Abstract
Studies on oocyte‐specific genes are important in understanding the genetic pathways essential for folliculogenesis, oogenesis and early embryogenesis. Although the molecular mechanisms regulating oocyte growth and embryo development in mammals have partially been unraveled by gene knockout studies, many aspects concerning reproduction remain to be determined. Development of mammalian embryos starts with the fusion of sperm and egg. After fertilization, the first major developmental transition, maternal to zygotic transition, occurs at the specific stages of preimplantation development in each mammal. The transition is called zygotic gene activation (ZGA) or embryonic genome activation. The ZGA is one of the most important events that occur during preimplantation development; however, the mechanism of the event remains unknown. Because the development until the transition is maintained by maternally inherited proteins and transcripts stored in the oocytes, it is highly likely that these products play an important role in the initiation of ZGA. Several maternal‐effects genes that are specifically expressed in oocytes have been identified and their involvement in preimplantation development has been revealed. Therefore, to study oocyte‐specific gene regulation would help not only to understand the precise mechanisms of mammalian development, but also to show the mechanisms of reproductive disorders, such as premature ovarian failure and infertility. (Reprod Med Biol 2006; 5: 175–182)
Keywords: embryo development, maternal‐effect gene, oocyte‐specific gene, oog1, oogenesis
INTRODUCTION
DEVELOPMENT OF MAMMALIAN embryos has been studied for more than 50 years. 1 The culture of mammalian embryos has been intensively examined in mice. The studies have lead to the successful in vitro culture of embryos, however, the precise mechanism of mammalian development has not yet been elucidated.
The development of one‐cell mouse embryos, except for embryos of some inbred strains and their F1 hybrids, is blocked at the two‐cell stage, a phenomenon that has been termed ‘the two‐cell block’. 2 , 3 , 4 Cross‐breeding experiments have shown that maternally inherited developmental information plays an important role in controlling early cleavage of the mouse embryo. 3 In addition, the transfer of cytoplasm from non‐blocked embryos into blocked embryos recovers the developmental competence of two‐cell embryos in vitro. 5 These results suggest that the gene products such as mRNAs and/or proteins stored in oocytes play important roles in the development of embryos. The developmental arrest in vitro can be overcome by modifying the culture conditions; the addition of ethylenediaminetetracetic acid (EDTA) 6 , 7 and deletion of phosphate 8 can eliminate the developmental arrest in vitro. It has also been shown that isolated mouse ampulla maintained in organ culture can overcome the two‐cell block in the mouse 9 and hamster. 10 These observations indicate that the maternal factors involved in the embryonic development are closely associated with developmental environment. Therefore, to study the gene functions in oocytes provides information about the relationship between maternal factors and embryonic development in mammals.
In the present review, the role of oocyte‐specific genes in early embryogenesis is discussed.
ZYGOTIC GENE ACTIVATION AND EMBRYO DEVELOPMENT IN MAMMALS
THE MATERNAL TO zygotic transition is the first major transition that occurs after fertilization. This developmental program is initially directed by maternally inherited proteins and transcripts, and the transcripts are mostly replaced by newly synthesized ones. In mammals, zygotic gene activation (ZGA) has been shown to be a two‐step process consisting of minor and major phases. 11 In the mouse, the minor ZGA phase is initiated at the late one‐cell stage (G2 phase) with very weak transcriptional activity. 12 , 13 , 14 , 15 , 16 , 17 , 18 Consequently, some of the proteins are synthesized at the early two‐cell stage (G1/S) for the next phase of ZGA, the major phase. 19 , 20 , 21 , 22 , 23 , 24 It has been reported that reporter genes microinjected into the pronuclei of one‐cell mouse embryos are transcribed during the minor ZGA phase. 17 , 25 , 26 , 27 , 28 Because the translation of maternal RNA is required for the initiation of ZGA, the proteins stored in oocytes are utilized in the early event of transcription at the late one‐cell stage. 29 The regulation of the initiation of the transcription and the following first mitotic events are mainly controlled by maternally inherited products. At the G2 phase of the second cell cycle, the major ZGA phase, which is characterized by an increase of transcriptional and translational activity, occurs and results in a dramatic change in the pattern of protein synthesis. 11 , 19 , 27 , 30 , 31 , 32 In many mammalian species, development of one‐cell embryos is blocked at various early stages in vitro. It has been reported that the time of developmental arrest in vitro coincides with the time of ZGA, 33 suggesting that there might be a relationship between the developmental arrest in culture and transcriptional activity of embryos.
ROLE OF OOCYTE‐SPECIFIC GENES
THE GENE PRODUCTS expressed specifically in oocytes play important roles in folliculogenesis, fertilization and preimplantation development (Fig. 1). 34 One of the most exciting molecules expressed in oocytes is a member of the transforming growth factor β (TGF‐β) superfamily, growth differentiation factor 9 (Gdf‐9), which is obligatory for proper folliculogenesis beyond the primary stage and fertility in female mice. 35 , 36 , 37 , 38 Another oocyte‐specific member of TGF‐β superfamily is bone morphogenetic protein 15 (Bmp15), which is a single copy gene on the X chromosome in mammals. 39 Ovarian follicles in sheep homozygous Bmp15 mutations do not normally grow beyond the primary follicle stage. 40 , 41 Bmp15 null mice are phenotypically different from sheep and have minimal ovarian histopathological defects and smaller litter sizes than wild type mice. 42 Genes encoding several other growth factors are also expressed in mammalian oocytes: Bmp6, 43 Tgf‐β2 44 and fibroblast growth factor 8 (Fgf8). 45 Recent studies have revealed key roles of the oocyte in folliculogenesis and have shown that bidirectional communication between the oocyte and somatic cells is essential for development of an egg so it can undergo fertilization and embryogenesis. 46 , 47 Although these growth factors and other unknown factors are known to be involved in folliculogenesis, many of their specific functions are not well characterized.
Figure 1.
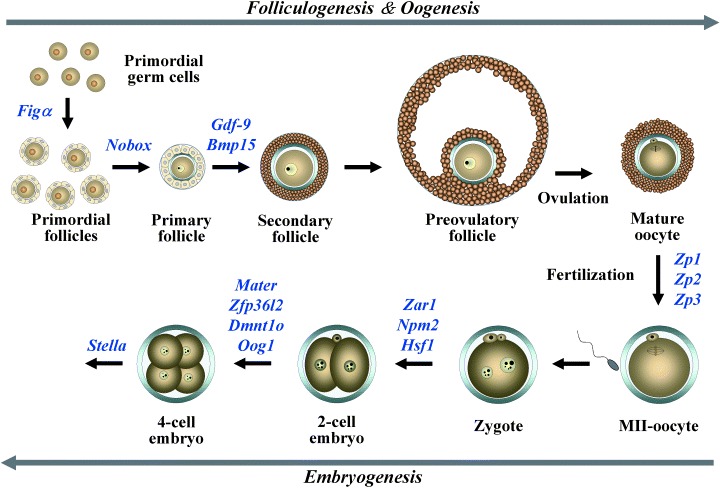
Summary of the expression of oocyte‐specific genes involved in folliculogenesis, oogenesis and early embryogenesis.
A factor in the germline, alpha (FIGα) is a basic helix‐loop‐helix (bHLH) transcription factor first detected in oocytes at 13.5 dpc. Female mice lacking Figα are unable to form primordial follicles which results in massive depletion of oocytes and sterility. 48 Figα has also been implicated in the coordinate expression of the three zona pellucida genes (Zp1, Zp2, Zp3) that encode the mouse egg coat. 49 , 50
MATERNAL‐EFFECT GENES IN EARLY DEVELOPMENT
DURING OOCYTE GROWTH and follicular development oocytes accumulate maternal‐effect factors necessary for early embryogenesis, which occurs in the absence of de novo transcription of either parental genome (Fig. 1). 27 Maternal‐effect genes, which are well documented in lower species such as Drosophila melanogaster and Xenopus laevis, encode transcripts or proteins in the egg during oogenesis that play pivotal roles after fertilization. 51 , 52 As described above, ZGA occurs at the 1‐ to two‐cell stages and is a critical event that is indispensable for further embryonic development in mice. 25 It is speculated that several hundred genes participate in the activation, 53 indicating that some of the maternal‐effect genes might be involved in the ZGA. However, relatively few maternal‐effect genes have been identified in mammals. Until now, eight maternal‐effect genes, such as Mater, Hsf1, Dnmt1o, Pms2, Zar1, Npm2, stella and Zfp36I2 have been identified in mice, and knockout models of these genes have shown that many of the maternal‐effect genes are involved in the early embryogenesis, especially at the 1‐ to two‐cell stages. 54 , 55 , 56 , 57 , 58 , 59 , 60 , 61
The maternal antigen that embryo required (MATER) was first identified as an ooplasm‐specific protein encoded by a single‐copy gene that is transcribed in growing oocytes. 62 Homozygous null Mater males and heterozygous females have normal fertility, although homozygous females are sterile. Although folliculogenesis, ovulation, fertilization and the first cleavage appear normal, early embryos lacking MATER are unable to progress beyond the two‐cell stage. 60 Heat‐shock factor‐1 (Hsf1) was also identified as a maternal‐effect gene. Embryos lacking HSF1 are blocked mainly at the one‐cell stage and show ultrastructural abnormality in the nuclei at the two‐cell stage. 55 Dnmt1o, an oocyte‐specific DNA methyltransferase, maintains genomic methylation during preimplantation development. 57 Although DNMT1o accumulates in nuclei of early growing oocytes, but is sequestered in the cytoplasm of mature oocytes, 63 it is required for the maintenance of the methylation pattern specifically at the 8‐cell stage. 57 Pms2 has also been shown to act as a maternal‐effect gene which functions in DNA mismatch repair. 56 Recently, oocyte‐specific gene, Zar1 (zygote arrest 1) and Npm2 (nucleoplasmin 2) have been identified using subtractive hybridization. 54 , 64 Homozygous null Zar1 female are sterile because the embryos from the female are arrested at the 1‐ to two‐cell stage. ZAR1 is detected after resumption of meiosis, it persists in one‐cell embryos and rapidly disappears at the two‐cell stage, suggesting a critical role in the oocyte‐to‐embryo transition. 61 Npm2 knockout females have fertility defects because of reduced cleavage to the two‐cell stage. In Npm2 null oocytes and zygotes, absence of coalesced nucleolar structures and loss of heterochromatin and deacetylated histone H3 are observed, suggesting that Npm2 is critical for nuclear and nucleolar organization and embryonic development. 54 Stella is a germ cell‐specific maternal‐effect gene and embryos without STELLA are compromised in preimplantation development and rarely reach the blastocyst stage. 58 The effects of lack of STELLA become evident shortly after fertilization, with progressively fewer embryos exhibiting normal development during preimplantation stages. A SAP‐like domain and a splicing factor motif‐like structure of Stella suggest possible roles in chromosomal organization or RNA processing. Zinc finger protein 36 like 2 (Zfp36l2) is also reported to be one of the maternal‐effect genes. 59 Zfp36l2 null females apparently cycle and ovulate normally, and their ova can be fertilized; however, the embryos do not progress beyond the two‐cell stage of development. ZFP36l2 belongs to an unusual family of zinc finger proteins containing tandem zinc‐binding motifs characterized by three cysteines followed by one histidine (CCCH). Through this zinc finger, the protein can bind to mRNA containing class II AU‐rich elements; binding is then followed by degradation of the target mRNA. 65
FUNCTION OF GERM CELL‐SPECIFIC GENES IN GAMETOGENESIS
RECENTLY, NOVEL GENES that are specifically expressed in the ovary and testis have been reported. Gasz is one of the newly identified germ cell‐specific genes, encoding a protein containing four ankyrin repeats (ANK), a sterile‐alpha motif (SAM) and a basic leucine zipper (bZIP) domain. 66 Mouse Gasz is shown to be expressed in oocytes at all stages of oogenesis, pachytene spermatocytes, round spermatids and preimplantation embryos at the mRNA and protein revels. Therefore, it is likely that GASZ functions as a cytoplasmic signaling molecule in germ cells, because the protein has a motif that is important for protein–protein interaction. Furthermore, Gasz orthologs are present in rats, cows, baboons, chimpanzees and humans, indicating that the gene has an evolutionally conserved function in germ cell. 67
Newborn ovary homeobox‐encoding gene (NOBOX) is a transcription factor containing a homeobox domain. 68 Nobox is expressed in germ cell cysts, and in primordial and growing oocytes during folliculogenesis (Fig. 1). Lack of NOBOX accelerates postnatal oocyte loss and abolishes the transition from primordial to growing follicles in mice. 69 Nobox −/– mice also show a downregulation of genes preferentially expressed in the oocyte including Oct4, Mos, Rfpl4, Fgf8, Zar1, Dnmt1o, Bmp15, H1oo and Gdf9, whereas ubiquitous genes such as Bmp4, Kit and Bax are unaffected. Thus, NOBOX might have a direct role in the regulation of the oocyte‐specific gene expression during folliculogenesis, oogenesis and early embryogenesis.
ROLE OF AN OOCYTE‐SPECIFIC GENE, OOG1 DURING MOUSE PREIMPLANTATION DEVELOPMENT
WE PREVIOUSLY IDENTIFIED an oocyte‐specific novel gene, Oogenesin (Oog1), that encodes 326 amino acids containing a leucine zipper and a leucine rich repeat which appears to be necessary for protein–protein interactions. 70 , 71 More interestingly, OOG1 localized in the nuclei at late one‐cell and early two‐cell stages, the time when the zygotic genome activation occurs in mice. The zygotic genome activation relies on transcripts and proteins stored in the oocyte during oogenesis. However, the molecular mechanisms governing these events are largely unknown. Recently, another group identified three additional Oog1‐like genes (Oog2, 3, 4) containing a leucine rich repeat, speculating that this family functions by mediating protein–protein interactions. 72 To identify the interacting proteins of OOG1, we carried out a yeast two‐hybrid screening using a GV oocyte cDNA library and found that RAL guanine nucleotide dissociation stimulator (RALGDS) is the potential binding partner of OOG1. 73 RALGDS is one of the Ras effector proteins, exchanging a GDP‐bound inactive state RAL to a GTP‐bound active state RAL in a RAS‐dependent manner. 74 , 75 We also showed that RAS‐binding domain (RBD) of RALGDS is indispensable for the interaction with OOG1 and that OOG1 interact with activated RAS. It has been reported that the leucine‐rich repeats binds to the GTP‐binding motif of G‐proteins. 76 It is probable that leucine‐rich repeats of OOG1, GTP‐binding motif of RAS and RBD of RALGDS play an important role in the interaction of these proteins. RALGDS transcript is detected in GV oocytes and preimplantation embryos until the end of the four‐cell stage and the protein is localized in the cytoplasm in oocytes and preimplantation embryos. Interestingly, the protein appears in the nucleus rather than the cytoplasm between late one‐cell and late two‐cell stages, suggesting that RALGDS‐OOG1 complex is formed after the activation of RAS and functions in the nucleus of the one‐ to two‐cell stage embryos. 73 In a colocalization experiment, it was shown that OOG1 expression is necessary for the nuclear localization of RALGDS in transfected HeLa cells. Because the expression profiles and localization of OOG1 and RALGDS are quite similar in the late one‐cell and early two‐cell stage embryos, 71 , 73 the interaction between OOG1 and RALGDS probably occurs in mouse embryos.
A NEW INSIGHT INTO GENE FUNCTION USING BIOFORMATICS
THE CURRENT DATABASES provide many kinds of biological information, including gene expression, gene mapping, DNA and protein sequences, and protein structure and function, for example: National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/), European Molecular Biology Laboratory (EMBL; http://www.ebi.ac.uk/embl/) and DNA Data Bank of Japan (DDBJ; http://www.ddbj.nig.ac.jp/Welcome-j.html). Furthermore, programs designed to search these databases, such as BLAST (http://www.ncbi.nlm.nih.gov/blast), are helpful for scientists using these large data sets. Therefore, the combined use of molecular genetics and bioinformatics is a powerful approach to investigate the function of the gene of interest.
Recently, it has been reported by the in silico approach that some of the oocyte‐specific genes, including Oogenesin and Mater (also known as Nalp5), are organized in clusters to map near the chromosome ends. 72 , 77 , 78 Although most oocyte‐specific genes organized in clusters are paralogous genes, they seem to have individual biological roles, that is, Nalp9A‐F that are clustered at vicinity regions of Nalp5 are not able to compensate the absence of Mater product in Mater (Nalp5)−/– mice. 60 , 77 Interestingly, using the Genome Browser of UCSC Genome Bioinformatics (http://genome.ucsc.edu/cgi-bin/hgGateway), we found that the MATER and OOG1 share a similar tertiary structure (Fig. 2). Because a MATER lacking‐embryo is unable to progress beyond the two‐cell stage, 73 OOG1 might have the same function as well as MATER during early embryogenesis.
Figure 2.
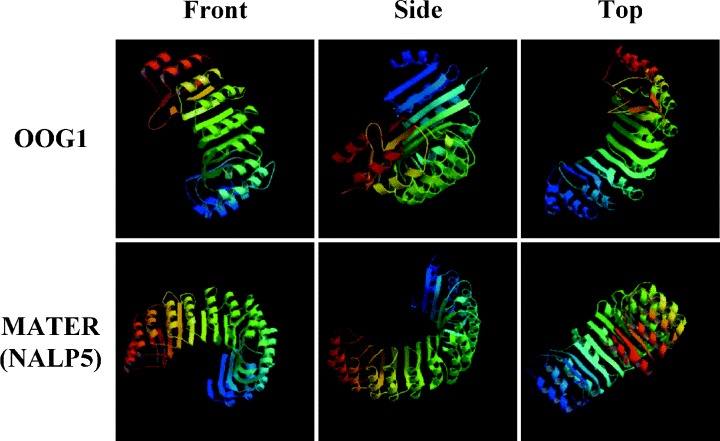
3D structures of OOG1 and MATER/NALP5. The structures of OOG1 and MATER were predicted by Modbase using the March 2005 mouse (Mus musculus) draft genome data and the August 2005 mouse draft genome data, respectively.
CONCLUSIONS
IDENTIFICATION AND CHARACTERIZATION of genes preferentially expressed in oocytes would be extremely useful in unraveling their oocyte specific functions in oogenesis, folliculogenesis, fertilization and early embryogenesis. Therefore, understanding the biological functions of oocyte‐specific genes using genetics, genomics, proteomics and bioinformatics would help to accelerate the elucidation of the mechanisms involved in mammalian development.
ACKNOWLEDGMENTS
THE PRESENT WORK was supported in part by a Grants‐in‐Aid for Scientific Research (No. 16380187) from the Japan Society for the Promotion of Science and a grant from The INAMORI foundation to N. M.
REFERENCES
- 1. Hammond J. Recovery and culture of tubal mouse ova. Nature 1949; 163: 28–29. [DOI] [PubMed] [Google Scholar]
- 2. Kaufman MH, Sachs L. Complete preimplantation development in culture of parthenogenetic mouse embryos. J Embryol Exp Morph 1976; 35: 179–190. [PubMed] [Google Scholar]
- 3. Goddard MJ, Pratt HP. Control of events during early cleavage of the mouse embryo: an analysis of the ‘two‐cell block’. J Embryol Exp Morph 1983; 73: 111–133. [PubMed] [Google Scholar]
- 4. Whitten WK, Biggers JD. Complete development in vitro of the pre‐implantation stages of the mouse in a simple chemically defined medium. J Reprod Fertil 1968; 17: 399–401. [DOI] [PubMed] [Google Scholar]
- 5. Muggleton‐Harris A, Whittingham DG, Wilson L. Cytoplasmic control of preimplantation development in vitro in the mouse. Nature 1982; 299: 460–462. [DOI] [PubMed] [Google Scholar]
- 6. Abramczuk J, Solter D, Koprowski H. The beneficial effect EDTA on development of mouse one‐cell embryos in chemically defined medium. Dev Biol 1977; 61: 378–383. [DOI] [PubMed] [Google Scholar]
- 7. Chatot CL, Ziomek CA, Bavister BD, Lewis JL, Torres I. An improved culture medium supports development of random‐bred one‐cell mouse embryos in vitro. J Reprod Fertil 1989; 86: 679–688. [DOI] [PubMed] [Google Scholar]
- 8. Haraguchi S, Naito K, Sato E. Phosphate exposure during the late one‐cell and early two‐cell stages induces a time‐specific decrease in cyclin B and cdc25B mRNAs in AKR/N mouse embryos in vitro. Zygote 1999; 7: 87–93. [DOI] [PubMed] [Google Scholar]
- 9. Biggers JD, Gwatkin RLB, Brinster RL. Development of mouse embryos in organ culture of fallopian tubes on a chemically defined medium. Nature 1962; 194: 747–749. [DOI] [PubMed] [Google Scholar]
- 10. Minami N, Bavister BD, Iritani A. Development of hamster two‐cell embryos in the isolated mouse oviduct in organ culture system. Gamete Res 1988; 19: 235–240. [DOI] [PubMed] [Google Scholar]
- 11. Bellier S, Chastant S, Adenot P, Vincent M, Renard JP, Bensaude O. Nuclear translocation and carboxyl‐terminal domain phosphorylation of RNA polymerase II delineate the two phases of zygotic gene activation in mammalian embryos. EMBO J 1997; 16: 6250–6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Latham KE, Solter D, Schultz RM. Acquisition of a transcriptionally permissive state during the one‐cell stage of mouse embryogenesis. Dev Biol 1992; 149: 457–462. [DOI] [PubMed] [Google Scholar]
- 13. Matsumoto K, Anzai M, Nakagata N, Takahashi A, Takahashi Y, Miyata K. Onset of paternal gene activation in early mouse embryos fertilized with transgenic mouse sperm. Mol Reprod Dev 1994; 39: 136–140. [DOI] [PubMed] [Google Scholar]
- 14. Ram PT, Schultz RM. Reporter gene expression in G2 of the one‐cell mouse embryo. Dev Biol 1993; 156: 552–556. [DOI] [PubMed] [Google Scholar]
- 15. Temeles GL, Ram PT, Rothstein JL, Schultz RM. Expression patterns of novel genes during mouse preimplantation embryogenesis. Mol Reprod Dev 1994; 37: 121–129. [DOI] [PubMed] [Google Scholar]
- 16. Bouniol C, Nguyen E, Debey P. Endogenous transcription occurs at the one‐cell stage in the mouse embryo. Exp Cell Res 1995; 218: 57–62. [DOI] [PubMed] [Google Scholar]
- 17. Christians E, Campion E, Thompson EM, Renard JP. Expression of the HSP 70.1 gene, a landmark of early zygotic activity in the mouse embryo, is restricted to the first burst of transcription. Development 1995; 121: 113–122. [DOI] [PubMed] [Google Scholar]
- 18. Aoki F, Worrad DM, Schultz RM. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev Biol 1997; 181: 296–307. [DOI] [PubMed] [Google Scholar]
- 19. Flach G, Johnson MH, Braude PR, Taylor RA, Bolton VN. The transition from maternal to embryonic control in the two‐cell mouse embryo. EMBO J 1982; 1: 681–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Latham KE, Solter D, Schultz RM. Activation of a two‐cell stage‐specific gene following transfer of heterologous nuclei into enucleated mouse embryos. Mol Reprod Dev 1991; 30: 182–186. [DOI] [PubMed] [Google Scholar]
- 21. Bensaude O, Babinet C, Morange M, Jacob F. Heat shock proteins, first major products of zygotic gene activity in mouse embryo. Nature 1983; 305: 331–333. [DOI] [PubMed] [Google Scholar]
- 22. Conover JC, Temeles GL, Zimmermann JW, Burke B, Schultz RM. Stage‐specific expression of a family of proteins that are major products of zygotic gene activation in the mouse embryo. Dev Biol 1991; 144: 392–404. [DOI] [PubMed] [Google Scholar]
- 23. Davis WJ, De Sousa PA, Schultz RM. Transient expression of translation initiation factor eIF‐4C during the two‐cell stage of the preimplantation mouse embryo: identification by mRNA differential display and the role of DNA replication in zygotic gene activation. Dev Biol 1996; 174: 190–201. [DOI] [PubMed] [Google Scholar]
- 24. Latham KE, Rambhatla L, Hayashizaki Y, Chapman VM. Stage‐specific induction and regulation by genomic imprinting of the mouse U2afbp‐rs gene during preimplantation development. Dev Biol 1995; 168: 670–676. [DOI] [PubMed] [Google Scholar]
- 25. Schultz RM. Regulation of zygotic gene activation in the mouse. Bioessays 1993; 15: 531–538. [DOI] [PubMed] [Google Scholar]
- 26. Majumder S, DePamphilis ML. A unique role for enhancers is revealed during early mouse development. Bioessays 1995; 17: 879–889. [DOI] [PubMed] [Google Scholar]
- 27. Nothias JY, Majumder S, Kaneko KJ, DePamphilis ML. Regulation of gene expression at the beginning of mammalian development. J Biol Chem 1995; 270: 22077–22080. [DOI] [PubMed] [Google Scholar]
- 28. Henery CC, Miranda M, Wiekowski M, Wilmut I, DePamphilis ML. Repression of gene expression at the beginning of mouse development. Dev Biol 1995; 169: 448–460. [DOI] [PubMed] [Google Scholar]
- 29. Hamatani T, Carter MG, Sharov AA, Ko MS. Dynamics of global gene expression changes during mouse preimplantation development. Dev Cell 2004; 6: 117–131. [DOI] [PubMed] [Google Scholar]
- 30. Howlett SK, Webb M, Maro B, Johnson MH. Meiosis II, mitosis I and the linking interphase: a study of the cytoskeleton in the fertilised mouse egg. Cytobios 1985; 43: 295–305. [PubMed] [Google Scholar]
- 31. Taylor KD, Piko L. Patterns of mRNA prevalence and expression of B1 and B2 transcripts in early mouse embryos. Development 1987; 101: 877–892. [DOI] [PubMed] [Google Scholar]
- 32. Latham KE, Garrels JI, Chang C, Solter D. Quantitative analysis of protein synthesis in mouse embryos. I. Extensive reprogramming at the one‐ and two‐cell stages. Development 1991; 112: 921–932. [DOI] [PubMed] [Google Scholar]
- 33. Telford NA, Watson AJ, Schultz GA. Transition from maternal to embryonic control in early mammalian development: a comparison of several species. Mol Reprod Dev 1990; 26: 90–100. [DOI] [PubMed] [Google Scholar]
- 34. Matzuk MM, Burns KH, Viveiros MM, Eppig JJ. Intercellular communication in the mammalian ovary: oocytes carry the conversation. Science 2002; 296: 2178–2180. [DOI] [PubMed] [Google Scholar]
- 35. Dong J, Albertini DF, Nishimori K, Kumar TR, Lu N, Matzuk MM. Growth differentiation factor‐9 is required during early ovarian folliculogenesis. Nature 1996; 383: 531–535. [DOI] [PubMed] [Google Scholar]
- 36. Matzuk MM. Revelations of ovarian follicle biology from gene knockout mice. Mol Cell Endocrinol 2000; 163: 61–66. [DOI] [PubMed] [Google Scholar]
- 37. Carabatsos MJ, Elvin J, Matzuk MM, Albertini DF. Characterization of oocyte and follicle development in growth differentiation factor‐9‐deficient mice. Dev Biol 1998; 204: 373–384. [DOI] [PubMed] [Google Scholar]
- 38. McGrath SA, Esquela AF, Lee SJ. Oocyte‐specific expression of growth/differentiation factor‐9. Mol Endocrinol 1995; 9: 131–136. [DOI] [PubMed] [Google Scholar]
- 39. Dube JL, Wang P, Elvin J, Lyons KM, Celeste AJ, Matzuk MM. The bone morphogenetic protein 15 gene is X‐linked and expressed in oocytes. Mol Endocrinol 1998; 12: 1809–1817. [DOI] [PubMed] [Google Scholar]
- 40. Smith P, O WS, Corrigan KA et al. Ovarian morphology and endocrine characteristics of female sheep fetuses that are heterozygous or homozygous for the inverdale prolificacy gene (fecX1). Biol Reprod 1997; 57: 1183–1192. [DOI] [PubMed] [Google Scholar]
- 41. Braw‐Tal R, McNatty KP, Smith P et al. Ovaries of ewes homozygous for the X‐linked Inverdale gene (FecXI) are devoid of secondary and tertiary follicles but contain many abnormal structures. Biol Reprod 1993; 49: 895–907. [DOI] [PubMed] [Google Scholar]
- 42. Yan C, Wang P, DeMayo J et al. Synergistic roles of bone morphogenetic protein 15 and growth differentiation factor 9 in ovarian function. Mol Endocrinol 2001; 15: 854–866. [DOI] [PubMed] [Google Scholar]
- 43. Lyons KM, Pelton RW, Hogan BL. Patterns of expression of murine Vgr‐1 and BMP‐2a RNA suggest that transforming growth factor‐beta‐like genes coordinately regulate aspects of embryonic development. Genes Dev 1989; 3: 1657–1668. [DOI] [PubMed] [Google Scholar]
- 44. Schmid P, Cox D, Van_der_Putten H, McMaster GK, Bilbe G. Expression of TGF‐beta s and TGF‐beta type II receptor mRNAs in mouse folliculogenesis: stored maternal TGF‐beta 2 message in oocytes. Biochem Biophys Res Commun 1994; 201: 649–656. [DOI] [PubMed] [Google Scholar]
- 45. Valve E, Penttila TL, Paranko J, Harkonen P. FGF‐8 is expressed during specific phases of rodent oocyte and spermatogonium development. Biochem Biophys Res Commun 1997; 232: 173–177. [DOI] [PubMed] [Google Scholar]
- 46. Carabatsos MJ, Sellitto C, Goodenough DA, Albertini DF. Oocyte‐granulosa cell heterologous gap junctions are required for the coordination of nuclear and cytoplasmic meiotic competence. Dev Biol 2000; 226: 167–179. [DOI] [PubMed] [Google Scholar]
- 47. Eppig JJ, Wigglesworth K, Pendola FL. The mammalian oocyte orchestrates the rate of ovarian follicular development. Proc Natl Acad Sci USA 2002; 99: 2890–2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Soyal SM, Amleh A, Dean J. FIGalpha, a germ cell‐specific transcription factor required for ovarian follicle formation. Development 2000; 127: 4645–4654. [DOI] [PubMed] [Google Scholar]
- 49. Epifano O, Liang LF, Familari M, Moos MC, Dean J. Coordinate expression of the three zona pellucida genes during mouse oogenesis. Development 1995; 121: 1947–1956. [DOI] [PubMed] [Google Scholar]
- 50. Liang L, Soyal SM, Dean J. FIGalpha, a germ cell specific transcription factor involved in the coordinate expression of the zona pellucida genes. Development 1997; 124: 4939–4947. [DOI] [PubMed] [Google Scholar]
- 51. Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell 1982; 30: 675–686. [DOI] [PubMed] [Google Scholar]
- 52. Morisato D, Anderson KV. Signaling pathways that establish the dorsal‐ventral pattern of the Drosophila embryo. Annu Rev Genet 1995; 29: 371–399. [DOI] [PubMed] [Google Scholar]
- 53. Dean J. Oocyte‐specific genes regulate follicle formation, fertility and early mouse development. J Reprod Immunol 2002; 53: 171–180. [DOI] [PubMed] [Google Scholar]
- 54. Burns KH, Viveiros MM, Ren Y et al. Roles of NPM2 in chromatin and nucleolar organization in oocytes and embryos. Science 2003; 300: 633–636. [DOI] [PubMed] [Google Scholar]
- 55. Christians E, Davis AA, Thomas SD, Benjamin IJ. Maternal‐effect of Hsf1 on reproductive success. Nature 2000; 407: 693–694. [DOI] [PubMed] [Google Scholar]
- 56. Gurtu VE, Verma S, Grossmann AH, Liskay RM, Skarnes WC, Baker SM. Maternal‐effect for DNA mismatch repair in the mouse. Genetics 2002; 160: 271–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Howell CY, Bestor TH, Ding F et al. Genomic imprinting disrupted by a maternal‐effect mutation in the Dnmt1 gene. Cell 2001; 104: 829–838. [DOI] [PubMed] [Google Scholar]
- 58. Payer B, Saitou M, Barton SC et al. Stella is a maternal‐effect gene required for normal early development in mice. Curr Biol 2003; 13: 2110–2117. [DOI] [PubMed] [Google Scholar]
- 59. Ramos SB, Stumpo DJ, Kennington EA et al. The CCCH tandem zinc‐finger protein Zfp36l2 is crucial for female fertility and early embryonic development. Development 2004; 131: 4883–4893. [DOI] [PubMed] [Google Scholar]
- 60. Tong ZB, Gold L, Pfeifer KE et al. Mater, a maternal‐effect gene required for early embryonic development in mice. Nat Genet 2000; 26: 267–268. [DOI] [PubMed] [Google Scholar]
- 61. Wu X, Viveiros MM, Eppig JJ, Bai Y, Fitzpatrick SL, Matzuk MM. Zygote arrest 1 (Zar1) is a novel maternal‐effect gene critical for the oocyte‐to‐embryo transition. Nat Genet 2003; 33: 187–191. [DOI] [PubMed] [Google Scholar]
- 62. Tong ZB, Nelson LM. A mouse gene encoding an oocyte antigen associated with autoimmune premature ovarian failure. Endocrinology 1999; 140: 3720–3726. [DOI] [PubMed] [Google Scholar]
- 63. Mertineit C, Yoder JA, Taketo T, Laird DW, Trasler JM, Bestor TH. Sex‐specific exons control DNA methyltransferase in mammalian germ cells. Development 1998; 125: 889–897. [DOI] [PubMed] [Google Scholar]
- 64. Xu Z, Kopf GS, Schultz RM. Involvement of inositol 1,4,5‐trisphosphate‐mediated Ca2+ release in early and late events of mouse egg activation. Development 1994; 120: 1851–1859. [DOI] [PubMed] [Google Scholar]
- 65. Lai WS, Carballo E, Thorn JM, Kennington EA, Blackshear PJ. Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin‐related zinc finger proteins to Au‐rich elements and destabilization of mRNA. J Biol Chem 2000; 275: 17827–17837. [DOI] [PubMed] [Google Scholar]
- 66. Yan W, Rajkovic A, Viveiros MM, Burns KH, Eppig JJ, Matzuk MM. Identification of Gasz, an Evolutionarily Conserved Gene Expressed Exclusively in Germ Cells and Encoding a Protein with Four Ankyrin Repeats, a Sterile‐alpha Motif, and a Basic Leucine Zipper. Mol Endocrinol 2002; 16: 1168–1184. [DOI] [PubMed] [Google Scholar]
- 67. Yan W, Ma L, Zilinski CA, Matzuk MM. Identification and characterization of evolutionarily conserved pufferfish, zebrafish, and frog orthologs of GASZ. Biol Reprod 2004; 70: 1619–1625. [DOI] [PubMed] [Google Scholar]
- 68. Suzumori N, Yan C, Matzuk MM, Rajkovic A. Nobox is a homeobox‐encoding gene preferentially expressed in primordial and growing oocytes. Mech Dev 2002; 111: 137–141. [DOI] [PubMed] [Google Scholar]
- 69. Rajkovic A, Pangas SA, Ballow D, Suzumori N, Matzuk MM. NOBOX deficiency disrupts early folliculogenesis and oocyte‐specific gene expression. Science 2004; 305: 1157–1159. [DOI] [PubMed] [Google Scholar]
- 70. Minami N, Sasaki K, Aizawa A, Miyamoto M, Imai H. Analysis of gene expression in mouse two‐cell embryos using fluorescein differential display: comparison of culture environments. Biol Reprod 2001; 64: 30–35. [DOI] [PubMed] [Google Scholar]
- 71. Minami N, Aizawa A, Ihara R, Miyamoto M, Ohashi A, Imai H. Oogenesin is a novel mouse protein expressed in oocytes and early cleavage‐stage embryos. Biol Reprod 2003; 69: 1736–1742. [DOI] [PubMed] [Google Scholar]
- 72. Dade S, Callebaut I, Mermillod P, Monget P. Identification of a new expanding family of genes characterized by atypical LRR domains. Localization of a cluster preferentially expressed in oocyte. FEBS Lett 2003; 555: 533–538. [DOI] [PubMed] [Google Scholar]
- 73. Tsukamoto S, Ihara R, Aizawa A et al. Oog1, an oocyte‐specific protein, interacts with Ras and Ras‐signaling proteins during early embryogenesis. Biochem Biophys Res Commun 2006; 343: 1105–1112. [DOI] [PubMed] [Google Scholar]
- 74. Albright CF, Giddings BW, Liu J, Vito M, Weinberg RA. Characterization of a guanine nucleotide dissociation stimulator for a ras‐related GTPase. EMBO J 1993; 12: 339–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kikuchi A, Demo SD, Ye ZH, Chen YW, Williams LT. RalGDS family members interact with the effector loop of ras p21. Mol Cell Biol 1994; 14: 7483–7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Haberland J, Gerke V. Conserved charged residues in the leucine‐rich repeat domain of the Ran GTPase activating protein are required for Ran binding and GTPase activation. Biochem J 1999; 343 Part 3: 653–662. [PMC free article] [PubMed] [Google Scholar]
- 77. Dade S, Callebaut I, Paillisson A, Bontoux M, Dalbies‐Tran R, Monget P. In silico identification and structural features of six new genes similar to MATER specifically expressed in the oocyte. Biochem Biophys Res Commun 2004; 324: 547–553. [DOI] [PubMed] [Google Scholar]
- 78. Paillisson A, Dade S, Callebaut I et al. Identification, characterization and metagenome analysis of oocyte‐specific genes organized in clusters in the mouse genome. BMC Genomics 2005; 6: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]


