intravenous leiomyomatosis typically have long term survival after successful removal of the tumor, and most patients have an unremarkable clinical course with a relatively low risk of pelvic recurrence or distant metastasis (10). The lung is the most common site of subsequent spread (15, 16).
Although the etiology of intravenous leiomyomatosis remains to be elucidated, two theories have been advanced. One theory suggests that intravenous leiomyomatosis originates from the vessel wall, while the other purports that intravenous leiomyomatosis invasion into the vessel wall occurs subsequent to extension from a uterine leiomyoma (17). Analyzing molecular genetic events underlying intravenous leiomyomatosis provides an opportunity to gain understanding of its pathogenesis.
The presence of a karyotype with a der(14) t(12;14)(q15;q24) in our two previously published intravenous leiomyomatosis cases (Table 1) correlating with the t(12;14)(q15;q24) cytogenetic subgroup in uterine leiomyoma suggests a potential pathogenetic relationship between intravenous leiomyomatosis and uterine leiomyoma based on dysregulation of the non-histone chromatin factor HMGA2 at 12q14.3 (18, 19). Given the proximity of MDM2 and CDK4 at 12q15 and 12q14.1, respectively, to the HMGA2 locus, and their roles in various mesenchymal tumors (20–23), alterations in their expression might underlie molecular mechanisms in intravenous leiomyomatosis. Because uterine leiomyoma and intravenous leiomyomatosis are histologically similar and usually present concomitantly in a patient, analyzing the common cytogenetic alterations of uterine leiomyoma in intravenous leiomyomatosis might provide insights into the biology of intravenous leiomyomatosis and its relationship with uterine leiomyoma.
Table 1.
GTG-banded karyotype and metaphase FISH characterization of intravenous leiomyomatosis cases
| Case No. | Karyotype | Metaphase FISH interpretation |
|---|---|---|
| 1 | 45,XX,del(12)(q?14q?15),hsr(14)(q2?1),-22[7].ish del(12)(q14.3q14.3)(5′HMGA2-,3′HMGA2-),hsr(14)amp(3′HMGA2) |
HMGA2 amplification on abnormal 14q, breakpoint 5′ (centromeric) to 3′HMGA2 |
| 21 | 45,XX,der(14)t(12;14)(q15;q24),-22.ish der(14)t(12;14) (RAD51L1+,5′HMGA2+,3′HMGA2+) |
3 copies of HMGA2, breakpoint 5′ (centromeric) to HMGA2 |
| 32 | 45,XX,-10,add(11)(q11),der(14)t(12;14)(q15;q24)[12]/45,XX,-10,add(11)(q11),t(12;14)(q15;q24)[3].ish der(14)t(12;14)(5′HMGA2+,3′HMGA2+)/t(12;14)(5′HMGA2-,3′HMGA2-;5′HMGA2+,3′HMGA2+) |
Mosaic karyotype with 3 and 2 copies of HMGA2, breakpoint 5′ (centromeric) to HMGA2 |
Herein, a series of 12 cases of intravenous leiomyomatosis was analyzed for immunohistochemical expression of HMGA2, MDM2, and CDK4, and interphase FISH analysis was performed to assess co-localization of probes at the HMGA2 and 14q24 loci. In addition, presence of an interstitial deletion of 7q was assessed because the deletion of 7q22 is one of the most common cytogenetic abnormalities in uterine leiomyoma (24). Finally, expression profiles of three cases of intravenous leiomyomatosis with myometrium, uterine leiomyoma, histological variants of leiomyomata (cellular, atypical and plexiform) and leiomyosarcoma were compared by hierarchical clustering analysis and differential gene expression was analyzed between intravenous leiomyomatosis cases and a set of nine uterine leiomyoma cases with t(12;14) (25).
MATERIALS AND METHODS
Cases diagnosed as intravenous leiomyomatosis were retrieved from archives of the Brigham and Women’s Hospital (nine cases) and Baystate Medical Center (three cases) under IRB approved protocols. Hematoxylin and eosin-stained slides were reviewed to confirm the diagnoses based upon published criteria (26–28). Two additional intravenous leiomyomatosis samples (CHTN 19480 and 52343) were obtained from the Cooperative Human Tissue Network (http://www.chtn.nci.nih.gov/) for expression profile analysis.
Immunohistochemical Analysis
Immunohistochemistry was performed following pressure cooker pretreatment for antigen retrieval. Intravenous leiomyomatosis tissue sections were subsequently incubated with primary anti-HMGA2 polyclonal antibody (59170AP, Biocheck Inc., Foster City, CA), and MDM2 (IF2 clone, EMD Chemicals, San Diego, CA) and CDK4 (DCS-31 clone, Invitrogen, Carlsbad, CA) antibodies for 40–60 minutes at 25° C. Following rinsing with Tris buffer solution, bound antibody was detected with the Envision Plus/Horseradish Peroxidase system (Dako, Carpinteria, CA). Tissue was then incubated using the Envision Plus secondary antibody for 30 minutes followed by diaminobenzidine for five minutes. Appropriate positive and negative controls were stained in parallel. Staining for HMGA2, MDM2 and CDK4 proteins was scored as 0 (no nuclear staining), 1+ (< 5% of nuclei positive), 2+ (5% to 25% of nuclei positive), 3+ (26% to 50% of nuclei positive), or 4+ (>50% of nuclei positive).
GTG-banded Karyotyping
Discarded tumor tissue from Case 1 was obtained aseptically immediately following resection, during intra-operative pathology consultation, was disaggregated for short-term culture, and chromosome analysis performed as previously described (29). GTG-banded karyotypes of Cases 2 and 3 were reported previously (Table 1) (18, 19). Case 3 had two masses (pelvic ST02-165 and adnexal ST02-166) with different cellular ratios of the same mosaic karyotypes (18). Specimens for Cases 4–12 were not available for tissue culture, and thus, karyotyping was unable to be performed.
Array Comparative Genomic Hybridization
DNA was available only for Case 3 (ST02-166) (18). Array comparative genomic hybridization (aCGH) compared DNA obtained from Case 3 to DNA from a pooled female DNA sample (Promega, Madison, WI) as previously described (30).
Metaphase Fluorescence In Situ Hybridization
Metaphase fluorescence in situ hybridization (FISH) analysis was performed for Case 1. Bacterial artificial chromosome (BAC) clones were selected using the University of California Santa Cruz Genome browser (http://genome.ucsc.edu) (February 2009 assembly). BAC DNAs were isolated following a standard protocol consisting of alkaline lysis, neutralization, and ethanol precipitation (Qiagen, Valencia, CA). BAC clone RP11-299L9 located at the 5′ end of HMGA2, including exons 1 and 2 (nucleotides 66,049,805-66,225,867, hg19), was labeled with SpectrumGreen (Abbott Molecular, Des Plaines, IL), and RP11-427K2 located at the 3′ end of HMGA2, including exons 4 and 5 (nucleotides 66,323,478-66,481,711, hg19), was labeled with SpectrumOrange (Abbott). Metaphase FISH analyses of Cases 2 and 3 were reported previously (Table 1) (18, 19).
Interphase Fluorescence In Situ Hybridization
FISH was performed on interphase nuclei of intravenous leiomyomatosis tissue sections for assessing the presence of t(12;14) and del(7)(q22). Tissue sections of Case 3 were not available for interphase FISH. Interphase FISH analysis was unsuccessful for two cases: Case 6 in the t(12;14) study and Case 11 in the del(7)(q22) study. Four-micron sections of formalin fixed paraffin embedded intravenous leiomyomatosis tissue on glass slides were baked overnight at 56° C, deparaffinized by three immersions in xylene, followed by dehydration in 100% alcohol. Air dried slides were immersed in 100 mM Tris-base, 50 mM EDTA (pH 7.0) buffer for 45 minutes at 100° C, rinsed in 1x phosphate buffered saline (PBS) for 5 minutes, and treated twice with 100 μl of Digest-All (Invitrogen) at 37°C for 20 minutes. Next, slides were rinsed in 1x PBS, fixed in 10% buffered formalin at room temperature for one minute, and rinsed again in 1x PBS. Slides were then dehydrated in an alcohol series of 70%, 90%, and 100% and air-dried prior to hybridization. 12–16 μg of fluorochrome-labeled BAC probes were applied to sections on the air-dried slides, followed by denaturation in the HYBrite apparatus (Abbott) at 95°C for three minutes and overnight incubation at 37°C. Hybridized slides were washed in 0.5x SSC at 72°C for five minutes followed by rinsing three times in PBS-Tween 0.025% and air drying in the dark before counterstaining with DAPI II anti-fade solution (Abbott). Ninety to 100 interphase nuclei were evaluated per case. For each nucleus, numbers of red and green hybridization signals were counted. To detect presence of the t(12;14), co-localization of hybridization signals (yellow signal or direct juxtaposition of red and green signals) for BAC probes RP11-366L20, located at the 3′ end of HMGA2 including exons 4 and 5 (nucleotides 66,246,519-66,425,264, hg19) (SpectrumGreen), and RP11-195L19, located at 14q24.1 (nucleotides 68,341,567-68,510,240, hg19) (SpectrumOrange), was assessed. For detection of del(7)(q22), loss of the hybridization signal of BAC probe RP11-374E17, located at 7q22.3, and presence of the control probe TelVysion 7p SpectrumGreen Probe/Hyb Set (Abbott), located within 300 kb of the end of 7p, were evaluated.
Statistical analysis comparing interphase FISH for t(12;14) and immunohistochemistry status of HMGA2 was performed using the two-sample t-test included in SYSTAT version 12.01.01 (SYSTAT Software, Chicago, IL.).
RNA Expression Profiling
RNA isolated from Case 3 and two additional samples obtained from the Cooperative Human Tissue Network (CHTN 19480 and 52343) were analyzed using standard protocols and statistical methods as previously described (25, 31–33). These three intravenous leiomyomatosis expression profiles were compared to those previously reported for myometrium, uterine leiomyoma, histological variants of leiomyomata (cellular, atypical and plexiform) and leiomyosarcoma (31–33). In addition, differential gene expression was analyzed between intravenous leiomyomatosis cases and a set of nine previously reported uterine leiomyomata with t(12;14) (25) to gain an understanding of the quasi-malignant behavior of intravenous leiomyomatosis despite their similar cytogenetic characteristics. Independent assessment of the differentially expressed genes by quantitative PCR could not be performed due to insufficient RNA.
RESULTS
Immunohistochemistry analysis of HMGA2 revealed that seven of 12 (58%) cases of intravenous leiomyomatosis (Cases 1–7) showed strong, diffuse (4+) nuclear staining and the remaining five (Cases 8–12) were negative (0 staining) (Fig. 1 and Table 2). Expression of MDM2 or CDK4 proteins by immunohistochemistry was negative in all 12 cases examined.
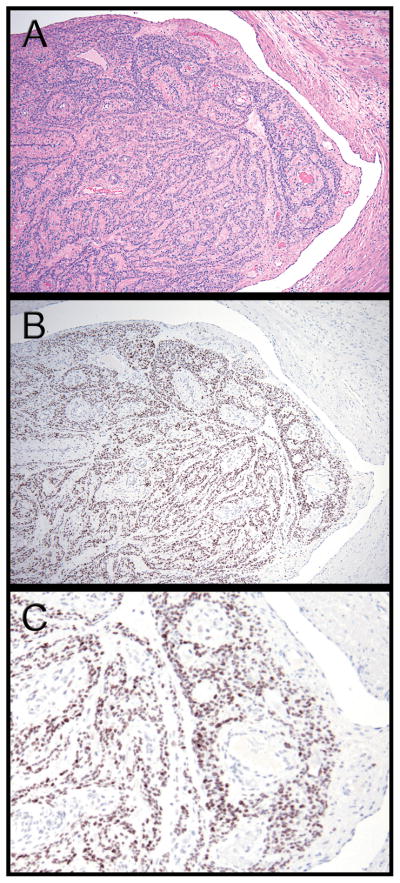
Fig. 1.
(A) Hematoxylin and eosin stained tissue section of Case 1 shows a plug-like tumor mass nearly occluding the residual cleft-like endothelial-lined vascular lumen with normal myometrium on the right. (B) Immunohistochemistry with a polyclonal HMGA2 antibody showing strong staining in intravenous leiomyomatosis tissue, but not in the adjacent myometrium. (C) Higher magnification image of panel B, in which one can appreciate that the HMGA2 staining is specific to nuclei in smooth muscle cells in lesional cells, but not in endothelial cells or smooth cells in the supporting normal blood vessels and adjacent myometrium.
Table 2.
HMGA2 immunohistochemistry and HMGA2 interphase FISH analysis
| Case no.1 | HMGA2 expression by immunohistochemistry | % nuclei with HMGA2 and 14q24 probe co-localization by interphase FISH2 |
|---|---|---|
| 1 | Positive (+4) | 823 |
| 2 | Positive (+4) | 944 |
| 3 | Positive (+4) | Not Determined5 |
| 4 | Positive (+4) | 90 |
| 5 | Positive (+4) | 906 |
| 6 | Positive (+4) | Not Determined |
| 7 | Positive (+4) | 90 |
| 8 | Negative (0) | 14 |
| 9 | Negative (0) | 13 |
| 10 | Negative (0) | 16 |
| 11 | Negative (0) | 9 |
| 12 | Negative (0) | 10 |
Cases ordered by HMGA2 immunohistochemistry expression status
When grouped by immunohistochemistry expression status, the mean percentages of FISH probe co-localization were significantly different (p=8.24×10−10): 89.2% (95% confidence interval: 83.8–94.6%) vs. 12.4% (95% confidence interval: 8.8–16.0%) for cases with positive and negative HMGA2 protein expression, respectively.
Interphase FISH showed HMGA2 amplification in 96/100 nuclei.
Interphase FISH showed that 69/100 nuclei had three hybridization signals for HMGA2, and was previously reported as ST00-142 to have der(14)t(12;14)(q15;q24) and three hybridization signals for HMGA2 in metaphase FISH (Table 1, published as ST00-142) (19).
Previously reported to have der(14)t(12;14)(q15;q24) and three hybridization signals for HMGA2 in metaphase FISH (Table 1, published as ST02-165) (18).
Interphase FISH showed that 20/100 nuclei had three hybridization signals for HMGA2.
Results of GTG-banded karyotype and metaphase FISH analyses of Case 1 were determined to be 45,XX,del(12)(q?14q?15),hsr(14)(q2?1),-22[7].ish del(12)(q14.3q14.3)(5′HMGA2-,3′HMGA2-),hsr(14)amp(3′HMGA2) (Table 1, Fig. 2), interpreted as a der(14) with amplification of the 3′ HMGA2 region and an interstitial deletion of 12q14.3 involving the entire HMGA2. This result is further supported by interphase FISH results consistent with a der(14) with amplification of the 3′ HMGA2 region and absence of 3′ HMGA2 region-hybridization to the del(12) (in Fig. 2, the 3′HMGA2 probe is labeled with SpectrumOrange in metaphase FISH and with SpectrumGreen in interphase FISH). Of note, this case had plexiform histological features (Fig. 1A).
Fig. 2.
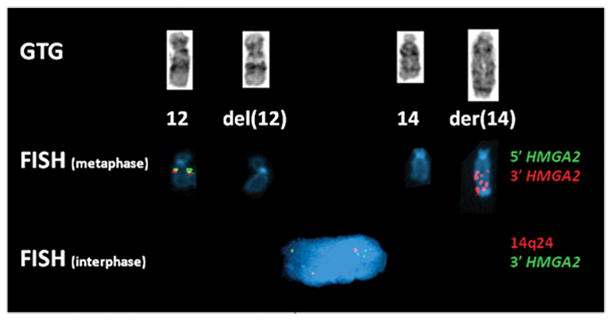
Partial GTG-banded karyotype, metaphase and interphase FISH of Case 1. Partial karyotype (top) shows a chromosome 12, del(12), chromosome 14 and der(14). In the metaphase FISH (middle), absence of hybridization of both HMGA2 probes (5′ green and 3′ orange) is observed on the del(12) whereas amplification of the 3′ HMGA2 (orange) is detected on the der(14). In the interphase FISH (bottom) the absence of 3′ HMGA2 signal (green, 5′ HMGA2 not performed) is observed and its amplification is detected on the der(14) next to the 14q24 signal (orange).
Similar chromosomal aberrations with der(14) have been reported for Cases 2 and 3 in our previous publications (18, 19). The GTG-banded karyotype of Case 2 had a der(14)t(12;14) (q15;q24) in all metaphases, and Case 3 had a mosaic karyotype including both der(14)t(12;14) (q15;q24) and t(12;14) (q15;q24). Of note, both Cases 1 and 2 had monosomy 22 (Table 1). Metaphase FISH analysis for Cases 2 and 3 revealed three hybridization signals for HMGA2 on two apparently structurally normal chromosomes 12 and on the der(14) in the breakpoint region with a pattern indicating the 12q breakpoint occurred 5′ (centromeric) to HMGA2 (Table 1) (18, 19). aCGH was able to be performed only for Case 3 (ST02-166) and the results correlated with the complex karyotype with multiple aneuploidies, in addition to a loss of material on chromosome 14 (14q24->14qter), confirming replacement of a chromosome 14 with a der(14)t(12;14) (Table 3).
Based on the high frequency of these chromosomal aberrations in intravenous leiomyomatosis, interphase FISH was performed using BAC probes for HMGA2 (at 12q14.3) and 14q24.1 loci on intravenous leiomyomatosis cases (Fig. 3 and Table 2). In Case 1, HMGA2 amplification was detected in 90/100 nuclei and co-localization of signals was found in 82/100 nuclei in parallel to metaphase FISH and GTG-banded karyotype analyses (Fig. 2). In Case 2, three hybridization signals for HMGA2 were found in 69/100 nuclei and co-localization of signals was detected in 94/100 nuclei consistent with the previously reported karyotype and metaphase FISH analyses revealing two copies of chromosome 12 and one copy of the der(14)(12;14) (19). 89.2% (95% confidence interval: 83.8–94.6%) of nuclei on immunohistochemically HMGA2-positive tumors analyzed by interphase FISH (5/7) showed co-localization of chromosome 12 and 14 signals, while only 12.4% (95% confidence interval: 8.8–16.0%) of nuclei on HMGA2-negative tumors analyzed by interphase FISH (5/5) showed co-localization of the probes. The correlation of immunohistochemistry and FISH status was significant at p=8.24×10−10. Of note, evidence of more than two hybridization signals for HMGA2 was found only in one additional case; 20/100 interphase nuclei in Case 5 showed three hybridization signals for HMGA2. This level was appreciably lower than that observed in Case 2 (69/100), and may represent mosaicism. Overall, four of six (66.6%) HMGA2-positive intravenous leiomyomatosis cases analyzed by FISH had three or more signals for HMGA2, an indication of the presence of an unbalanced der(14) rather than a balanced t(12;14). It should, however, be noted that the interphase FISH results with only co-localization of chromosome 12 and 14 signals without multiplication of HMGA2 signal may still be due to the presence of a der(14), if there were concomitantly an interstitial deletion of chromosome 12 similar to the del(12) seen in Case 1 (Fig. 2).
Fig. 3.
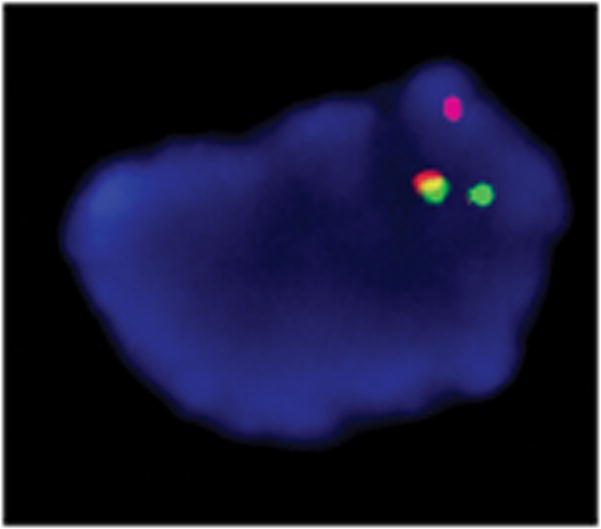
Representative image for co-localization of hybridization signals of BAC probes RP11-366L20 at 12q14.3 (green), spanning the 3′ HMGA2, and RP11-195L9 at 14q24.1 (orange) in Case 4.
Interphase FISH for deletion of 7q22 was also performed to assess whether the most commonly observed chromosomal aberration reported in uterine leiomyoma (24) also presents as a nonrandom chromosomal abnormality in intravenous leiomyomatosis. None of 10 cases able to be evaluated was interpreted to have a del(7)(q22) based on loss of hybridization for the probe.
Hierarchical cluster analysis was performed to compare expression profiles of three intravenous leiomyomatosis cases with previously reported leiomyosarcomas, myometria, uterine leiomyomata, and histological variants of uterine leiomyoma (cellular, atypical and plexiform) (31–33). Intravenous leiomyomatosis cases clustered together on a single node along with a case of metastatic leiomyosarcoma (LMS 906), despite the presence of t(12;14)(q15;q24) and HMGA2 over-expression in one plexiform leiomyoma case (ST06-015F) and one uterine leiomyoma case (ST99-240) (Fig. 4). To investigate further the difference between intravenous leiomyomatosis cases and uterine leiomyoma with t(12;14), differentially expressed genes (p<0.05) were assessed between intravenous leiomyomatosis cases and an independent set of nine t(12;14) uterine leiomyoma cases which had previous transcriptional profiling (25) (Fig. 5). Twenty-four out of 33 genes found to be significant for differential expression by this analysis are reported to be up or down-regulated in cancer as potential oncogenes or tumor suppressor genes, or contributors to cancer progression. Of note, MDM2 was found to be down-regulated in the intravenous leiomyomatosis cases, supporting further the negative immunohistochemistry results for MDM2 (see Supplementary Table).
Fig. 4.
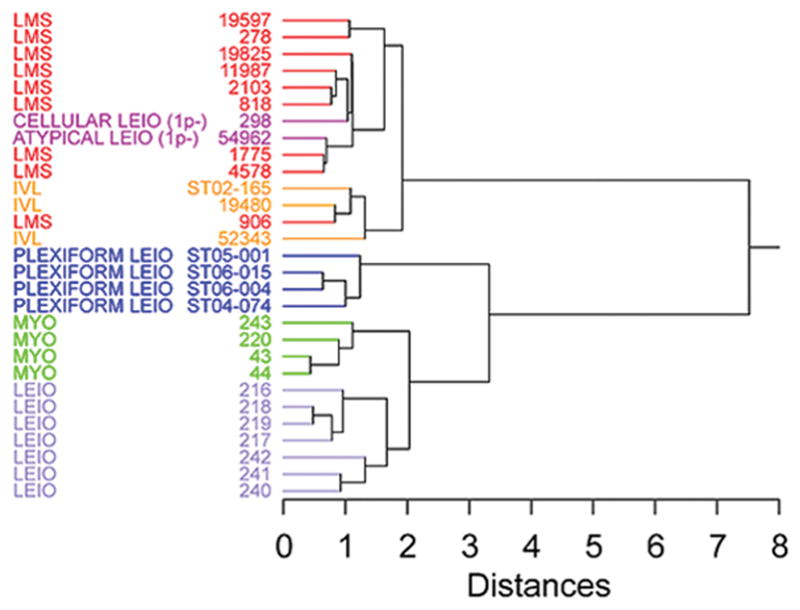
Hierarchical clustering of mRNA expression profiling of three cases of intravenous leiomyomatosis (IVL) compared to the profiles of leiomyosarcoma (LMS), leiomyoma of the usual histologic type (LEIO), histologic variants of leiomyomata (cellular, atypical and plexiform), and myometrium (MYO).
Fig. 5.
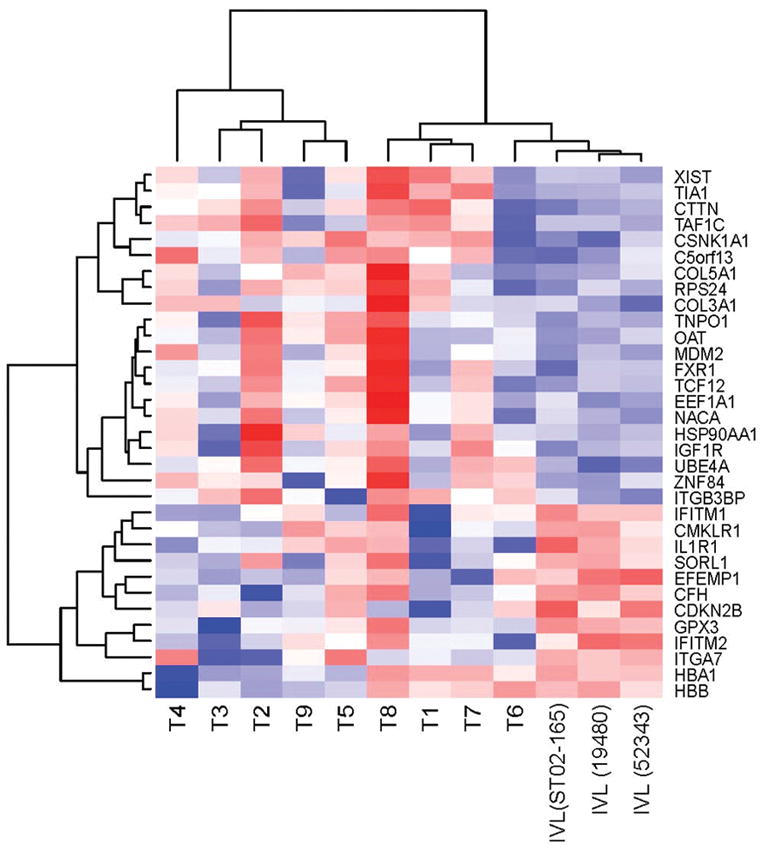
Differentially expressed genes (p<0.05) in three intravenous leiomyomatosis cases (IVL) in comparison to uterine leiomyomata with t(12;14) (T1 to T9) (red: up-regulated, blue: down-regulated).
DISCUSSION
Intravenous leiomyomatosis is a histologically benign smooth muscle tumor, developing within the uterine and pelvic veins, that extends in severe cases into the inferior vena cava and chambers of the right heart (10–12). Similar to uterine leiomyoma, intravenous leiomyomatosis is composed of a bland proliferation of fascicles of smooth muscle, and in many cases, is associated with a typical uterine leiomyoma on pathologic examination. Although an uncommon phenomenon, analyzing the molecular mechanisms of intravenous leiomyomatosis may provide valuable insights into the histological events that underlie the transition from a non-invasive uterine leiomyoma to an invasive intravenous leiomyomatosis.
Chromosomal rearrangements at 12q14-15 are frequent in various mesenchymal tumors including breast fibroadenoma, cutaneous lipoma, pulmonary chondroid hamartoma, salivary gland pleomorphic adenoma, vulvar aggressive angiomyxoma, and uterine leiomyoma (3, 34–39). It is well established that these rearrangements are clustered around a genomic region that includes HMGA2 (formerly HMGIC, 12q14.3), as well as MDM2 (12q15) and CDK4 (12q14.1) (20, 32, 36, 40). Also, overexpression of HMGA2, MDM2, and CDK4 proteins associated with these chromosomal alterations has been shown in some mesenchymal tumors including Müllerian adenosarcoma (20, 41, 42). MDM2 and CDK4 proto-oncogenes have an important role in permitting override of the G1-S cell cycle checkpoint, and up-regulation of these genes leads to increased cell proliferation and survival (43–45). HMGA2 is a non-histone DNA architectural factor involved in transcriptional regulation that likely affects a variety of downstream targets involved in differentiation and proliferation (46–48). It plays a role in normal mesenchymal growth, and developmental perturbations result in abnormal fat and skeletal changes determining overall adiposity and height in both mice and humans (32, 47, 49–51). The genetic alteration leading to the downstream effect of HMGA2 expression varies among the aforementioned mesenchymal tumors (42, 52–54). For example, fusion gene formation predominates in lipomas resulting in translocation of the three AT-hook DNA binding domains of HMGA2 to a variety of chromosome partners (36), whereas in uterine leiomyoma the chromosomal aberration 5′ of the HMGA2 locus is preferentially a translocation between chromosomes 12 and 14. HMGA2 coding sequence remains almost uniformly intact with dysregulated HMGA2 expression, presumably due to either a transposition of a promoter or positive regulatory element on chromosome 14 adjacent to HMGA2 on chromosome 12, or removal of a negative regulatory element from chromosome 12 (55).
The present study analyzed expression of HMGA2, MDM2, and CDK4 proteins in 12 intravenous leiomyomatosis cases based on the previous finding of der(14)t(12;14)(q15;q24) in intravenous leiomyomatosis (18, 19). The frequency of HMGA2 protein expression in intravenous leiomyomatosis (58%) was higher than that reported in uterine leiomyoma (32%) (56); MDM2 and CDK4 expression was not detected in any cases. These findings suggest that HMGA2 protein expression might contribute to the underlying mechanisms for intravenous leiomyomatosis development, particularly considering similar results reported for uterine leiomyoma, plexiform leiomyomata, vulvar aggressive angiomyxoma, and other mesenchymal tumors (20, 32, 35, 42, 57–59), in addition to a potential role of HMGA2 as a tumor driver for metastasis and invasion (50, 60, 61).
Only Case 1 was suitable for GTG-banded karyotyping and metaphase FISH analyses in addition to the previously published Cases 2 and 3 (18, 19). Cases 1 and 2 showed loss of chromosome 22 consistent with a recent publication reporting aCGH in nine intravenous leiomyomatosis cases with deletions of 22 as the most frequent aberration (66.7%) (62). Case 1 had an interstitial deletion of the 12q14.3 region leading to loss of HMGA2 at this site concurrent with co-amplification of HMGA2 on the long arm of chromosome 14, consistent with interphase FISH results (Tables 1 and 2, Fig. 2). The aCGH result for Case 3 supported further the replacement of a chromosome 14 with a der(14)t(12;14) with loss of the 14q24->14qter region. To assess cytogenetic correlation with HMGA2 protein expression, interphase FISH was performed on intravenous leiomyomatosis cases using probes for both HMGA2 and 14q24 loci. All of the analyzed HMGA2-positive cases (five out of seven) had hybridization patterns with co-localization of the probes. Three cases had more than two hybridization signals for HMGA2. Overall, based on the high frequency (89.2%, 95% confidence interval: 83.8–94.6%) of co-localization of FISH signals in HMGA2-positive cases compared to that of HMGA2-negative cases (12.4%) and the relatively lower frequency of supernumerary HMGA2 copies, the t(12;14) leading to HMGA2 expression might be considered as the primary pathogenetic event, and acquisition of extra HMGA2 copies a secondary, but not necessarily critical event in intravenous leiomyomatosis pathogenesis. This finding is further supported by the aforementioned intravenous leiomyomatosis microarray study reporting amplification of the HMGA2 locus in only two out of nine cases (22%) (62). We propose that loss of der(12)t(12;14) followed by reduplication of an apparently normal copy of chromosome 12 (resulting in three copies of HMGA2 or, alternatively, potential loss of heterozygosity on chromosomes 12 and 14) might play a role in the pathogenesis of intravenous leiomyomatosis. Interphase FISH was also performed for evaluating presence of a deletion of 7q, the most common cytogenetic finding in uterine leiomyoma (24); none of the cases showed loss of hybridization signal for the 7q22 probe, further highlighting the role of HMGA2 in intravenous leiomyomatosis pathogenesis.
Emerging evidence suggests at least four molecular subclasses for uterine leiomyoma: MED12 mutation, FH inactivation, HMGA2 overexpression, and COL4A6–COL4A5 deletion. MED12 and HMGA2 aberrations are found to be mutually exclusive with very distinct gene expression profiles, suggesting two separate pathways of uterine leiomyoma formation. Taken together, they may account for 80%–90% of all uterine leiomyoma cases. Chromosome 7q alterations have been reported to co-occur with both MED12 and HMGA2 aberrations, indicating a secondary event in uterine leiomyoma pathogenesis (63, 64). The absence of MED12 mutations in previously reported intravenous leiomyomatosis cases (62) and the lack of 7q deletion detection in the current study, provide further evidence for the resemblance of intravenous leiomyomatosis to uterine leiomyoma with HMGA2 aberrations and the role of HMGA2 in the primary pathogenesis of intravenous leiomyomatosis.
Rearrangements of 12q14-15, typically t(12;14)(q14.3;q24), occur in approximately 7.5% of all uterine leiomyoma and 20% of karyotypically abnormal uterine leiomyoma (65). It has been shown that the presence of t(12;14) in uterine leiomyoma leads to elevated expression of HMGA2 (25, 57–59). Also, uterine leiomyoma with rearrangements of 12q14-15 are larger in size than those with either interstitial 7q22 deletions or normal karyotypes (66, 67), suggesting a marked growth advantage of cells with dysregulated HMGA2. Tumor size might be directly related to increased expression of HMGA2, which has been identified as a delayed early response gene promoting progression to S phase in response to growth factors in various cell types, by overcoming the requirement for mitogenic stimulation (68–71).
Intravenous leiomyomatosis is usually considered together with a group of tumors including disseminated peritoneal leiomyomatosis and benign metastasizing leiomyoma, which resemble uterine leiomyoma at both gross and microscopic levels but are found in unusual anatomical locations. A common molecular mechanism underlying these tumors might help to understand better intravenous leiomyomatosis pathogenesis, as well as the malignant potential of some uterine leiomyomata. In that regard, a study analyzing a disseminated peritoneal leiomyomatosis case occurring after laparoscopic morcellation for uterine leiomyoma was observed to have a t(12;14)(q15;q24), del(3)(q23q26.33), and r(1)(p34.3q41), all of which are characteristic cytogenetic findings of uterine leiomyoma (30). Like the findings of the current study, deletion of 7q22 was not detected in this disseminated peritoneal leiomyomatosis case. Another study analyzing a paratesticular leiomyoma reported a karyotype of 46,XY,der(5)t(5;14)(q31;q24),der(14)t(12;14)(q15;q24)[25] with HMGA2 over-expression providing further evidence for the importance of HMGA2 and t(12;14) in the pathogenesis of the unusual variants of uterine leiomyoma (72).
Vulvar aggressive angiomyxoma also have frequent aberrant HMGA2 expression due to 12q14-15 rearrangements. Although not observed to metastasize or invade vessels, this vulvar mesenchymal neoplasm is locally and destructively invasive. In contrast to uterine leiomyoma and intravenous leiomyomatosis, a wide range of translocation partners are observed in aggressive angiomyxomas (35, 73–75). Müllerian adenosarcomas of the uterus have frequent amplification of 12q that leaves HMGA2 intact and leads to over-expression (42). The molecular basis by which the malignant phenotypes of these tumors is determined remains to be elucidated, but is presumably superimposed upon the effect of aberrant HMGA2 expression.
Despite the common properties of intravenous leiomyomatosis and uterine leiomyoma at both histopathologic and cytogenetic levels as described above, uterine leiomyoma is considered benign whereas intravenous leiomyomatosis has a quasi-malignant behavior characterized by prominent vascular invasion. Another possible mechanism that might explain the different phenotype is the state of the cell in which HMGA2 dysregulation occurs (76). This phenomenon is identified as “the cell of origin effect”. If the founder cell transformed in intravenous leiomyomatosis is different than uterine leiomyoma, it might be morphologically similar, but have a different biologic potential supported by the result of the cluster analysis with the closer relationship of intravenous leiomyomatosis to leiomyosarcoma in the current study. Although differences in transcription profiles between uterine leiomyoma and intravenous leiomyomatosis may provide a molecular explanation for vascular invasiveness, a clear indication of key genes that would explain this difference in phenotype was not readily apparent in our analysis, although there are some suggestive genes (Supplementary Table). Of potential relevance, it should be noted that despite the common t(12;14) abnormality, intravenous leiomyomatosis cases also had additional chromosomal abnormalities detected by GTG-banded karyotyping and aCGH (Tables 1 and 2), which might contribute to the unusual behavior of intravenous leiomyomatosis. Further study is warranted to delineate the molecular mechanism(s) underlying the intravenous leiomyomatosis phenotype.
In conclusion, the significant association detected between HMGA2 expression and t(12;14)(q15;q24) in intravenous leiomyomatosis cases is likely to play an important role in intravenous leiomyomatosis tumorigenesis, but the quasi-malignant behavior of intravenous leiomyomatosis might be attributed to additional genetic alterations as suggested by transcriptional and aCGH analyses.
Supplementary Material
Acknowledgments
This research was supported by HD060530 (to C.C.M.). The authors thank the Harvard-Partners Center for Genetics and Genomics for kindly providing the BAC clones and the Cytogenetics Core of the Dana-Farber Harvard Cancer Center (P30 CA006516) for assisting with the aCGH analysis.
Footnotes
DISCLOSURE/CONFLICT OF INTEREST
The authors declare no conflicts of interest.
References
- 1.Cramer SF, Patel A. The frequency of uterine leiomyomas. Am J Clin Pathol. 1990;94:435–438. doi: 10.1093/ajcp/94.4.435. [DOI] [PubMed] [Google Scholar]
- 2.Nibert M, Heim S. Uterine leiomyoma cytogenetics. Genes Chromosomes Cancer. 1990;2:3–13. doi: 10.1002/gcc.2870020103. [DOI] [PubMed] [Google Scholar]
- 3.Rein MS, Friedman AJ, Barbieri RL, et al. Cytogenetic abnormalities in uterine leiomyomata. Obstet Gynecol. 1991;77:923–926. [PubMed] [Google Scholar]
- 4.Wilkinson N, Rollason TP. Recent advances in the pathology of smooth muscle tumours of the uterus. Histopathology. 2001;39:331–341. doi: 10.1046/j.1365-2559.2001.01300.x. [DOI] [PubMed] [Google Scholar]
- 5.Evans HL, Chawla SP, Simpson C, Finn KP. Smooth muscle neoplasms of the uterus other than ordinary leiomyoma. A study of 46 cases, with emphasis on diagnostic criteria and prognostic factors. Cancer. 1988;62:2239–2247. doi: 10.1002/1097-0142(19881115)62:10<2239::aid-cncr2820621028>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 6.Bader LV. Multicentric plexiform tumourlets of the uterus: case report. Pathology. 1971;3:167–170. [PubMed] [Google Scholar]
- 7.Bell SW, Kempson RL, Hendrickson MR. Problematic uterine smooth muscle neoplasms. A clinicopathologic study of 213 cases. Am J Surg Pathol. 1994;18:535–558. [PubMed] [Google Scholar]
- 8.Fletcher JA, Morton CC, Pavelka K, Lage JM. Chromosome aberrations in uterine smooth muscle tumors: potential diagnostic relevance of cytogenetic instability. Cancer Res. 1990;50:4092–4097. [PubMed] [Google Scholar]
- 9.Sreekantaiah C, Davis JR, Sandberg AA. Chromosomal abnormalities in leiomyosarcomas. Am J Pathol. 1993;142:293–305. [PMC free article] [PubMed] [Google Scholar]
- 10.Fukaya Y, Iida F, Morimoto M, et al. A case report on successful removal of intravenous leiomyomatosis extending in the right ventricle. Surgery. 1991;110:909–911. [PubMed] [Google Scholar]
- 11.Mitsuhashi A, Nagai Y, Sugita M, Nakajima N, Sekiya S. GnRH agonist for intravenous leiomyomatosis with cardiac extension. A case report. J Reprod Med. 1999;44:883–886. [PubMed] [Google Scholar]
- 12.Nam MS, Jeon MJ, Kim YT, et al. Pelvic leiomyomatosis with intracaval and intracardiac extension: a case report and review of the literature. Gynecol Oncol. 2003;89:175–180. doi: 10.1016/s0090-8258(02)00138-5. [DOI] [PubMed] [Google Scholar]
- 13.Valdes Devesa V, Conley CR, Stone WM, Collins JM, Magrina JF. Update on intravenous leiomyomatosis: report of five patients and literature review. Eur J Obstet Gynecol Reprod Biol. 2013;171:209–213. doi: 10.1016/j.ejogrb.2013.09.031. [DOI] [PubMed] [Google Scholar]
- 14.Ling FT, David TE, Merchant N, Yu E, Butany JW. Intracardiac extension of intravenous leiomyomatosis in a pregnant woman: A case report and review of the literature. Can J Cardiol. 2000;16:73–79. [PubMed] [Google Scholar]
- 15.Lo KW, Lau TK. Intracardiac leiomyomatosis. Case report and literature review. Arch Gynecol Obstet. 2001;264:209–210. doi: 10.1007/s004040000115. [DOI] [PubMed] [Google Scholar]
- 16.Matsumoto K, Yamamoto T, Hisayoshi T, Asano G. Intravenous leiomyomatosis of the uterus with multiple pulmonary metastases associated with large bullae-like cyst formation. Pathol Int. 2001;51:396–401. doi: 10.1046/j.1440-1827.2001.01205.x. [DOI] [PubMed] [Google Scholar]
- 17.Konrad P, Mellblom L. Intravenous leiomyomatosis. Acta Obstet Gynecol Scand. 1989;68:371–376. doi: 10.3109/00016348909028675. [DOI] [PubMed] [Google Scholar]
- 18.Dal Cin P, Quade BJ, Neskey DM, et al. Intravenous leiomyomatosis is characterized by a der(14)t(12;14)(q15;q24) Genes Chromosomes Cancer. 2003;36:205–206. doi: 10.1002/gcc.10159. [DOI] [PubMed] [Google Scholar]
- 19.Quade BJ, Dal Cin P, Neskey DM, Weremowicz S, Morton CC. Intravenous leiomyomatosis: molecular and cytogenetic analysis of a case. Mod Pathol. 2002;15:351–356. doi: 10.1038/modpathol.3880529. [DOI] [PubMed] [Google Scholar]
- 20.Italiano A, Bianchini L, Keslair F, et al. HMGA2 is the partner of MDM2 in well-differentiated and dedifferentiated liposarcomas whereas CDK4 belongs to a distinct inconsistent amplicon. Int J Cancer. 2008;122:2233–2241. doi: 10.1002/ijc.23380. [DOI] [PubMed] [Google Scholar]
- 21.Dei Tos AP, Doglioni C, Piccinin S, et al. Coordinated expression and amplification of the MDM2, CDK4, and HMGI-C genes in atypical lipomatous tumours. J Pathol. 2000;190:531–536. doi: 10.1002/(SICI)1096-9896(200004)190:5<531::AID-PATH579>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 22.van Roggen JF, van Unnik JA, Briaire-de Bruijn IH, Hogendoorn PC. Aggressive angiomyxoma: a clinicopathological and immunohistochemical study of 11 cases with long-term follow-up. Virchows Arch. 2005;446:157–163. doi: 10.1007/s00428-004-1135-9. [DOI] [PubMed] [Google Scholar]
- 23.Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer. 2013;13:83–96. doi: 10.1038/nrc3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xing YP, Powell WL, Morton CC. The del(7q) subgroup in uterine leiomyomata: genetic and biologic characteristics. Further evidence for the secondary nature of cytogenetic abnormalities in the pathobiology of uterine leiomyomata. Cancer Genet Cytogenet. 1997;98:69–74. doi: 10.1016/s0165-4608(96)00406-2. [DOI] [PubMed] [Google Scholar]
- 25.Hodge JC, Kim TM, Dreyfuss JM, et al. Expression profiling of uterine leiomyomata cytogenetic subgroups reveals distinct signatures in matched myometrium: transcriptional profilingof the t(12;14) and evidence in support of predisposing genetic heterogeneity. Hum Mol Genet. 2012;21:2312–2329. doi: 10.1093/hmg/dds051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clement PB, Young RH, Scully RE. Intravenous leiomyomatosis of the uterus. A clinicopathological analysis of 16 cases with unusual histologic features. Am J Surg Pathol. 1988;12:932–945. [PubMed] [Google Scholar]
- 27.Mulvany NJ, Slavin JL, Ostor AG, Fortune DW. Intravenous leiomyomatosis of the uterus: a clinicopathologic study of 22 cases. Int J Gynecol Pathol. 1994;13:1–9. doi: 10.1097/00004347-199401000-00001. [DOI] [PubMed] [Google Scholar]
- 28.Norris HJ, Parmley T. Mesenchymal tumors of the uterus. V. Intravenous leiomyomatosis. A clinical and pathologic study of 14 cases. Cancer. 1975;36:2164–2178. doi: 10.1002/cncr.2820360935. [DOI] [PubMed] [Google Scholar]
- 29.Fletcher JA, Pinkus GS, Weidner N, Morton CC. Lineage-restricted clonality in biphasic solid tumors. Am J Pathol. 1991;138:1199–1207. [PMC free article] [PubMed] [Google Scholar]
- 30.Ordulu Z, Dal Cin P, Chong WW, et al. Disseminated peritoneal leiomyomatosis after laparoscopic supracervical hysterectomy with characteristic molecular cytogenetic findings of uterine leiomyoma. Genes Chromosomes Cancer. 2010;49:1152–1160. doi: 10.1002/gcc.20824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Christacos NC, Quade BJ, Dal Cin P, Morton CC. Uterine leiomyomata with deletions of Ip represent a distinct cytogenetic subgroup associated with unusual histologic features. Genes Chromosomes Cancer. 2006;45:304–312. doi: 10.1002/gcc.20291. [DOI] [PubMed] [Google Scholar]
- 32.Hodge JC, Quade BJ, Rubin MA, et al. Molecular and cytogenetic characterization of plexiform leiomyomata provide further evidence for genetic heterogeneity underlying uterine fibroids. Am J Pathol. 2008;172:1403–1410. doi: 10.2353/ajpath.2008.071102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quade BJ, Wang TY, Sornberger K, et al. Molecular pathogenesis of uterine smooth muscle tumors from transcriptional profiling. Genes Chromosomes Cancer. 2004;40:97–108. doi: 10.1002/gcc.20018. [DOI] [PubMed] [Google Scholar]
- 34.Medeiros F, Erickson-Johnson MR, Keeney GL, et al. Frequency and characterization of HMGA2 and HMGA1 rearrangements in mesenchymal tumors of the lower genital tract. Genes Chromosomes Cancer. 2007;46:981–990. doi: 10.1002/gcc.20483. [DOI] [PubMed] [Google Scholar]
- 35.Nucci MR, Weremowicz S, Neskey DM, et al. Chromosomal translocation t(8;12) induces aberrant HMGIC expression in aggressive angiomyxoma of the vulva. Genes Chromosomes Cancer. 2001;32:172–176. doi: 10.1002/gcc.1179. [DOI] [PubMed] [Google Scholar]
- 36.Schoenmakers EF, Wanschura S, Mols R, et al. Recurrent rearrangements in the high mobility group protein gene, HMGI-C, in benign mesenchymal tumours. Nat Genet. 1995;10:436–444. doi: 10.1038/ng0895-436. [DOI] [PubMed] [Google Scholar]
- 37.Dal Cin P, Kools P, De Jonge I, et al. Rearrangement of 12q14-15 in pulmonary chondroid hamartoma. Genes Chromosomes Cancer. 1993;8:131–133. doi: 10.1002/gcc.2870080211. [DOI] [PubMed] [Google Scholar]
- 38.Rohen C, Caselitz J, Stern C, et al. A hamartoma of the breast with an aberration of 12q mapped to the MAR region by fluorescence in situ hybridization. Cancer Genet Cytogenet. 1995;84:82–84. doi: 10.1016/0165-4608(95)00060-7. [DOI] [PubMed] [Google Scholar]
- 39.Staats B, Bonk U, Wanschura S, et al. A fibroadenoma with a t(4;12) (q27;q15) affecting the HMGI-C gene, a member of the high mobility group protein gene family. Breast Cancer Res Treat. 1996;38:299–303. doi: 10.1007/BF01806149. [DOI] [PubMed] [Google Scholar]
- 40.Fejzo MS, Yoon SJ, Montgomery KT, et al. Identification of a YAC spanning the translocation breakpoints in uterine leiomyomata, pulmonary chondroid hamartoma, and lipoma: physical mapping of the 12q14-q15 breakpoint region in uterine leiomyomata. Genomics. 1995;26:265–271. doi: 10.1016/0888-7543(95)80210-d. [DOI] [PubMed] [Google Scholar]
- 41.Binh MB, Sastre-Garau X, Guillou L, et al. MDM2 and CDK4 immunostainings are useful adjuncts in diagnosing well-differentiated and dedifferentiated liposarcoma subtypes: a comparative analysis of 559 soft tissue neoplasms with genetic data. Am J Surg Pathol. 2005;29:1340–1347. doi: 10.1097/01.pas.0000170343.09562.39. [DOI] [PubMed] [Google Scholar]
- 42.Howitt BE, Sholl LM, Dal Cin P, et al. Targeted genomic analysis of Mullerian adenosarcoma. J Pathol. 2014 doi: 10.1002/path.4442. [DOI] [PubMed] [Google Scholar]
- 43.Ortega S, Malumbres M, Barbacid M. Cyclin D-dependent kinases, INK4 inhibitors and cancer. Biochim Biophys Acta. 2002;1602:73–87. doi: 10.1016/s0304-419x(02)00037-9. [DOI] [PubMed] [Google Scholar]
- 44.Sherr CJ. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 45.Singer S, Socci ND, Ambrosini G, et al. Gene expression profiling of liposarcoma identifies distinct biological types/subtypes and potential therapeutic targets in well-differentiated and dedifferentiated liposarcoma. Cancer Res. 2007;67:6626–6636. doi: 10.1158/0008-5472.CAN-07-0584. [DOI] [PubMed] [Google Scholar]
- 46.Grosschedl R, Giese K, Pagel J. HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet. 1994;10:94–100. doi: 10.1016/0168-9525(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 47.Ligon AH, Moore SD, Parisi MA, et al. Constitutional rearrangement of the architectural factor HMGA2: a novel human phenotype including overgrowth and lipomas. Am J Hum Genet. 2005;76:340–348. doi: 10.1086/427565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolffe AP. Architectural transcription factors. Science. 1994;264:1100–1101. doi: 10.1126/science.8178167. [DOI] [PubMed] [Google Scholar]
- 49.Lettre G. Genetic regulation of adult stature. Curr Opin Pediatr. 2009;21:515–522. doi: 10.1097/MOP.0b013e32832c6dce. [DOI] [PubMed] [Google Scholar]
- 50.Zaidi MR, Okada Y, Chada KK. Misexpression of full-length HMGA2 induces benign mesenchymal tumors in mice. Cancer Res. 2006;66:7453–7459. doi: 10.1158/0008-5472.CAN-06-0931. [DOI] [PubMed] [Google Scholar]
- 51.Zhou X, Benson KF, Ashar HR, Chada K. Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature. 1995;376:771–774. doi: 10.1038/376771a0. [DOI] [PubMed] [Google Scholar]
- 52.Fedele M, Palmieri D, Fusco A. HMGA2: A pituitary tumour subtype-specific oncogene? Mol Cell Endocrinol. 2010;326:19–24. doi: 10.1016/j.mce.2010.03.019. [DOI] [PubMed] [Google Scholar]
- 53.Fedele M, Battista S, Manfioletti G, et al. Role of the high mobility group A proteins in human lipomas. Carcinogenesis. 2001;22:1583–1591. doi: 10.1093/carcin/22.10.1583. [DOI] [PubMed] [Google Scholar]
- 54.Cleynen I, Van de Ven WJ. The HMGA proteins: a myriad of functions (Review) Int J Oncol. 2008;32:289–305. [PubMed] [Google Scholar]
- 55.Quade BJ, Weremowicz S, Neskey DM, et al. Fusion transcripts involving HMGA2 are not a common molecular mechanism in uterine leiomyomata with rearrangements in 12q15. Cancer Res. 2003;63:1351–1358. [PubMed] [Google Scholar]
- 56.Klotzbucher M, Wasserfall A, Fuhrmann U. Misexpression of wild-type and truncated isoforms of the high-mobility group I proteins HMGI-C and HMGI(Y) in uterine leiomyomas. Am J Pathol. 1999;155:1535–1542. doi: 10.1016/S0002-9440(10)65469-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Klemke M, Meyer A, Nezhad MH, et al. Overexpression of HMGA2 in uterine leiomyomas points to its general role for the pathogenesis of the disease. Genes Chromosomes Cancer. 2009;48:171–178. doi: 10.1002/gcc.20627. [DOI] [PubMed] [Google Scholar]
- 58.Gattas GJ, Quade BJ, Nowak RA, Morton CC. HMGIC expression in human adult and fetal tissues and in uterine leiomyomata. Genes Chromosomes Cancer. 1999;25:316–322. [PubMed] [Google Scholar]
- 59.Gross KL, Neskey DM, Manchanda N, et al. HMGA2 expression in uterine leiomyomata and myometrium: quantitative analysis and tissue culture studies. Genes Chromosomes Cancer. 2003;38:68–79. doi: 10.1002/gcc.10240. [DOI] [PubMed] [Google Scholar]
- 60.Morishita A, Zaidi MR, Mitoro A, et al. HMGA2 is a driver of tumor metastasis. Cancer Res. 2013;73:4289–4299. doi: 10.1158/0008-5472.CAN-12-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zha L, Zhang J, Tang W, et al. HMGA2 elicits EMT by activating the Wnt/beta-catenin pathway in gastric cancer. Dig Dis Sci. 2013;58:724–733. doi: 10.1007/s10620-012-2399-6. [DOI] [PubMed] [Google Scholar]
- 62.Buza N, Xu F, Wu W, et al. Recurrent chromosomal aberrations in intravenous leiomyomatosis of the uterus: high-resolution array comparative genomic hybridization study. Hum Pathol. 2014;45:1885–1892. doi: 10.1016/j.humpath.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 63.Mehine M, Makinen N, Heinonen HR, Aaltonen LA, Vahteristo P. Genomics of uterine leiomyomas: insights from high-throughput sequencing. Fertil Steril. 2014;102:621–629. doi: 10.1016/j.fertnstert.2014.06.050. [DOI] [PubMed] [Google Scholar]
- 64.Mashal RD, Fejzo ML, Friedman AJ, et al. Analysis of androgen receptor DNA reveals the independent clonal origins of uterine leiomyomata and the secondary nature of cytogenetic aberrations in the development of leiomyomata. Genes Chromosomes Cancer. 1994;11:1–6. doi: 10.1002/gcc.2870110102. [DOI] [PubMed] [Google Scholar]
- 65.Meloni AM, Surti U, Contento AM, Davare J, Sandberg AA. Uterine leiomyomas: cytogenetic and histologic profile. Obstet Gynecol. 1992;80:209–217. [PubMed] [Google Scholar]
- 66.Hennig Y, Deichert U, Bonk U, et al. Chromosomal translocations affecting 12q14-15 but not deletions of the long arm of chromosome 7 associated with a growth advantage of uterine smooth muscle cells. Mol Hum Reprod. 1999;5:1150–1154. doi: 10.1093/molehr/5.12.1150. [DOI] [PubMed] [Google Scholar]
- 67.Rein MS, Powell WL, Walters FC, et al. Cytogenetic abnormalities in uterine myomas are associated with myoma size. Mol Hum Reprod. 1998;4:83–86. doi: 10.1093/molehr/4.1.83. [DOI] [PubMed] [Google Scholar]
- 68.Ayoubi TA, Jansen E, Meulemans SM, Van de Ven WJ. Regulation of HMGIC expression: an architectural transcription factor involved in growth control and development. Oncogene. 1999;18:5076–5087. doi: 10.1038/sj.onc.1202881. [DOI] [PubMed] [Google Scholar]
- 69.Lanahan A, Williams JB, Sanders LK, Nathans D. Growth factor-induced delayed early response genes. Mol Cell Biol. 1992;12:3919–3929. doi: 10.1128/mcb.12.9.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li D, Lin HH, McMahon M, Ma H, Ann DK. Oncogenic raf-1 induces the expression of non-histone chromosomal architectural protein HMGI-C via a p44/p42 mitogen-activated protein kinase-dependent pathway in salivary epithelial cells. J Biol Chem. 1997;272:25062–25070. doi: 10.1074/jbc.272.40.25062. [DOI] [PubMed] [Google Scholar]
- 71.Kamb A, Gruis NA, Weaver-Feldhaus J, et al. A cell cycle regulator potentially involved in genesis of many tumor types. Science. 1994;264:436–440. doi: 10.1126/science.8153634. [DOI] [PubMed] [Google Scholar]
- 72.Gorunova L, Bjerkehagen B, Heim S. Paratesticular leiomyoma with a der(14)t(12;14)(q15;q24) Cancer Genet. 2011;204:465–468. doi: 10.1016/j.cancergen.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 73.Micci F, Panagopoulos I, Bjerkehagen B, Heim S. Deregulation of HMGA2 in an aggressive angiomyxoma with t(11;12)(q23;q15) Virchows Arch. 2006;448:838–842. doi: 10.1007/s00428-006-0186-5. [DOI] [PubMed] [Google Scholar]
- 74.Rabban JT, Dal Cin P, Oliva E. HMGA2 rearrangement in a case of vulvar aggressive angiomyxoma. Int J Gynecol Pathol. 2006;25:403–407. doi: 10.1097/01.pgp.0000209572.54457.7b. [DOI] [PubMed] [Google Scholar]
- 75.Rawlinson NJ, West WW, Nelson M, Bridge JA. Aggressive angiomyxoma with t(12;21) and HMGA2 rearrangement: report of a case and review of the literature. Cancer Genet Cytogenet. 2008;181:119–124. doi: 10.1016/j.cancergencyto.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ince TA, Richardson AL, Bell GW, et al. Transformation of different human breast epithelial cell types leads to distinct tumor phenotypes. Cancer Cell. 2007;12:160–170. doi: 10.1016/j.ccr.2007.06.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.