Abstract
Aim:
The present study was conducted to detect and identify the virulence genes in Pasteurella multocida isolates of porcine origin from Assam.
Materials and Methods:
A total of 21 porcine P. multocida isolates were subjected to capsular typing and detection of virulence-associated genes (pfhA, tbpA, hgbB, toxA, oma87, ompH, and nanB) using various polymerase chain reaction (PCR) methods reported elsewhere. Further, pathogenicity of the porcine isolates of P. multocida was studied in mice. For each strain of P. multocida selected for pathogenicity trial, the group of mice was injected intraperitoneally (i/p) with 0.1 ml of the inoculum prepared from respective field isolates, containing 109 organisms per ml.
Results:
Capsular typing of the isolates by multiplex PCR showed two capsular types, type A (66.66%) and type D (33.33%). All the isolates were positive for outer membrane protein genes, oma87 and ompH genes. Iron acquisition genes, tbpA and hgbB, were detected in 14.28% and 19.04% of the isolates. The dermonecrotoxin encoding gene, toxA, was present in 23.80% of the isolates. Filamentous hemagglutinin encoding gene, pfhA, was detected in 28.57%. The virulence gene distribution pattern of the isolates indicates the important role of the genes in disease pathogenesis.
Conclusion:
From the present study, it can be concluded that toxA gene is an important marker gene for defining the pathogenic potential of P. multocida strains in swine.
Keywords: capsular type, Pasteurella multocida, porcine, virulence-associated genes
Introduction
Pasteurella multocida belonging to family Pasteurellaceae is a ubiquitous organism affecting multiple host species, thus causing several diseases such as hemorrhagic septicemia in cattle and buffalo, enzootic bronchopneumonia in cattle, sheep, and goats, atrophic rhinitis in swine, fowl cholera in poultry, and snuffles in rabbits [1,2]. It is one of the most fascinating Gram-negative, opportunistic animals and human pathogens with worldwide distribution.
The organism is grouped into 5 capsular serogroups (A, B, D, E, and F) with host specificity and disease induction [3]. Both toxigenic and non-toxigenic strains of serogroups A and D are associated with diseases in swine [4]. The pathogenicity of P. multocida is associated with various virulence factors which include diverse adhesions, dermonecrotic toxin, iron acquisition proteins, sialidases, and outer membrane proteins [3,5-7]. These virulence factors help in colonization and invasion of the host, avoid host defense mechanisms, injury to host tissues, and stimulate host inflammatory response. The association of virulence factors with specific serogroups of P. multocida and its disease status in animals was also reported by Ewers et al. [8]. Since the pathogenic behavior of P. multocida could be predicted both by the virulence factors and the serogroups, evaluation of these virulence factors is important.
The present study investigates the distribution pattern of virulence-associated genes (VGAs) in P. multocida isolates of porcine origin.
Materials and Methods
Ethical approval
Ethical approval for the study was obtained from IAEC, Assam Agricultural University (AAU), Khanapara campus vide approval No. 770/ac/CPCSEA/FVSc/AAU/IAEC/10-11/79 dated 09.09.2011.
Source of P. multocida isolates
Twenty-one P. multocida isolates maintained in the ICAR Network Project on Hemorrhagic Septicemia, Department of Microbiology, College of Veterinary Science, Assam Agricultural University, Khanapara, Guwahati, were used for the study. The reference strain (P52) was obtained from the Division of Bacteriology and Mycology, ICAR-Indian Veterinary Research Institute, Izatnagar, Bareilly, Uttar Pradesh.
Revival and confirmation of P. multocida isolates
The isolates were reconfirmed by following standard bacteriological techniques and by P. multocida species-specific polymerase chain reaction (PM-PCR) as per the method described by Townsend et al. [9] using specific primer pairs (Table-1) [7,9-13]. PCR was done in 25 μl reaction mixture by mixing 3.0 μl DNA template with 12.5 μl master mix (2X, Qiagen, Germany) and forward and reverse primers (10 pmol each). The PCR amplification was performed in a thermocycler (Applied Biosystems, USA) using the thermal conditions as initial denaturation at 94°C for 4 min, 35 cycles of 94°C for 45 s, 55°C for 45 s, 72°C for 45 s, followed by final extension at 72°C for 6 min.
Table-1.
Sequences of the oligonucleotides used in the P. multocida multiplex capsular and virulence-associated genes typing assay of P. multocida.
| Gene | Primer | Sequence (5’-3’) | Amplicon size | References |
|---|---|---|---|---|
| KMT1 | KMT1T7 Fwd | ATCCGCTATTTACCCAGTGG | 460 bp | Townsend et al. [9] |
| KMT1SP6 Rev | GCTGTAAACGAACTCGCCAC | |||
| hyaD-hyaC | CAPA Fwd | TGCCAAAATCGCAGTCAG | 1044 bp | Townsend et al. [10] |
| CAPA Rev | TTGCCATCATTGTCAGTG | |||
| bcbD | CAPB Fwd | CATTTATCCAAGCTCCACC | 760 bp | Townsend et al. [10] |
| CAPB Rev | GCCCGAGAGTTTCAATCC | |||
| dcbF | CAPD Fwd | TTACAAAAGAAAGACTAGGAGCCC | 657 bp | Townsend et al. [10] |
| CAPD Rev | CATCTACCCACTCAACCATATCAG | |||
| toxA | Forward | TCT TAG ATG AGC GAC AAG G | 846 bp | Shayegh et al. [11] |
| Reverse | GAA TGC CAC ACC TCT ATA G | |||
| hgbB | Forward | TCT TTG AGT ACG GCT TGA C | 540 bp | Shayegh et al. [11] |
| Reverse | CTT ACG TCA GTA ACA CTC G | |||
| tbpA | Forward | TGG TTG GAA ACG GTA AAG C | 728 bp | Shayegh et al. [11] |
| Reverse | TAA CGT GTA CGG AAA AGC C | |||
| pfhA | Forward | AGC TGA TCA AGT GGT GAA C | 275 bp | Shayegh et al. [11] |
| Reverse | TGG TAC ATT GGT GAA TGC TG | |||
| nanB | Forward | CAT TGC ACC TAA CAC CTC T | 555 bp | Tang et al. [12]; Ewer et al. [7] |
| Reverse | GGA CAC TGA TTG CCC TGA A | |||
| Oma87 | Forward | GGC AGC GAG CAA CAG ATA ACG | 838 bp | Tang et al. [12]; Ewer et al. [7] |
| Reverse | TGT TCG TCA AAT GTC GGG TGA | |||
| ompH | Forward | GCG TTT CAT TCA AAG CAT CTC | 1000 bp | Luo et al. [13] |
| Reverse | ATG ACC GCG TAA CGA CTT TC |
P. multocida=Pasteurella multocida
Capsular typing
Capsular PCR typing for all the isolates was done using a multiplex PCR as per the method described by Townsend et al. [10] with the reaction condition as illustrated in Table-2 [7,11].
Table-2.
Thermal cycling condition for the detection of virulence-associated genes of P. multocida.
| PCR steps | Gene | ||||||
|---|---|---|---|---|---|---|---|
| toxA | tbpA | hgbB | pfhA | ompH | oma87 | nanB | |
| Initial denaturation | 95°C 5 min | 95°C 5 min | 95°C 5 min | 95°C 5 min | 94°C 3 min | 94°C 3 min | 94°C 3 min |
| Denaturation | 94°C 45 s | 94°C 45 s | 94°C 45 s | 94°C 45 s | 94°C 30 s | 94°C 30 s | 94°C 30 s |
| Annealing | 54°C 50 s | 54°C 50 s | 54°C 50 s | 54°C 50 s | 57°C 30 s | 55°C 30 s | 56°C 30 s |
| Extension | 72°C 50 s | 72°C 50 s | 72° C 50 s | 72°C 50 s | 72°C 60 s | 72°C 60 s | 72°C 45 s |
| Number of cycles | 35 | 35 | 35 | 35 | 25 | 25 | 30 |
| Final extension | 72°C 10 min | ||||||
| Hold | 4°C | ||||||
| References | Shayegh et al. [11] | Shayegh et al. [11] | Shayegh et al. [11] | Shayegh et al. [11] | Ewer et al. [7] | Ewer et al. [7] | Ewer et al. [7] |
P. multocida=Pasteurella multocida, PCR=Polymerase chain reaction
Virulence gene detection
P. multocida isolates were tested for the presence of various VGAs, for example, pfhA, tbpA, hgbB, toxA, oma87, ompH, and nanB by simplex PCR as per the method of Ewers et al. [8], using specific primers (Table-1) with the standard thermal conditions (Table-2). The amplified products were electrophoresed in 1.5% agarose gel in 1X tris acetate EDTA buffer at 60 V for 1 h with ethidium bromide stain and visualized with ultraviolet light by Gel Documentation System (Kodak, Biostep, Germany).
Pathogenicity study of P. multocida isolates
Pathogenicity of the porcine isolates of P. multocida was studied in mice following the method described by Curtis [14] with slight modification. For each strain of P. multocida selected for pathogenicity trial, the group of mice was injected intraperitoneally (i/p) with 0.1 ml of the inoculum prepared from respective field isolates, containing 109 organisms per ml [15].
Results
All the bacterial isolates were revived in blood agar. The small, smooth, circular, glistening, and dewdrop-like colonies with a very characteristic odor and non-hemolytic colonies on blood agar plate (data not shown) were found to be Gram-negative coccobacilli and identified as P. multocida. The organisms were further confirmed based on specific amplification of KMT1 gene by PCR yielding an expected product size of 460 bp (Figure-1) which was detected in all the isolates.
Figure-1.
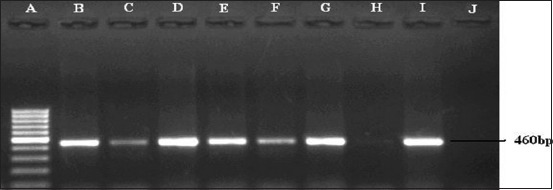
Pasteurella multocida species-specific polymerase chain reaction for the detection of KMT1 gene (460 bp) of P. multocida. Lane A: 100 bp DNA Ladder. Lane B to H: Samples positive for kmt gene. Lane J: Negative control.
On capsular PCR typing, of 21 P. multocida isolates, serogroup D-specific gene (657 bp, Figure-2) was detected in seven (33.33%) isolates, while serogroup A-specific gene (1044 bp, Figure-2) was detected in 14 (66.66%) isolates. The reference strain gave an amplified product of 760 bp band size corresponding to capsular type B.
Figure-2.
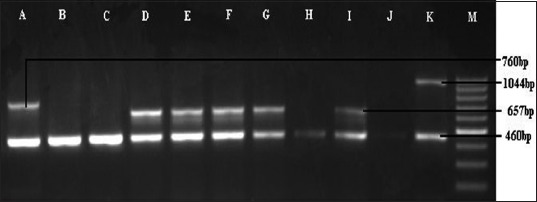
Multiplex-polymerase chain reaction for capsular typing of Pasteurella multocida. Lane A: Reference strain (P52). Lane D, E, F, G, and I: Sample positive for cap D. Lane K: Sample positive for cap A. Lane M: 100 bp DNA Ladder.
On virulence gene detection PCR (Table-3), it was observed that the outer membrane genes (oma87 and ompH) were found to be present in all the 21 isolates of porcine origin used in the present study and the reference strain giving expected band size of 838 and 1077 bp, respectively (Figures-3 and 4). The hemoglobin binding gene (hgbB, Figure-5) was found in four (57.14%) isolates of serotype D only, while tbpA (Figure-6) gene was detected in four (21.42%) serotype A isolates only.
Table-3.
Virulence-associated gene detection in different serotypes of swine Pasteurella multocida by PCR and their pathogenicity in mice.
| S. No. | Isolate No. | Serotype | Virulence genes | Pathogenicity | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| toxA | nanB | tbpA | pfhA | hgbB | oma87 | ompH | ||||
| 1 | P2 | A | + | + | (83.33) | |||||
| 2 | P3 | A | + | + | ||||||
| 3 | P4 | A | + | + | (66.66) | |||||
| 4 | P6 | A | + | + | (83.33) | |||||
| 5 | P16 | A | + | + | ||||||
| 6 | P19 | A | + | + | ||||||
| 7 | P20 | A | + | + | ||||||
| 8 | P22 | A | + | + | (50) | |||||
| 9 | P5 | A | + | + | + | (83.33) | ||||
| 10 | P7 | A | + | + | + | |||||
| 11 | P13 | A | + | + | + | |||||
| 12 | P14 | A | + | + | + | + | + | (100) | ||
| 13 | P18 | A | + | + | + | + | + | (83.33) | ||
| 14 | P10 | A | + | + | + | + | (100) | |||
| Sub total | 14 | 3 (21.42) | 0 | 3 (21.42) | 5 (35.71) | 0 | 14 (100) | 14 (100) | 8 (57.14) | |
| 15 | P1 | D | + | + | ||||||
| 16 | P9 | D | + | + | ||||||
| 17 | P15 | D | + | + | + | + | + | (100) | ||
| 18 | P17 | D | + | + | + | |||||
| 19 | P21 | D | + | + | + | (50) | ||||
| 20 | P8 | D | + | + | + | (33.33) | ||||
| 21 | P11 | D | + | + | + | + | + | (100) | ||
| Sub total | 7 | 2 (28.57) | 2 (28.57) | 0 | 1 (14.28) | 4 (57.14) | 7 (100) | 7 (100) | 4 (57.14) | |
| 22 | P12 | B (P52) | 0 | 0 | 1 | 1 | 0 | 1 | 1 | (100) |
| Grand total | 22 | 5 (22.72) | 2 (9.09) | 4 (18.18) | 7 (31.81) | 4 (18.18) | 22 (100) | 22 (100) | 13 (59.09) | |
PCR=Polymerase chain reaction
Figure-3.
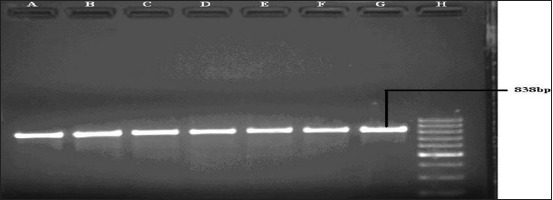
Polymerase chain reaction for the detection of oma87gene (838 bp) of Pasteurella multocida. Lane A to G: Samples positive for oma87 gene. Lane H: 100bp DNA Ladder.
Figure-4.
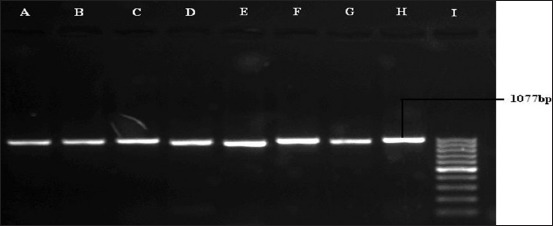
Polymerase chain reaction for the detection of ompH gene (1077 bp) of Pasteurella multocida. Lane A to H: Samples positive for ompH gene, Lane I: 100 bp DNA Ladder.
Figure-5.
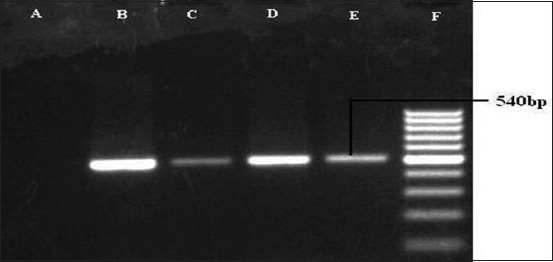
Polymerase chain reaction for the detection of hgbB gene (540 bp) of Pasteurella multocida. Lane A: Samples negative for hgbB gene. Lane B, C, D, and E: Samples positive for hgbB gene. Lane F: 100 bp DNA Ladder.
Figure-6.
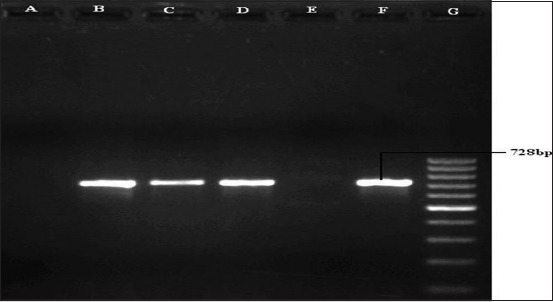
Polymerase chain reaction for the detection of tbpA gene (728 bp) of Pasteurella multocida. Lane B, C, D, and F: Samples positive for tbpA gene. Lane A and E: Samples negative for tbpA gene. Lane G: 100 bp DNA Ladder.
On amplification of filamentous hemagglutinin gene (pfhA), of the 21 isolates, six exhibited the presence of the same by yielding an expected amplicon size of 275 bp (Figure-7), of which 5 isolates belong to serotype A (35.71%) and 1 to serotype D (14.28%). The sialidase coding gene (nanB) was detected only in 2 (28.57%) isolates of serotype D (Figure-8).
Figure-7.
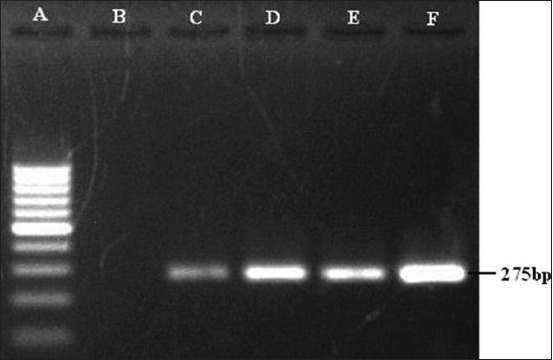
Polymerase chain reaction for the detection of pfhA gene (275 bp) of Pasteurella multocida. Lane A: 100 bp DNA Ladder, Lane B: Samples negative for pfhA gene, Lane C to F: Samples positive for pfhA gene.
Figure-8.
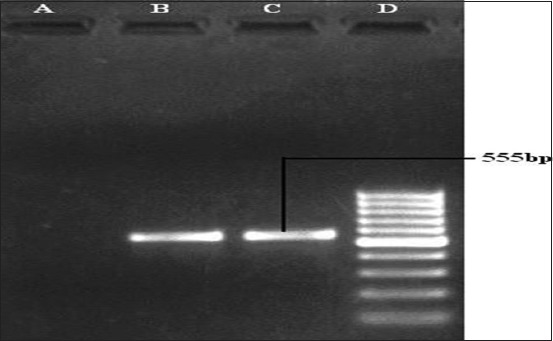
Polymerase chain reaction for the detection of nanB gene (555 bp) of Pasteurella multocida. Lane A: Samples negative for nanB gene. Lane B and C: Samples positive for nanB gene. Lane D: 100 bp DNA Ladder.
Toxigenic gene (toxA) was detected in five isolates, of which three were serotype A (21.42%), while 2 isolates of serotype D (28.57%). toxA gene was absent in the reference strain P52 strain (Figure-9).
Figure-9.
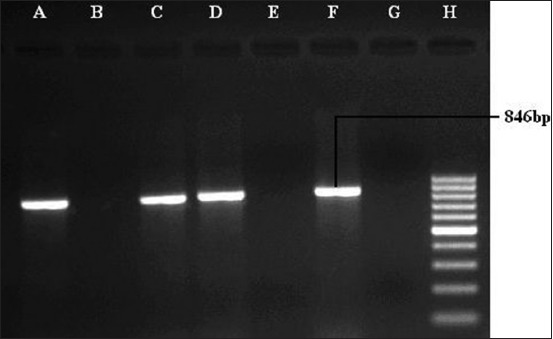
Polymerase chain reaction for the detection of toxA gene (846 bp) of Pasteurella multocida. Lane A, C, D, and F: Samples positive for toxA gene. Lane B, E, and G: Samples negative for toxA gene. Lane H: 100 bp DNA Ladder.
Discussion
The present paper describes as the first report of virulence gene profiles of porcine P. multocida from Assam, India. Pasteurellosis is a common disease of pigs worldwide with specific serotype and pathotype associated with the respiratory disease [16-18]. However, the distribution pattern of the serotypes and pathotypes can vary considerably from region to region and over time in a given region [12,19]. The present study results in the detection of more percentage of serotype A than serotype D. The isolates used in the present study were mainly from cases of the respiratory syndrome. A higher percentage of serotype A than serotype D in P. multocida isolates from indigenous pigs in central Kalorey et al. [20] and Northeast India [19,21] have been reported.
Variable distribution of the virulence genes was observed among the serotypes of P. multocida during the present study (Table-3). However, no correlation could be established with respect to the presence of virulence gene and serotypes of the Wide spread. Widespread distribution of hgbB gene among the porcine P. multocida strains and its regular detection in capD strains compared to capA strains of P. multocida was also reported [7,22]. On the contrary to the present detection of tbpA gene, Ewers et al. [7] could detect the gene exclusively in bovine, sheep, and buffalo isolates and not in P. multocida pig strains. The present observation of 18.18% positive strains of P. multocida isolates possessing the tbpA gene could be probably due to interspecies transmission of tbpA gene-positive P. multocida, as also suggested by Kumar et al. [23]. However, further study needs to be conducted before giving conclusive remark on interspecies transmission.
The present finding of low percent positivity of pfhA gene in strains of serogroup D compared with serogroup A also supports the observations of association of filamentous hemagglutinin gene pfhA with serogroups A, B, E, and F [7,12,22].
The occurrence of OMP gene in porcine strains of P. multocida and its equal distribution among capsular serogroups (type A, type D, and other serotypes) was also reported [7,12,22]. P. multocida OMPs have been identified as potent immunogens [24] as reported by Rajkhowa et al. [25] and play a significant role in the pathogenesis of pasteurellosis [26]. Hazarika et al. [27] also reported that the sonicated and bacterin vaccines prepared from pig strains of P. multocida conferred 100% protection against homologous as well as heterologous strains compared to 66.66 and 86.66% protection with vaccine prepared from reference strain (P52). From the present study, it can be opined that the OMP gene has a great role in pathogenesis of P. multocida infection irrespective of the serotypes. Considering the immunogenic potential of the OMPs, both the genes can be explored in the development of a suitable vaccine against swine pasteurellosis. However, before giving any conclusive remarks on distribution and its applicability as vaccine candidate, a detailed study will be required involving a large number of isolates recovered from pigs of different parts of the Northeastern region.
The 13 (59.09%) pathogenic isolates were found to possess different combinations of virulence genes. Pathogenic serotype A isolates with different virulence gene combinations produce 83.33-100% mortality in inoculated mice when compared to isolates with the presence of only OMP genes (50-83.33%). Similarly, serotype D isolates with different virulence gene combinations produce 33.33-100% mortality in inoculated mice. No reports could be traced out from the available literature with respect to the correlation between presence virulence gene in the P. multocida isolates and their pathogenicity in mice. However, it can be opined from the present observation that OMP alone is not responsible for pathogenesis of the disease and association of other VGAs with OMP gene might play a key role in pathogenesis. The present study indicates that toxA gene plays an important role in pathogenesis of P. multocida in pigs, which is supported by the findings of many workers [28-30]. Among the other virulence genes, tbpA gene was found to be closely associated with pathogenesis of the P. multocida. Contrary to the present findings, Ewers et al. [7] reported the presence of tbpA gene only in bovine strains of P. multocida and could not detect in porcine strains. Although they observed a significant association between pfhA and toxA with clinically diseased swine, toxA alone was found to be associated with the disease status independently. A perusal of the literature reveals that there is no report on the detection of tbpA gene from porcine strains, the detection of the gene in highly pathogenic porcine isolate is an important finding, and its role in pathogenesis needs further investigation. During the present study, some of the P. multocida isolates could not produce mortality in mice following inoculation with P. multocida isolate having virulence gene, either alone or in combination. This might be due to repeated subculturing of the isolates in laboratory media that result in the suppression of gene function or due to gene mutation, resulting in non-expression of the genes in vivo [31,32]. Detection of a high proportion of toxigenic capsular type A P. multocida from pigs was also reported by other workers [11,20,30,33]. Detection of toxin gene in both the type A and D isolates of P. multocida of the region in the present study indicates its important role in the disease pathogenesis mechanism.
Conclusion
From the present study, it can be concluded that toxA gene is an important marker gene for defining the pathogenic potential of P. multocida strains in swine. However, other virulence genes are also found to be distributed well among pathogenic strains of P. multocida. Among the other virulence genes, tbpA gene was found to be closely associated with the pathogenesis of the P. multocida. The association of the gene in disease producing mechanism needs further evaluation.
Authors’ Contributions
This study was a part of LBD’s research work during his PhD program. LBD carried out the experiment. SKD, RKS, and DPB designed the experiment. SKD, RKS, DPB, SM, and RAH provided necessary guidelines. DPB drafted the final manuscript. All authors have read and approved the final manuscript.
Acknowledgments
The authors would like to thank the Indian Council of Agricultural Research (ICAR) for providing financial help to carry out the research under Network Project on Hemorrhagic Septicemia (NWP on HS). Grant no. F.No 1(23)/02-IA-I dated 9th Feb, 2004.
Competing Interests
The authors also declare that there are no conflicts.
References
- 1.Hatfaludi T, Al-Hasani K, Boyce J.D, Adler B. Outer membrane proteins ofPasteurella multocida. Vet. Microbiol. 2010;144:1–17. doi: 10.1016/j.vetmic.2010.01.027. [DOI] [PubMed] [Google Scholar]
- 2.Sarangi L.N, Priyadarshini A, Kumar S, Thomas P, Gupta S.K, Nagaleekar V.K, Singh V.P. Virulence genotyping ofPasteurella multocidaisolated from multiple hosts from India. Sci. World J. 2014;2014:1–10. doi: 10.1155/2014/814109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harper M, Boyce J.D, Adler B. Pasteurella multocidapathogenesis:125 years after Pasteur. FEMS Microbiol Lett. 2006;265:1–10. doi: 10.1111/j.1574-6968.2006.00442.x. [DOI] [PubMed] [Google Scholar]
- 4.Ujvári B, Szeredi L, Pertl L, Tóth G, Erdélyi K.K, Jánosi S, Molnár T, Magyar T. First detection ofPasteurella multocidaType B:2 in Hungary associated with systemic pasteurellosis in backyard pigs. Acta Vet. Hung. 2015;63:141–156. doi: 10.1556/AVet.2015.012. [DOI] [PubMed] [Google Scholar]
- 5.Hunt M.L, Adler B, Townsend K.M. The molecular biology ofPasteurella multocida. Vet. Microbiol. 2000;72:3–25. doi: 10.1016/s0378-1135(99)00183-2. [DOI] [PubMed] [Google Scholar]
- 6.Hunt M.L, Boucher D.J, Boyce J.D, Adler B. In vivo-expressed genes of Pasteurella multocida. Infect. Immun. 2001;69:3004–3012. doi: 10.1128/IAI.69.5.3004-3012.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ewers C, Lu¨bke-Becker A, Bethe A, Kiebling S, Filter M, Wieler L.H. Virulence genotype ofPasteurella multocidastrains isolated from different hosts with various disease status. Vet. Microbiol. 2006;114:304–317. doi: 10.1016/j.vetmic.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 8.Ewers C, Lu¨bke-Becker A, Wieler L.H. Pasteurella:Insights into the virulence determinants of a heterogenous bacterium. Berl. Munch. Tierarztl. Wochenschr. 2004;9-10:367–386. [PubMed] [Google Scholar]
- 9.Townsend K.M, Frost A.J, Lee C.W, Papadimitriou J.M, Dawkins H.J. Development of PCR assays for species- and type-specific identification ofPasteurella multocidaisolates. J. Clin. Microbiol. 1998;36:1096–1100. doi: 10.1128/jcm.36.4.1096-1100.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Townsend K.M, Boyce J.D, Chung J.Y.A, Frost J, Adler B. Genetic organization ofPasteurella multocida caploci and development of a multiplex capsular PCR typing system. J. Clin. Microbiol. 2001;39:924–929. doi: 10.1128/JCM.39.3.924-929.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shayegh J, Atashpaz S, Hejazi M.S. Virulence genes profile and typing of ovinePasteurella multocida. Asian J Anim. Vet. Adv. 2008;3:206–213. [Google Scholar]
- 12.Tang X, Zhao Z, Hu J, Wu B, Cai X, He Q, Chen H. Isolation, antimicrobial resistance, and virulence genes ofPasteurella multocidastrains from Swine in China. J Clin. Microbiol. 2009;47:951–958. doi: 10.1128/JCM.02029-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo Y, Glisson J.R, Jackwood M.W, Hancock R.E, Bains M, Cheng I.H, Wang C. Cloning and characterization of the major outer membrane protein gene (ompH) of Pasteurella multocida X-73. J Bacteriol. 1997;179:7856–7864. doi: 10.1128/jb.179.24.7856-7864.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Curtis P.E. In: Pasteurella multocida. Isolation and Identification of Microorganisms of Medical and Veterinary Importance. Collins C.H, Grange J.M, editors. London: Academic Press; 1985. [Google Scholar]
- 15.Wijewardana T.G. Haemorrhagic septicaemia. Diagnostic and Vaccine Production Procedures. FAO Regional Reference Laboratory (Asian Region) Paradeniya, Sri Lanka: Veterinary Research Institute, Department of Animal Production & Health; 1992. [Google Scholar]
- 16.Pors S.E, Hansen M.S, Bisgaard M, Jensen H.E, Iburg T.M. Immuno histochemical study of porcine lung lesions associated withPasteurella multocida. Vet. J. 2013;197:483–488. doi: 10.1016/j.tvjl.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tigga M, Ghosh R.C, Malik P, Choudhary B.K, Tigga P, Nagar D.K. Isolation, characterization, antibiogram and pathology of isolated from pigs. Vet. World. 2014;7:363–368. [Google Scholar]
- 18.Choudhary M, Ghosh R.C, Malik P, Choudhary B.K, Nety S. Pathological changes associated with natural outbreak of swine pasteurellosis. J. Pure Appl. Microbiol. 2017;11:237–240. [Google Scholar]
- 19.Varte Z, Dutta T.K, Roychoudhury P, Begum J, Chandra R. Isolation, identification, characterization and antibiogram ofPasteurella multocidaisolated from pigs in Mizoram with special reference to progressive atrophic rhinitis. Vet. World. 2014;7:95–99. [Google Scholar]
- 20.Kalorey D.R, Yuvaraj S, Vanjari S.S, Gunjal P.S, Dhanawade N.B, Barbuddhe S.B, Bhandarkar A.G. PCR analysis ofPasteurella multocidaisolates from an outbreak of pasteurellosis in Indian pigs. J. Comp. Immunol. Microbiol. Infect. Dis. 2008;31:459–465. doi: 10.1016/j.cimid.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 21.George S, Rajbongshi G, Deuri S, Khatoon A, Barman N.N. Simultaneous occurrence of pneumonic pasteurellosis and swine fever in an organised pig farm in Assam. North East Vet. 2012;12:5–8. [Google Scholar]
- 22.Bethe A, Wieler L.H, Selbitz Hans J, Ewers C. Genetic diversity of porcinePasteurella multocidastrains from the respiratory tract of healthy and diseased swine. Vet. Microbiol. 2009;139:97–105. doi: 10.1016/j.vetmic.2009.04.027. [DOI] [PubMed] [Google Scholar]
- 23.Kumar H, Mahajan V, Sharma S, Alka Singh R, Arora A.K, Banga H.S, Verma S, Kaur K, Kaur P, Meenakshi Sandhu K.S. Concurrent pasteurellosis and classical swine fever in Indian pigs. J. Swine Health. Prod. 2007;15:279–283. [Google Scholar]
- 24.Singh R, Tewari K, Packiriswamy N, Marla S, Rao V.D.P. Molecular characterization and computational analysis of the major outer membrane protein (omph) gene ofPasteurella multocidap52. Vet. Arhiv. 2011;81:211–222. [Google Scholar]
- 25.Rajkhowa S, Shakuntala I, Pegu S.R, Das R.K, Das A. Detection ofPasteurella multocidaisolates from local pigs of India by polymerase chain reaction and their antibiogram. Trop. Anim. Health Prod. 2012;44:1497–1503. doi: 10.1007/s11250-012-0094-4. [DOI] [PubMed] [Google Scholar]
- 26.Srivastava S.K. Outer membrane protein ofPasteurella multocidaserotype B:2 are immunogenic and antiphagocytic. Ind. J. Exp. Biol. 1998;36:530–532. [PubMed] [Google Scholar]
- 27.Hazarika M.P, Barman N.N, George S, Sharma R.K. Characterization ofPasteurella multocidaisolated from pneumonic pigs of Assam. Indian J. Anim. Res. 2011;44:265–269. [Google Scholar]
- 28.Jong M.F. In: Diseases of Swine. 9th ed. Straw B.E, Zimmerman J.J, d' Allaire S, Taylor D.J, editors. Ames, Iowa, USA: Iowa State University Press; 2006. pp. 577–602. [Google Scholar]
- 29.Martineau G.P, Broes A, de Jong M.F. Experimental reproduction of atrophic rhinitis withPasteurella multocidaon gnotobiotic and conventional piglets. Proc. Int. Pig Vet. Soc. Cong. (Subject Rhinitis) 1982;6:88. [Google Scholar]
- 30.Rutter J.M, Rojas X. Atrophic rhinitis in gnotobiotic piglets:Differences in the pathogenicity ofPasteurella multocidain combined infection withBordetella bronchoseptica. Vet. Rec. 1982;110:531–535. [Google Scholar]
- 31.Borowski S.M, Silva S.C, Schrank I, Cardoso M. Toxin detection inPasteurella multocidastrains isolated from swine lungs in the state of Rio Grande do Sul, Brazil. Arq. Fac. Vet. UFRGS. 2001;29:79–85. [Google Scholar]
- 32.Stępniewska K, Markowska-Daniel I. Phenotypic and genotypic characterization ofPasteurella multocidastrains isolated from pigs in Poland. Bull. Vet. Inst. Pulawy. 2013;57:29–3. doi: 10.2478/pjvs-2014-0009. [DOI] [PubMed] [Google Scholar]
- 33.Cardoso T.F, Laguna G.J, Callejo M, Vela A.I, Carrasco L, Fernandez G.J.F, Maldonado A, Luque I. Septicaemic pasteurellosis in free-range pigs associated with an unusual biovar 13 ofPasteurella multocida. Vet. Microbiol. 2013;167:690–694. doi: 10.1016/j.vetmic.2013.08.005. [DOI] [PubMed] [Google Scholar]


