Abstract
Genome editing methods have commonly relied on the initial introduction of double-stranded DNA breaks (DSBs), resulting in stochastic insertions, deletions, and translocations at the target genomic locus. To achieve gene correction, these methods typically require the introduction of exogenous DNA repair templates and low-efficiency homologous recombination processes. In this perspective we describe alternative, mechanistically motivated strategies to perform chemistry on the genome of unmodified cells without introducing DSBs. One such strategy, base editing, uses chemical and biological insights to directly and permanently convert one target base pair to another. Despite its recent introduction, base editing has already enabled a number of new capabilities and applications in the genome editing community. We summarize these advances here and discuss the new possibilities that this method has unveiled, concluding with a brief analysis of future prospects for genome and transcriptome editing without double-stranded DNA cleavage.
Introduction
Traditional approaches to edit genomes in living cells introduce a double-stranded DNA break (DSB) at a desired genomic locus. Genomic DNA near the DSB can be replaced with an exogenous donor DNA template by using the endogenous homology-directed repair (HDR) pathway. HDR requires that the donor DNA be homologous to the targeted locus to precisely and predictably result in gene modification.1 Non-homologous end joining (NHEJ), microhomology-mediated end joining (MMEJ), and single-strand annealing (SSA) are complementary endogenous DSB repair pathways that ultimately result in the accumulation of stochastic insertions and deletions (indels) or translocations at the site of the DSB.2–5 Notably, DSBs are typically repaired more efficiently through these pathways than through HDR, especially in non-mitotic cells.
CRISPR systems are RNA-guided protein endonucleases that have stimulated a renaissance in the field of genome editing, allowing researchers to introduce a DSB at nearly any location in the genome with unprecedented ease. In CRISPR systems, Cas endonuclease proteins form effector complexes with “guide RNA” molecules, which program the resultant complex to localize to and cleave a target nucleic acid sequence (the protospacer) through canonical Watson-Crick base-pairing. Protospacer sequences are constrained by the requirement for a nearby protospacer-adjacent motif (PAM), a Cas protein-dependent short nucleic acid sequence.6 Reprogramming the complex to recognize and cut an alternative DNA or RNA sequence simply requires replacing the spacer of the guide RNA with the new sequence of interest.7, 8 The advent and implementation of the CRISPR-Cas systems for use in eukaryotic genome editing9–11 has led to an explosion of advances in the life sciences over a remarkably short time.12
Despite significant progress to improve HDR-mediated genome editing outcomes in cultured cells, largely through the inhibition of NHEJ, synchronization of cells, and donor template designs,13–17 current strategies to precisely modify eukaryotic genomes using HDR under therapeutically relevant conditions remain inefficient, especially in unmodified, non-replicating cells, and frequently result in stochastic mixtures of genome modifications. Motivated by these limitations, we and others have developed alternative approaches to genome editing that do not rely on HDR. Base editing, a novel genome editing strategy, integrates concepts drawn from chemical biology and genome editing to enable the direct chemical conversion of one target genomic DNA base into another at an intended locus without inducing DSB formation.18–21 In this perspective article, we discuss base editing and other strategies for genome editing that do not require DSBs, with an emphasis on recent applications and prospects for the future of this emerging field.
Oligonucleotide-Directed Mutagenesis
Site-directed mutagenesis is a powerful and enabling molecular cloning technology. This method integrates a short DNA oligonucleotide homologous to a region of interest but containing a desired mutation,22 enabling DNA modification in a locus-specific manner. This strategy was successfully adapted for genome editing in yeast by transforming S. cerevisiae with oligonucleotides harboring the desired modifications, introducing multiple mutations with efficiencies reaching 0.1%.23 Mechanistic analysis later revealed that in vivo oligonucleotide-directed mutagenesis (ODM) did not require DSB formation, but rather involved annealing of the oligonucleotide with the genomic DNA during replication to form an Okazaki fragment-like intermediate, followed by incorporation of the mutation via the mismatch repair (MMR) pathway.24
Unsurprisingly, ODM efficiencies are governed by the initial annealing between the oligonucleotide and the targeted genomic locus.25 To improve in vivo hybridization efficiencies, researchers used “chimeraplasts”, chimeric DNA-RNA oligonucleotides containing hairpin-capped ends (Figure 1).26 The DNA segment is complementary to the gene of interest and bears the desired mutation(s), while the RNA segments improve hybridization with genomic DNA.27 The resulting genomic DNA:chimeric oligonucleotide heteroduplex is recognized by the endogenous cellular mismatch repair machinery and repaired, albeit infrequently, using the chimeraplast as a template instead of the genomic DNA.28
Figure 1.
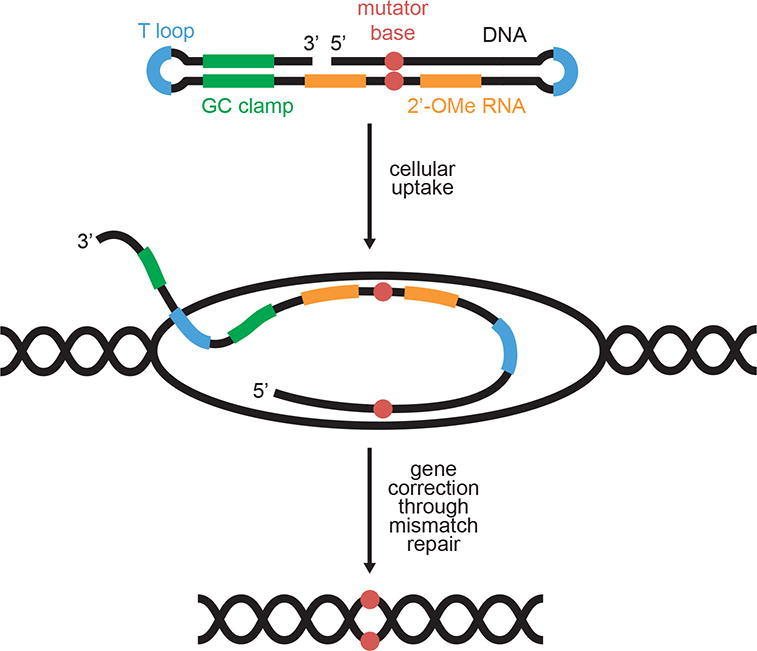
Genome editing using oligonucleotide-directed mutagenesis (ODM). The chimeraplast (top) is a DNA-RNA hybrid double hairpin containing a mutated sequence (red) homologous to the target locus of interest. 2′-OMe RNA (orange) is incorporated in the homology region to increase the binding affinity between the chimeraplast and genomic DNA. A GC clamp (green) enhances nuclease resistance, while hairpin loops (blue) at the very ends of the chimeraplast prevent concatamerization. Following cellular uptake, the chimeraplast anneals to the genomic DNA, forming mismatches at the mutator base. Endogenous cellular mismatch repair pathway will then repair the mismatches at low frequencies using the chimeraplast as a template instead of the genomic DNA.
Further optimization has resulted in the successful use of chimeraplasts in eukaryotic cells in vitro26, 29–31 and in vivo.32, 33 Unfortunately, the generality of this approach is limited by its dependence on many factors, including the target sequence, transcription level of the target genetic locus, target cell type, cellular replication state, and abundance of the Rad51 and MSH2 proteins.34–36 Collectively, these dependencies result in large variations in efficiency.37, 38 Such unpredictable outcomes have made ODM (in its current form) unsuitable for some research applications and most therapeutic uses. However, ODM has been extensively optimized for use in plant genome editing despite requiring single-nucleotide polymorphism screening to discover the desired genetic change(s). Such editing pipelines have facilitated genome modification in tobacco,39, 40 corn,41, 42 rice,43 wheat,44 and rapeseed.45 Notably, ODM has been used to develop a non-transgenic plant breeding technology that has led to a commercial, non-transgenic herbicide-resistant canola.46
To combat the inconsistencies with the ODM method, Storici and coworkers developed the two-step double selection method “delitto perfetto”.47 The first step uses homologous recombination (HR) to incorporate a cassette containing a counterselectable marker and a reporter gene into a genomic locus of interest without explicitly creating DSBs. Cells that have successfully incorporated the cassette are isolated using the reporter gene. Next, an oligonucleotide that includes the mutation of interest is delivered into the cells and incorporated into the genome while excising the cassette, again via HR. Counterselection yields those cells with the desired mutation. This method has been successfully used in both yeast48 and mammalian cells,49, 50 but due to its reliance on multiple HR events and double selection, its use is restricted to those cell types that express proteins such as Rad52 that are required for HR, and its intrinsic efficiency is low.
Base Editing
We recently reported the development of base editing, a genome editing methodology that allows the precise, irreversible conversion of one base pair to another in a programmable manner, obviating the need for DSB formation or HDR.18 Other laboratories have since developed and reported related systems, demonstrating the robustness and scope of the base editing strategy.19 Our initial base editors used a single-stranded DNA-specific cytidine deaminase enzyme tethered to a catalytically impaired Cas9 protein (dCas9) to convert a C•G base pair to a T•A base pair at a target locus of interest (Figure 2a). The deaminase-dCas9 fusion catalyzes the hydrolytic deamination of cytosine to uridine within a small (~3- to 5-nucleotide) window of the target protospacer by exploiting a short segment of accessible single-stranded DNA in the “R-loop” of the Cas9:guide RNA:DNA ternary complex.51 To enhance conversion of the U•G intermediate to the desired T•A product in eukaryotic cells, we fused an 83-amino acid uracil glycosylase inhibitor (UGI) to the deaminase-dCas9 construct to inhibit uracil excision following deamination (Figure 2b). In addition, we reverted an inactivating catalytic mutation in the HNH domain of dCas9 to enable DNA nicking of the G-containing strand opposite the newly formed uracil, thereby inducing host MMR to repair the U•G mismatch into a U•A pair (Figure 2b).52 The combination of these innovations resulted in BE3, a single protein consisting of a tripartite fusion between the Rattus norvegicus APOBEC1 cytidine deaminase, Streptococcus pyogenes Cas9n(D10A), and Bacillus subtilis bacteriophage PBS2 UGI (Figure 2a). BE3 results in permanent C•G-to-T•A correction and does not rely on DSB formation, yielding higher base editing efficiencies than HDR (typically, 15–75% for BE3) and much lower indel frequencies than nuclease-mediated approaches (typically < 5% for BE3). In addition, multiple studies have shown that BE3 has fewer off-target editing events than Cas9.18, 53, 54 This chemoselective genomic modification can be efficiently achieved in living cells and organisms54–56 without resorting to the introduction of synonymous mutations via HDR to limit Cas9 reengagement,13 or subjecting cells to perturbative conditions that enhance HDR efficiency.14–16
Figure 2.
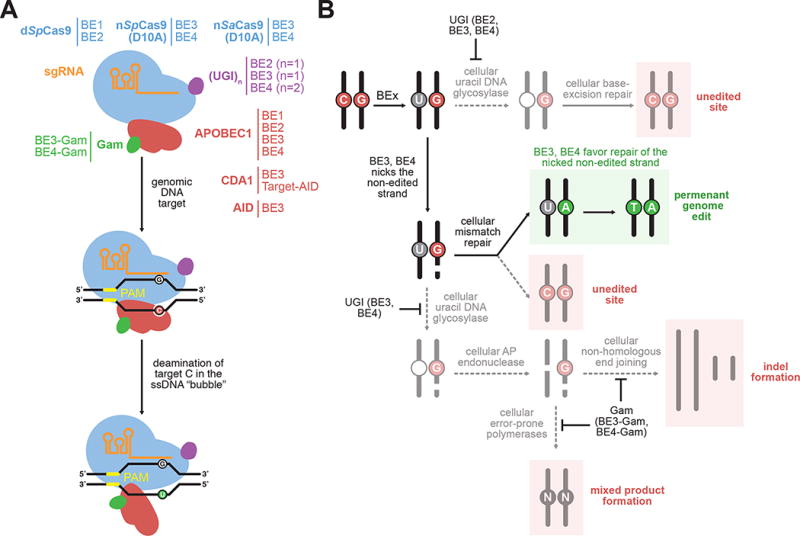
Overview of C•G-to-T•A base editing. (A) The components of C•G-to-T•A base editors. The sgRNA (orange) complexes with a Cas9 homolog (blue) to direct the base editor to a genomic locus of interest. A ssDNA-specific cytidine deaminase enzyme (red) catalytically deaminates cytidine nucleobases within the single-stranded portion of the R-loop to uracil nucleobases. The addition of one or more copies of uracil glycosylase inhibitor (UGI, purple) and the bacteriophage Mu DSB-binding protein Gam (green) help to maximize resolution of the U•G intermediate to a T•A base pair. (B) Possible outcomes following formation of the U•G base editing intermediate. The base excision repair protein uracil DNA glycosylase (UNG) can excise uracil from the U•G mismatch, resulting in an abasic site that is ultimately converted back to a C•G base pair. The UGI component of base editors (in BE2, BE3, and BE4) suppresses uracil excision. The BE3 and BE4 editors incorporate a Cas9 nickase to selectively nick the G-containing strand opposite the converted uracil. This nick guides the mismatch repair machinery to preferentially replace the G-containing strand, resulting in the desired T•A base pair. Excision of uracil from the nick-containing intermediate by UNG, followed by abasic site processing by AP endonuclease can yield a DSB. The DSB can be processed by NHEJ to produce indels, or by error-prone polymerases to yield mixed products such as G•C or A•T base pairs.
In addition to BE3, convergent strategies for genome editing have been developed by the community to address the inherent limitations of using DSBs for gene correction. Target-AID is a BE3-like base editor that uses an alternative architecture and deaminase domain, resulting in a C•G-to-T•A genome editing agent with a shifted deamination window compared to BE3.19 CRISPR-X21 and Targeted AID-induced Mutagenesis (TAM)20 are techniques that implement dCas9-cytidine deaminase fusions for targeted saturation mutagenesis of gRNA-dictated genomic regions. In the absence of any base excision repair inhibitor such as UGI, the U•G intermediate is converted to an abasic site by the base excision repair enzyme uracil DNA glycosylase, and can be further resolved by error-prone DNA repair processes to form G•C and A•T base pairs, rather than T•A products (Figure 2b).57 CRISPR-X and TAM exploit this alternate repair outcome to stochastically mutate the targeted C•G base pairs, enabling the generation of genetically encoded mutant libraries.
Despite its relatively recent addition as a genome editing tool, base editing has already enabled new applications. BE3 was used to develop CRISPR-STOP and iSTOP, general methods to generate gene knockouts through the introduction of stop codons.58, 59 As base editing largely avoids the introduction of indels, the resulting gene knockout edits are predictable and the formation of potentially cytotoxic or confounding DSB intermediates and their byproducts is avoided. A ligand-responsive base editor has also been engineered by incorporating a small-molecule activated self-cleaving element into the guide RNA used by BE3, enabling robust spatiotemporal control over base editing activity.60 In addition, BE3 variants that collectively expand the targeting scope of base editing have been developed, including variants with altered PAM requirements (increasing the number of potential genomic sites amenable to base editing by 2.5-fold),61 narrowed editing windows (enabling high-resolution discrimination of neighboring cytidines in the protospacer),61 and reduced off-target editing (producing higher confidence genome editing selectivity).54 We recently engineered a fourth generation base editor, BE4, with improved uracil glycosylase inhibition that halves the frequency with which the target C•G is converted to base pairs other than the desired T•A product (Figure 2).57 Fusion of this optimized construct to the DSB-binding protein Gam from bacteriophage Mu yielded BE4-Gam, a base editor that retains the optimized properties of BE4 while further reducing indel formation (Figure 2).57
Base editing has been used in a wide range of organisms. As the first examples of successful base editing in plants, BE3 was used to edit the OsNRT1.1B, OsSLR1, OsPDS and OsSBEIIb genes in rice, with C•G-to-T•A editing efficiencies between 12–20%.62, 63 These studies were complemented by additional work using BE3 to edit multiple genes in rice, wheat and corn with C•G-to-T•A efficiencies as high as 44%.64 Furthermore, Target-AID was used to introduce herbicide-resistant C•G-to-T•A point mutations in rice at efficiencies up to 32%, and modify genes involved in plant hormone signaling in tomato with up to 21% of progeny containing homozygous, heterozygous, or biallelic base substitutions (including various transversions and transitions).65 Recently, BE3 mRNA microinjection was shown to catalyze the introduction of C•G-to-T•A mutations in the Dmd and Tyr genes of mouse embryos with frequencies approaching 100%.55 Additionally, purified BE3:sgRNA complexes have been used for DNA-free genome editing in zebrafish embryos and live mice inner ears.54 These exciting studies illustrate the robustness of C•G-to-T•A base in a variety of organisms ranging from microbes to plants to mammals.
Future Directions
The major limitation of base editing thus far has been an inability to achieve other types of base-to-base conversions beyond C•G-to-T•A mutations. Numerous DNA-modifying enzymes, most notably methyltransferases, have been harnessed in combination with Cas9 as epigenome editing tools,66–68 yet the repertoire of naturally occurring enzymes capable of chemically modifying DNA bases in ways that alter their base pairing properties is quite limited. In contrast to the dearth of DNA-modifying catalysts, RNA is extensively post-transcriptionally modified by a variety of naturally occurring enzymes.69–71 The development of programmable RNA base editors has the potential to modulate the biological activity of RNAs with exquisite temporal resolution and thus enable novel therapeutics without the risk of permanently modifying the native genetic information.
Indeed, the double-stranded RNA adenosine deaminase editing enzyme ADAR2 (which deaminates adenosine to inosine, which is read as guanine by both translational and splicing enzymes)72 has already been repurposed for sequence-specific A-to-I RNA editing in mammalian cells.73–78 Researchers attached the catalytic domain of ADAR2 (which acts only on dsRNA) to an antisense RNA oligonucleotide using either a SNAP-tag75, 77, 78 or the λ-phage N protein-boxB RNA hairpin interaction.74, 76 Co-expression of these fusion proteins with their corresponding antisense RNA results in localization of ADAR2 to mRNA that is complementary to the antisense oligonucleotide. A-to-I editing then occurs at adenosines within the resulting double-stranded RNA region. Using a similar strategy, the wild-type ADAR2 enzyme was redirected to mRNA regions of interest using an antisense RNA oligonucleotide fused to the natural ADAR2 RNA substrate.73 The recent discovery of the RNA-guided endoribonuclease C2c2 (Cas13a)79 will likely enable additional opportunities for RNA-guided sequence-specific RNA editing. A combination of chemistry, protein engineering, directed evolution, and/or molecular biology could be used to transform these (and other) enzymes into new classes of genome editing tools for the transient, sequence-specific modification of RNAs.
Conclusion
Methods to convert one target base pair to a different base pair without requiring DSBs are promising technologies for the study and potential treatment of genetic diseases. By circumventing the introduction of cytotoxic and mutagenic DSBs, the cell cycle-dependent HDR repair pathway, and the stochastic NHEJ and related repair pathways, DSB-free genome editing methods such as base editing facilitate precise and controllable genome editing in a robust and general manner. Recently studies illustrate the potential of these approaches to address the limitations that arise from DSBs, and demonstrate the promise of applying chemical principles to the field of genome editing.
Acknowledgments
This work was supported by DARPA HR0011-17-2-0049, U.S. National Institutes of Health (NIH) R01 EB022376, R35 GM118062, and RM1 HG009490, and by the Howard Hughes Medical Institute.
Footnotes
Competing Financial Interests
A.C.K. and D.R.L. have filed provisional patent applications on base editing through Harvard University. D.R.L. is a consultant and co-founder of Editas Medicine and Beam Therapeutics, companies that are developing genome-editing therapeutics.
References
- 1.Jasin M. Genetic manipulation of genomes with rare-cutting endonucleases. Trends in Genetics. 1996;12:224–228. doi: 10.1016/0168-9525(96)10019-6. [DOI] [PubMed] [Google Scholar]
- 2.McVey M, Lee SE. MMEJ repair of double-strand breaks (director’s cut): deleted sequences and alternative endings. Trends in Genetics. 2008;24:529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jeggo PA. 5 DNA Breakage and Repair. Advances in Genetics. 1998;38:185–218. doi: 10.1016/s0065-2660(08)60144-3. [DOI] [PubMed] [Google Scholar]
- 4.Fishman-Lobell J, Rudin N, Haber JE. Two alternative pathways of double-strand break repair that are kinetically separable and independently modulated. Molecular and Cellular Biology. 1992;12:1292–1303. doi: 10.1128/mcb.12.3.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rudin N, Haber JE. Efficient repair of HO-induced chromosomal breaks in Saccharomyces cerevisiae by recombination between flanking homologous sequences. Molecular and Cellular Biology. 1988;8:3918–3928. doi: 10.1128/mcb.8.9.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang F, Doudna JA. CRISPR–Cas9 Structures and Mechanisms. Annual Review of Biophysics. 2017;46:505–529. doi: 10.1146/annurev-biophys-062215-010822. [DOI] [PubMed] [Google Scholar]
- 7.Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadan AH, Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 8.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-Guided Human Genome Engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science (New York, NY) 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Komor AC, Badran AH, Liu DR. CRISPR-Based Technologies for the Manipulation of Eukaryotic Genomes. Cell. 2017;168:20–36. doi: 10.1016/j.cell.2016.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwart D, Paquet D, Teo S, Tessier-Lavigne M. Precise and efficient scarless genome editing in stem cells using CORRECT. Nat Protocols. 2017;12:329–354. doi: 10.1038/nprot.2016.171. [DOI] [PubMed] [Google Scholar]
- 14.Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotech. 2015;33:538–542. doi: 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chu VT, Weber T, Wefers B, Wurst W, Sander S, Rajewsky K, Kuhn R. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotech. 2015;33:543–548. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- 16.Lin S, Staahl BT, Alla RK, Doudna JA. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife. 2014;3:e04766. doi: 10.7554/eLife.04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richardson CD, Ray GJ, DeWitt MA, Curie GL, Corn JE. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotech. 2016;34:339–344. doi: 10.1038/nbt.3481. [DOI] [PubMed] [Google Scholar]
- 18.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishida K, Arazoe T, Yachie N, Banno S, Kakimoto M, Tabata M, Mochizuki M, Miyabe A, Araki M, Hara KY, Shimatani Z, Kondo A. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science. 2016;353:aaf8729. doi: 10.1126/science.aaf8729. [DOI] [PubMed] [Google Scholar]
- 20.Ma Y, Zhang J, Yin W, Zhang Z, Song Y, Chang X. Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells. Nat Meth. 2016;13:1029–1035. doi: 10.1038/nmeth.4027. [DOI] [PubMed] [Google Scholar]
- 21.Hess GT, Fresard L, Han K, Lee CH, Li A, Cimprich KA, Montgomery SB, Bassik MC. Directed evolution using dCas9-targeted somatic hypermutation in mammalian cells. Nat Meth. 2016;13:1036–1042. doi: 10.1038/nmeth.4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hutchison CA, Phillips S, Edgell MH, Gillam S, Jahnke P, Smith M. Mutagenesis at a specific position in a DNA sequence. Journal of Biological Chemistry. 1978;253:6551–6560. [PubMed] [Google Scholar]
- 23.Moerschell RP, Tsunasawa S, Sherman F. Transformation of yeast with synthetic oligonucleotides. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:524–528. doi: 10.1073/pnas.85.2.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamamoto T, Moerschell RP, Wakem LP, Komar-Panicucci S, Sherman F. Strand-Specificity in the Transformation of Yeast with Synthetic Oligonucleotides. Genetics. 1992;131:811–819. doi: 10.1093/genetics/131.4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson JH, Leung WY, Bosco G, Dieu D, Haber JE. The frequency of gene targeting in yeast depends on the number of target copies. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:177–181. doi: 10.1073/pnas.91.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoon K, Cole-Strauss A, Kmiec EB. Targeted gene correction of episomal DNA in mammalian cells mediated by a chimeric RNA.DNA oligonucleotide. Proceedings of the National Academy of Sciences. 1996;93:2071–2076. doi: 10.1073/pnas.93.5.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kmiec EB, Cole A, Holloman WK. The REC2 gene encodes the homologous pairing protein of Ustilago maydis. Molecular and Cellular Biology. 1994;14:7163–7172. doi: 10.1128/mcb.14.11.7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Metz R, DiCola M, Kurihara T, Bailey A, Frank B, Roecklein B, Blaese M. Mode of Action of RNA/DNA Oligonucleotides. Chest. 2002;121:91S–97S. doi: 10.1378/chest.121.3_suppl.91s. [DOI] [PubMed] [Google Scholar]
- 29.Cole-Strauss A, Yoon K, Xiang Y, Byrne BC, Rice MC, Gryn J, Holloman WK, Kmiec EB. Correction of the Mutation Responsible for Sickle Cell Anemia by an RNA-DNA Oligonucleotide. Science. 1996;273:1386–1389. doi: 10.1126/science.273.5280.1386. [DOI] [PubMed] [Google Scholar]
- 30.Kren BT, Cole-Strauss A, Kmiec EB, Steer CJ. Targeted nucleotide exchange in the alkaline phosphatase gene of HuH-7 cells mediated by a chimeric RNA/DNA oligonucleotide. Hepatology. 1997;25:1462–1468. doi: 10.1002/hep.510250626. [DOI] [PubMed] [Google Scholar]
- 31.Alexeev V, Yoon K. Stable and inheritable changes in genotype and phenotype of albino melanocytes induced by an RNA-DNA oligonucleotide. Nat Biotech. 1998;16:1343–1346. doi: 10.1038/4322. [DOI] [PubMed] [Google Scholar]
- 32.Alexeev V, Igoucheva O, Domashenko A, Cotsarelis G, Yoon K. Localized in vivo genotypic and phenotypic correction of the albino mutation in skin by RNA-DNA oligonucleotide. Nat Biotech. 2000;18:43–47. doi: 10.1038/71901. [DOI] [PubMed] [Google Scholar]
- 33.Bartlett RJ, Stockinger S, Denis MM, Bartlett WT, Inverardi L, Le TT, Man Nt, Morris GE, Bogan DJ, Metcalf-Bogan J, Kornegay JN. In vivo targeted repair of a point mutation in the canine dystrophin gene by a chimeric RNA/DNA oligonucleotide. Nat Biotech. 2000;18:615–622. doi: 10.1038/76448. [DOI] [PubMed] [Google Scholar]
- 34.Igoucheva O, Alexeev V, Pryce M, Yoon K. Transcription affects formation and processing of intermediates in oligonucleotide-mediated gene alteration. Nucleic Acids Research. 2003;31:2659–2670. doi: 10.1093/nar/gkg360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu L, Rice MC, Drury M, Cheng S, Gamper H, Kmiec EB. Strand Bias in Targeted Gene Repair Is Influenced by Transcriptional Activity. Molecular and Cellular Biology. 2002;22:3852–3863. doi: 10.1128/MCB.22.11.3852-3863.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brachman EE, Kmiec EB. DNA replication and transcription direct a DNA strand bias in the process of targeted gene repair in mammalian cells. Journal of Cell Science. 2004;117:3867. doi: 10.1242/jcs.01250. [DOI] [PubMed] [Google Scholar]
- 37.Olga I, Vitali A, Kyonggeun Y. Oligonucleotide-Directed Mutagenesis and Targeted Gene Correction: A Mechanistic Point of View. Current Molecular Medicine. 2004;4:445–463. doi: 10.2174/1566524043360465. [DOI] [PubMed] [Google Scholar]
- 38.van der Steege G, Schuilenga-Hut PHL, Buys CHCM, Scheffer H, Pas HH, Jonkman MF. Persistent failures in gene repair. Nat Biotech. 2001;19:305–306. doi: 10.1038/86664. [DOI] [PubMed] [Google Scholar]
- 39.Beetham PR, Kipp PB, Sawycky XL, Arntzen CJ, May GD. A tool for functional plant genomics: Chimeric RNA/DNA oligonucleotides cause in vivo gene-specific mutations. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:8774–8778. doi: 10.1073/pnas.96.15.8774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kochevenko A, Willmitzer L. Chimeric RNA/DNA Oligonucleotide-Based Site-Specific Modification of the Tobacco Acetolactate Syntase Gene. Plant Physiology. 2003;132:174–184. doi: 10.1104/pp.102.016857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu T, Peterson DJ, Tagliani L, St Clair G, Baszczynski CL, Bowen B. Targeted manipulation of maize genes in vivo using chimeric RNA/DNA oligonucleotides. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:8768–8773. doi: 10.1073/pnas.96.15.8768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu T, Mettenburg K, Peterson DJ, Tagliani L, Baszczynski CL. Engineering herbicide-resistant maize using chimeric RNA/DNA oligonucleotides. Nat Biotech. 2000;18:555–558. doi: 10.1038/75435. [DOI] [PubMed] [Google Scholar]
- 43.Okuzaki A, Toriyama K. Chimeric RNA/DNA oligonucleotide-directed gene targeting in rice. Plant Cell Reports. 2004;22:509–512. doi: 10.1007/s00299-003-0698-2. [DOI] [PubMed] [Google Scholar]
- 44.Dong C, Beetham P, Vincent K, Sharp P. Oligonucleotide-directed gene repair in wheat using a transient plasmid gene repair assay system. Plant Cell Reports. 2006;25:457–465. doi: 10.1007/s00299-005-0098-x. [DOI] [PubMed] [Google Scholar]
- 45.Ruiter R, Van Den Brande I, Stals E, Delauré S, Cornelissen M, D’Halluin K. Spontaneous mutation frequency in plants obscures the effect of chimeraplasty. Plant Molecular Biology. 2003;53:715–729. doi: 10.1023/B:PLAN.0000019111.96107.01. [DOI] [PubMed] [Google Scholar]
- 46.Lombardo L, Coppola G, Zelasco S. New Technologies for Insect-Resistant and Herbicide-Tolerant Plants. Trends in Biotechnology. 2016;34:49–57. doi: 10.1016/j.tibtech.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 47.Storici F, Lewis LK, Resnick MA. In vivo site-directed mutagenesis using oligonucleotides. Nat Biotech. 2001;19:773–776. doi: 10.1038/90837. [DOI] [PubMed] [Google Scholar]
- 48.Storici F, Resnick MA. Methods in Enzymology. Academic Press; 2006. The Delitto Perfetto Approach to In Vivo Site‐Directed Mutagenesis and Chromosome Rearrangements with Synthetic Oligonucleotides in Yeast; pp. 329–345. [DOI] [PubMed] [Google Scholar]
- 49.Ruff P, Koh KD, Keskin H, Pai RB, Storici F. Aptamer-guided gene targeting in yeast and human cells. Nucleic Acids Research. 2014;42:e61–e61. doi: 10.1093/nar/gku101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Katz SS, Gimble FS, Storici F. To Nick or Not to Nick: Comparison of I-SceI Single- and Double-Strand Break-Induced Recombination in Yeast and Human Cells. PLOS ONE. 2014;9:e88840. doi: 10.1371/journal.pone.0088840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang F, Taylor DW, Chen JS, Kornfeld JE, Zhou K, Thompson AJ, Nogales E, Doudna JA. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science (New York, NY) 2016;351:867–871. doi: 10.1126/science.aad8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18:85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- 53.Kim D, Lim K, Kim ST, Yoon S-h, Kim K, Ryu SM, Kim JS. Genome-wide target specificities of CRISPR RNA-guided programmable deaminases. Nat Biotech. 2017;35:475–480. doi: 10.1038/nbt.3852. [DOI] [PubMed] [Google Scholar]
- 54.Rees HA, Komor AC, Yeh WH, Caetano-Lopes J, Warman M, Edge ASB, Liu DR. Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery. Nat Commun. 2017;8:15790. doi: 10.1038/ncomms15790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim K, Ryu SM, Kim ST, Baek G, Kim D, Lim K, Chung E, Kim S, Kim JS. Highly efficient RNA-guided base editing in mouse embryos. Nat Biotech. 2017;35:435–437. doi: 10.1038/nbt.3816. [DOI] [PubMed] [Google Scholar]
- 56.Zhang Y, Qin W, Lu X, Xu J, Huang H, Bai H, Li S, Lin S. Programmable base editing of zebrafish genome using a modified CRISPR-Cas9 system. Nature Communications. 2017;8:118. doi: 10.1038/s41467-017-00175-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Komor AC, Zhao KT, Packer MS, Gaudelli NM, Waterbury AL, Koblan LW, Kim YB, Badran AH, Liu DR. Improved Base Excision Repair Inhibition and Bacteriophage Mu Gam Protein Yields C:G-to-T:A Base Editors with Higher Efficiency and Product Purity. Science Advances. 2017;3:eaao4774. doi: 10.1126/sciadv.aao4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuscu C, Parlak M, Tufan T, Yang J, Szlachta K, Wei X, Mammadov R, Adli M. CRISPR-STOP: gene silencing through base-editing-induced nonsense mutations. Nat Meth. 2017;14:710–712. doi: 10.1038/nmeth.4327. [DOI] [PubMed] [Google Scholar]
- 59.Billon P, Bryant EE, Joseph SA, Nambiar TS, Hayward SB, Rothstein R, Ciccia A. CRISPR-Mediated Base Editing Enables Efficient Disruption of Eukaryotic Genes Through Induction of Stop Codons (iSTOP) Molecular Cell. 2017;67:1–12. doi: 10.1016/j.molcel.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tang W, Hu JH, Liu DR. Aptazyme-embedded guide RNAs enable ligand-responsive genome editing and transcriptional activation. 2017;8:15939. doi: 10.1038/ncomms15939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim YB, Komor AC, Levy JM, Packer MS, Zhao KT, Liu DR. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat Biotech. 2017;35:371–376. doi: 10.1038/nbt.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lu Y, Zhu JK. Precise Editing of a Target Base in the Rice Genome Using a Modified CRISPR/Cas9 System. Mol Plant. 2017;10:523–525. doi: 10.1016/j.molp.2016.11.013. [DOI] [PubMed] [Google Scholar]
- 63.Li J, Sun Y, Du J, Zhao Y, Xia L. Generation of Targeted Point Mutations in Rice by a Modified CRISPR/Cas9 System. Mol Plant. 2017;10:526–529. doi: 10.1016/j.molp.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 64.Zong Y, Wang Y, Li C, Zhang R, Chen K, Ran Y, Qiu JL, Wang D, Gao C. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat Biotech. 2017;35:438–440. doi: 10.1038/nbt.3811. [DOI] [PubMed] [Google Scholar]
- 65.Shimatani Z, Kashojiya S, Takayama M, Terada R, Arazoe T, Ishii H, Teramura H, Yamamoto T, Komatsu H, Miura K, Ezura H, Nishida K, Ariizumi T, Kondo A. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat Biotech. 2017;35:441–443. doi: 10.1038/nbt.3833. [DOI] [PubMed] [Google Scholar]
- 66.Vojta A, Dobrinić P, Tadić V, Bočkor L, Korać P, Julg B, Klasić M, Zoldoš V. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Research. 2016;44:5615–5628. doi: 10.1093/nar/gkw159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McDonald JI, Celik H, Rois LE, Fishberger G, Fowler T, Rees R, Kramer A, Martens A, Edwards JR, Challen GA. Reprogrammable CRISPR/Cas9-based system for inducing site-specific DNA methylation. Biology Open. 2016;5:866–874. doi: 10.1242/bio.019067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S, Shu J, Dadon D, Young Richard A, Jaenisch R. Editing DNA Methylation in the Mammalian Genome. Cell. 2016;167:233–247.e217. doi: 10.1016/j.cell.2016.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sun WJ, Li JH, Liu S, Wu J, Zhou H, Qu LH, Yang JH. RMBase: a resource for decoding the landscape of RNA modifications from high-throughput sequencing data. Nucleic Acids Research. 2016;44:D259–D265. doi: 10.1093/nar/gkv1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM, Helm M, Bujnicki JM, Grosjean H. MODOMICS: a database of RNA modification pathways—2013 update. Nucleic Acids Research. 2013;41:D262–D267. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, Vendeix FAP, Fabris D, Agris PF. The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Research. 2011;39:D195–D201. doi: 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Alseth I, Dalhus B, Bjørås M. Inosine in DNA and RNA. Current Opinion in Genetics & Development. 2014;26:116–123. doi: 10.1016/j.gde.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 73.Fukuda M, Umeno H, Nose K, Nishitarumizu A, Noguchi R, Nakagawa H. Construction of a guide-RNA for site-directed RNA mutagenesis utilising intracellular A-to-I RNA editing. 2017;7:41478. doi: 10.1038/srep41478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Montiel-Gonzalez MF, Vallecillo-Viejo I, Yudowski GA, Rosenthal JJC. Correction of mutations within the cystic fibrosis transmembrane conductance regulator by site-directed RNA editing. Proceedings of the National Academy of Sciences. 2013;110:18285–18290. doi: 10.1073/pnas.1306243110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stafforst T, Schneider MF. An RNA–Deaminase Conjugate Selectively Repairs Point Mutations. Angewandte Chemie International Edition. 2012;51:11166–11169. doi: 10.1002/anie.201206489. [DOI] [PubMed] [Google Scholar]
- 76.Montiel-González MF, Vallecillo-Viejo IC, Rosenthal Joshua JC. An efficient system for selectively altering genetic information within mRNAs. Nucleic Acids Research. 2016;44:e157–e157. doi: 10.1093/nar/gkw738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schneider MF, Wettengel J, Hoffmann PC, Stafforst T. Optimal guideRNAs for re-directing deaminase activity of hADAR1 and hADAR2 in trans. Nucleic Acids Research. 2014;42:e87–e87. doi: 10.1093/nar/gku272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vogel P, Schneider MF, Wettengel J, Stafforst T. Improving Site-Directed RNA Editing In Vitro and in Cell Culture by Chemical Modification of the GuideRNA. Angewandte Chemie International Edition. 2014;53:6267–6271. doi: 10.1002/anie.201402634. [DOI] [PubMed] [Google Scholar]
- 79.Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DBT, Shmakov S, Makarova KS, Semenova E, Minakhin L, Severinov K, Regev A, Lander ES, Koonin EV, Zhang F. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016 doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]


