Abstract
Aim:
The aim of this study was to detect Brucella spp. DNA in milk samples collected from seronegative cows using the real-time polymerase chain reaction (PCR) assay for diagnosis of brucellosis in seronegative dairy cows to prevent transmission of disease to humans and to reduce economic losses in animal production.
Materials and Methods:
In this study, 65 milk samples were investigated for the detection of Brucella spp. The detection of the IS711 gene in all samples was done by real-time PCR assay by comparative cycle threshold method.
Results:
The results show that of the 65 DNA samples tested, 2 (3.08%) were positive for Brucella infection. The mean cyclic threshold values of IS711 real-time PCR test were 37.97 and 40.48, indicating a positive reaction.
Conclusion:
The results of the present study indicated that the real-time PCR appears to offer several advantages over serological tests. For this reason, the real-time PCR should be validated on representative numbers of Brucella-infected and free samples before being implemented in routine diagnosis in human and animal brucellosis for controlling this disease.
Keywords: Brucella spp, milk, real-time polymerase chain reaction, seronegative cows
Introduction
Brucellosis is one of the most common and economically important zoonoses globally [1]. Bacteria of the genus Brucella spp. are coccobacilli, Gram-negative, aerobic, non-spore forming, non-motile, and non-capsulated [2]. To date, twelve different Brucella species have been described [3,4]. Each one may infect different host species, but each Brucella species has a preference for its host species: Brucella melitensis (sheep and goats), Brucella abortus (cattle), Brucella suis (pigs), Brucella ovis (rams), Brucella canis (dogs), Brucella microti (rodents - Microtus arvalis), Brucella neotomae (rodents - Neotoma lepida), Brucella pinnipedialis (pinnipeds), Brucella ceti (cetacea), and Brucella inopinata (originally isolated from a human patient, but its preferential host is not known) [5-7]. The two most recently described species are B. papionis, which was isolated from two baboons with retained placenta [4], and Brucella vulpis which was isolated in Austria from the mandibular lymph nodes of two red foxes [3].
Zoonotic transmission occurs most frequently through unpasteurized milk products in urban settings, while occupational exposure of farmers, veterinarians, or laboratory workers can result from direct contact with infected animals or tissues or fluids associated with abortion [8].
Brucellosis in humans almost always originates from an animal reservoir [9]. To reduce the incidence of many zoonotic infections among humans, the pathogens must be controlled in the animal population [10].
Diagnosis is usually based on serological tests and/or cultivation. Serological assays are rapid, sensitive, and easy to perform but lack specificity due to cross-reactions with other bacteria, particularly with Yersinia enterocolitica O:9, that result from O chains antigenic similarity [11,12]. Conventional cultural isolation and identification of the agent is the gold standard test for Brucella abortus but time consuming, laborious and also need skills as well as biosafety level-3 laboratory and measures to prevent zoonosis [13].
Due to isolation problems, the significance of molecular-based detection techniques is increasing. Polymerase chain reaction (PCR) techniques for the diagnosis of Brucella spp. are used successfully both from different clinical samples and pure cultures [14].
There are also real-time or quantitative PCR (qPCR) assays that have been developed for rapid and safe detection of Brucella, including assays targeting the bcsp31 gene or the IS711 insertion sequence [15].
In this study, we discuss the extraction and purification of column DNA from raw milk using a specific kit, and we tried to use a reliable molecular procedure that could increase the sensitivity and specificity of the detection of Bruclla spp. DNA in bovine milk which detects insertion of the IS711 sequence of B. melitensis, B. abortus, B. canis, B. ovis, and B. suis. IS711 is characteristic of Brucella spp. and appears in variable number (5-38 copies) [16].
Materials and Methods
Ethical approval
In this investigation, we did not use live animals. Milk samples were collected during routine milking by traditional hand-stripping. Therefore, no ethical approval was needed in the present study.
Samples of milk
A total of 65 milk samples were taken from seronegative cows (ages 3-8 years) belonging to 3 dairy farms in the Wilaya of Batna: 2 in the district of Djerma and 1 in the district of Ain Yagout. The first farm contains 21 cows, the second 25 cows, and the third 19 cows.
The milk samples were collected aseptically from teats of the udder which had previously been cleaned with water and soap, and then, the surface was sterilized with 70% ethanol. For each animal, the first jet of milk was removed, and then 15 mL of milk were collected from 4 quarters in sterile Falcon tubes previously identified. The samples were immediately transported to the laboratory at +4°C for DNA extraction.
This study was conducted at the Laboratory of Microbiology and Molecular Biology of Constantine Biotechnology Research Center, Algeria, during July 2017.
Pre-treatment of milk
Initially, the milk undergone a pre-treatment as follows: A test sample of 800 µL of the milk was centrifuged for 5 min at 10,000 g, then the supernatant was removed, and the pellet was resuspended in 300 µL of phosphate-buffered saline.
Extraction of DNA
Genomic DNA was extracted from raw milk using a BioExtract® Column purification kit according to the manufacturer protocol (Cat N° BEC050).
The extracted DNA is then quantified using Nanodrop® ND 8000.
The DNA was then stored at 4°C until use.
Real-time PCR amplification
Real-time PCR was used to detect the presence of Brucella spp. DNA in milk samples as described in the kit BactoReal® kit Brucella spp. (REF: DVEB02113). This test has been developed and validated for ABI PRISM® 7500 (Fast) instrument (Applied Biosystems). This test allows rapid and sensitive detection of DNA Brucella spp. from purified milk samples.
BactoReal® Kit Brucella spp. detects insertion of the IS711 sequence of B. melitensis, B. abortus, B. canis, B. ovis, and B. suis. IS711 is characteristic of Brucella spp. and appears in variable number (5-38 copies).
A positive internal control of the system for the detection at Cy5 (667 nm) makes it possible to demonstrate the inhibitions of the PCR, resulting in false-negative results when interpreting the results due to the inhibition of PCR in real time.
The reaction mixture
The real-time PCR assay was performed by an ABI PRISM® 7500 instrument thermal cycler (Applied Biosystems) using 96-well optical barcode plates (Ref: 4306737) and Adhesive Optical Films Starter Kit (Ref: 4311971) following the manufacturer’s instructions (Ingenetix, Austria). The real-time PCR amplification was carried out in a reaction mixture of 20 µL composed of 5 µL of sample (or genomic DNA) containing matrix DNA and 15 µL of Master Mix consisting of 3.0 µL of water, 10 µL of DNA mix reaction (1 µL of Brucella Mix Assay) for the detection of Brucella spp., and 1 µL of CR mix assay (Primers and Probes (Cy5) for IPC detection).
At least one negative control (water), one positive control (B. abortus), and one negative extraction included by PCR were ensured.
According to Ingenetix recommendations, all PCR analyzes are done in duplicate to optimize the probability of detecting pathogens and facilitates the interpretations of results.
Programming of the temperature profile
The reaction was carried out in a DNA thermocycler (Applied Biosystems) at a preliminary denaturing temperature of the DNA in dry at 95°C, followed by 45 cycles consisting of 5 s at 95°C for denaturation of DNA, and 1 min at 60°C for polymerase-mediated primer extension.
Results
The results show that of the 65 DNA samples tested, 2 (3.08%) were positive and 63 (96.92%) were negative for brucellosis, based on real-time PCR testing used. The mean cyclic threshold (Ct) values of IS711 real-time PCR test were 37.97 and 40.48, indicating a positive reaction.
The curve (Figure-1) shows a Target 1 (FAM Signal) multiplication which means that the Brucella spp. DNA was amplified and the sample interpreted as a positive. However, the Ct is 37.97 (low value) which means the presence of Brucella spp. in small quantities.
Figure-1.
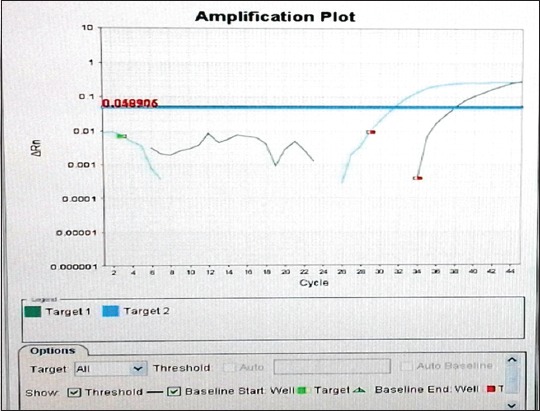
Logarithmic curve of amplification of one of the two positive samples. Target 1: FAM fluorescence signal of Brucella spp. DNA. Target 2: Fluorescence signal of the internal positive control (Cy5).
The curve (Figure-2) shows a Target 1 multiplication for positive controls (undiluted and diluted), as well as for 2 samples only. For the other samples, the Target 1 multiplication is not detectable (below the threshold value). The curves above the threshold value are Target 2 (Cy5) CPI multiplications.
Figure-2.
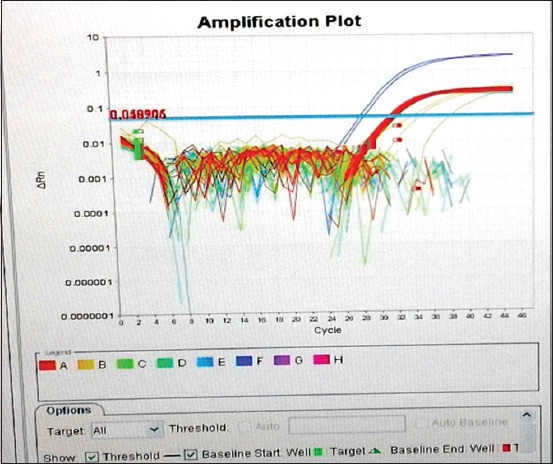
Logarithmic amplification curve of all samples (all plates even positive and negative controls).
Discussion
Brucellosis is an ancient and one of the world’s most widespread zoonotic diseases affecting both public health and animal production [17]. In livestock, the disease results in significant economic losses due to reproductive impairment caused by abortion, stillbirth or weak calves and neonatal mortality, and infertility [5]. In humans, Brucella spp. infection causes a febrile disease that may be associated with a broad spectrum of symptoms, and it may be fatal in some cases [18].
In the present study, we investigated the presence of Brucella DNA in the milk samples, and we chose to analyze all samples with primers targeting the IS711 insertion sequence because IS711 is a specific and highly sensitive method for the safe detection of the genus Brucella [15].
This study found that, of the 65 milk samples of seronegative cows, 3.08% (n=2) were qPCR positive for Brucella DNA with Ct-values ranging between 37.97 and 40.48. The results obtained in this study confirm that bacterial excretion of milk was low but sufficient to induce infection. The consumption of non-pasteurized dairy products from Brucella-infected animals is the most frequent route of human infection in general. Hence, pasteurization of milk will reduce Brucella transmission to humans [19].
The prevalence of brucellosis in Algeria in this study was lesser as compared to an earlier study obtained by Islam et al. [20], Rajala et al. [21], and Hinić et al. [22]. They found a similar level of positive cases among their examined herds with 11.23%, 11.8%, and 11.1%, respectively. The discrepancy between the serology and PCR results observed in the current study might indicate that the true number of Brucella-infected cattle within the study area could be underestimated by serology screening. False serological negative results have been reported previously [11,23,24] and one explanation could be that antibody titers reduce over time [25]. Hence, seronegative animals in the current study, which tested positive by qPCR, could have been exposed to Brucella and turned seronegative after a certain time period. Alternatively, if sampling at an early stage of the infection, i.e., within the first 14 days, the humoral immune response has not yet induced detectable levels of antibodies in the host [26]. Furthermore, individuals infected in utero or in the early post-natal period can become latently infected and hence never become seropositive [27]. Approximately 3.5% of infected cows are estimated to deliver latent-infected offspring [28]. In addition, MacMillan [29] reported that the Rose Bengal Test antigen could deteriorate when repeatedly cycled between refrigerator and room temperature during use. However, that serological testing has limits, especially after the disease has entered the chronic phase, when the organism is harbored intracellularly, often in the supramammary lymph nodes and the udder. Indeed, because the most important aspect of Brucella ecology is their ability to establish an intracellular replicative niche and remain protected from the host immune responses [2].
If this is the case, it might partially explain the discrepancy between the serology and qPCR results observed in the current study. Hypothetically, the discrepancy between the serology and qPCR results could be caused by previous vaccination against brucellosis as reported from a study in Egypt where cattle vaccinated with RB51 tested negative by serology tests but positive by qPCR [30]. However, in the current study, the samples of milk were taken from dairy cows submitted a control program of 1995 that none of the cattle had been vaccinated against brucellosis in Algeria.
Conclusion
Real-time PCR appears to offer several advantages over serological test in detecting the presence of extensive infection in cow’s milk samples. The detection was about 0.25 pg DNA (equivalent to 25-50 cells). As amplicon production is monitored and measured in real-time, results are available within 30 min [24]. This insertion sequence IS711 appears in several copies (10-40) all over the genome, but distribution and number differ in Brucella species [31]. For this reason, it is highly recommended to use this real-time PCR procedure to identify Brucella in all types of milk samples. Furthermore, we advise to use this procedure as a regular screening test in farms animals.
Authors’ Contributions
RS prepared the study design, carried out the research, and analyzed the data under the supervision of HTM and MB. RS and MA executed the extraction of DNA. RS and BK executed the real-time PCR. The manuscript was drafted and revised by RS under the guidance of HTM and MB. All authors read and approved the final manuscript.
Acknowledgments
The authors would like to thank the technical staff at the Laboratory of Microbiology and Molecular Biology of Constantine Biotechnology Research Center for their Collaboration, in particular, the Researcher Hamza Rahab, the Engineer Abderrahmane Selmania, and the responsible of Laboratory of Microbiology, Assia Ikhlef. The authors declare that they did not have any funding source or grant to support this research work.
Competing Interests
The authors declare that they have no competing interests.
References
- 1.McDermott J, Grace D, Zinsstag J. Economics of brucellosis impact and control in low-income countries. Rev. Sci. Tech. Int. Off. Epizoot. 2013;32(1):249–261. doi: 10.20506/rst.32.1.2197. [DOI] [PubMed] [Google Scholar]
- 2.Bargen K, Gorvel J.P, Salcedo S.P. Internal affairs:Investigating theBrucellaintracellular lifestyle. FEMS Microbiol. Rev. 2012;36:533–562. doi: 10.1111/j.1574-6976.2012.00334.x. [DOI] [PubMed] [Google Scholar]
- 3.Scholz H.C, Revilla-Fernández S, Al Dahouk S, Hammerl J.A, Zygmunt M.S, Cloeckaert A, Koylass M, Whatmore A.M, Blom J, Vergnaud G, Witte A, Aistleitner K, Hofer E. Brucella vulpissp. Nov, isolated from mandibular lymph nodes of red foxes (Vulpesvulpes) Int. J. Syst. Evol. Microbiol. 2016;66:2090–2098. doi: 10.1099/ijsem.0.000998. [DOI] [PubMed] [Google Scholar]
- 4.Whatmore A.M, Davison N, Cloeckaert A, Al Dahouk S, Zygmunt M.S, Brew S.D, Perrett L.L, Koylass M.S, Vergnaud G, Quance C, Scholz H.C, Dick E.J, Hubbard G, Schlabritz-Loutsevitch N.E. Brucella papionissp. Nov, isolated from baboons (Papiospp.) Int. J. Syst. Evol. Microbiol. 2014;64:4120–4128. doi: 10.1099/ijs.0.065482-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xavier M.N, Costa E.A, Paixão T.A, Santos R.L. GenusBrucellaand clinical manifestation. Ciênc. Rural. 2009;39(7):2252–2260. [Google Scholar]
- 6.Scholz H.C, Nöckler K, Göllner C, Bahn P, Vergnaud G, Tomaso H, Al Dahouk S, Huber B, Busse H.J, De B.K. Brucella inopinatasp. Nov, isolated from a breast implant infection. Int. J. Syst. Evol. Microbiol. 2010;60(4):801–808. doi: 10.1099/ijs.0.011148-0. [DOI] [PubMed] [Google Scholar]
- 7.De Jong M.F, Tsolis R.M. Brucellosis and Type IV secretion. Future Microbiol. 2012;7:47–58. doi: 10.2217/fmb.11.136. [DOI] [PubMed] [Google Scholar]
- 8.Olsen S, Palmer M. Advancement of knowledge ofBrucellaover the past 50 years. Vet. Pathol. 2014;51(6):1076–1089. doi: 10.1177/0300985814540545. [DOI] [PubMed] [Google Scholar]
- 9.Godfroid J, Al Dahouk S, Pappas G, Roth F, Matope G, Muma J, Marcot-Ty T, Pfeiffer D, Skjerve E. A one health surveillance and control of brucellosis in developing countries:Moving away from improvisation. Comp. Immunol. Microbiol. Infect. Dis. 2013;36:241–248. doi: 10.1016/j.cimid.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 10.WHO. The Control of Neglected Zoonotic Diseases - A Route to Poverty Alleviation. Report of a Joint WHO/DFID-AHP Meeting with the Participation of FAO and OIE. Geneva: WHO; 2005. [Accessed on 11-02-2018]. Available from: http://www.who.int/zoonoses/Report_Sept06.pdf . [Google Scholar]
- 11.Godfroid J, Saegerman C, Wellemans V, Walravens K, Letesson J.J, Tibor A, McMillan A, Spencer S, Sanna M, Bakker D. How to substantiate eradication of bovine brucellosis when aspecific serological reactions occur in the course of brucellosis testing. Vet. Microbiol. 2002;90:461–477. doi: 10.1016/s0378-1135(02)00230-4. [DOI] [PubMed] [Google Scholar]
- 12.Nilsen K, Smith P, Widdison J, Gall D, Kelly L, Kelly W, Nicoletti P. Serological relationship between cattle exposed toBrucella abortus Yersinia enterocolitica:9 andEscherichia coliO157:H7. Vet. Microbiol. 2004;100:25–30. doi: 10.1016/j.vetmic.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 13.Taleski V. An overview of introducing various laboratory tests for diagnostic of human brucellosis in the Republic of Macedonia. Macedonian J. Med. Sci. 2010;3:239–245. [Google Scholar]
- 14.Kumar S, Tuteja U, Sarika K, Singh D.K, Kumar A, Kumar O. Rapid multiplex PCR assay for the simultaneous detection of theBrucellagenusB. abortus, B. melitensisandB. suis. J. Microbiol. Biotechnol. 2011;21:89–92. doi: 10.4014/jmb.1007.07051. [DOI] [PubMed] [Google Scholar]
- 15.Bounaadja L, Albert D, Chénais B, Hénault S, Zygmunt M.S, Poliak S, Garin-Bastuji B. Real-time PCR for identification ofBrucellaspp.:A comparative study of IS711, bcsp31 and per target genes. Vet. Microbiol. 2009;137:156–164. doi: 10.1016/j.vetmic.2008.12.023. [DOI] [PubMed] [Google Scholar]
- 16.Kit manufactures BactoReal®KitBrucellaspp. Ingenetix GmbH Austria. 2014 [Google Scholar]
- 17.Ariza J, Bosilkovski M, Cascio A, Colmenero J.D, Corbel M.J, Falagas M.E, Memish Z.A, Roushan M.R.H, Rubinstein E, Sipsas N.V, Sipsas N.V, Solera J, Young E.J, Pappas G. Perspectives for the treatment of brucellosis in the 21stcentury:The ioannina recommendations. PLoS Med. 2007;4(12):e317. doi: 10.1371/journal.pmed.0040317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cutler S.J, Whatmore A.M, Commander N.J. Brucellosis-new aspects of an old disease. J. Appl. Microbiol. 2005;98(6):1270–1281. doi: 10.1111/j.1365-2672.2005.02622.x. [DOI] [PubMed] [Google Scholar]
- 19.Rubach M.P, Halliday J.E, Cleaveland S, Crump J.A. Brucellosis in low-income and middle-income countries. Curr. Opin. Infect. Dis. 2013;26:404–412. doi: 10.1097/QCO.0b013e3283638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Islam M.R.U, Pratap G.M, Kaur S.P, Filia G, Mohan S.H, Ahmed S.T. Comparative evaluation of indirect enzyme linked immunosorbent assay, rose bengal plate test, microagglutination test, and polymerase chain reaction for diagnosis of brucellosis in buffaloes. Turk. J. Vet. Anim. Sci. 2013;37:306–310. [Google Scholar]
- 21.Rajala E.L, Hoffman T, Fretin D, Godfroid J, Sattorov N, Boqvist S, Lundkvist A, Magnusson U. Detection and characterization ofBrucellaspp. PLoS Negl. Trop. Dis. 2017;11(3):e0005367. doi: 10.1371/journal.pntd.0005367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hinić V, Brodard I, Thomann A, Holub M, Miserez R, Abril C. IS711-based real-time PCR assays as a tool for detection ofBrucellaspp. in wild boars and comparison with bacterial isolation and serology. BMC Vet. Res. 2009;5:22. doi: 10.1186/1746-6148-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maills A, Rautureau S, Le Horgne J, Poignet-Leroux B, d' Amoux C, Dennetière G, Faure M, Lavigne J.P, Bru J.P, Garin-Bastuji B. Re-emergence of brucellosis in cattle in France and risk for Human health. [Accessed on 13-07-2017];Euro Surveill. 2012 17(30):20227. Available from: http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20227 . [PubMed] [Google Scholar]
- 24.Al Dahouk S, Tomaso H, Nöckler K, Neubauer H, Frangoulidis D. Laboratory based diagnosis of brucellosis - a review of the literature. Part II:Serological tests for brucellosis. Clin. Lab. 2003;49(11-12):577–589. [PubMed] [Google Scholar]
- 25.Godfroid J, Nielsen K, Saegerman C. Diagnosis of brucellosis in livestock and wildlife. Croat. Med. J. 2010;51:296–305. doi: 10.3325/cmj.2010.51.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gardner I.A, Stryhn H, Lind P, Collins M.T. Conditional dependence between tests affects the diagnosis and surveilance of animal diseases. Prev. Vet. Med. 2000;45:107–122. doi: 10.1016/s0167-5877(00)00119-7. [DOI] [PubMed] [Google Scholar]
- 27.Corbel M.J. Brucellosis in humans and animals, World Health Organization in collaboration with the Food and Agriculture Organization of the United Nations and World Organization for Animal Health. 2006. [Accessed on 12-10-2016]. Available from: http://www.who.int/csr/resources/publications/Brucellosis.pdf .
- 28.Saegerman C, Berkvens D, Godfroid J, Walravens K. Bovine brucellosis. In: Lefévre P.C, Blancou J, Chermette R, Uilenberg G, editors. Infectious and Parasitic Disease of Livestock. Paris, France: Lavoisier and Commonwealth Agricultural Bureau - International; 2010. pp. 971–1001. [Google Scholar]
- 29.MacMillan A. In: Conventional serological test. Nielsen K, Duncan J.R, editors. Boca Raton: Animal Brucellosis CRC Press; 1990. pp. 153–197. [Google Scholar]
- 30.Gwida M, El-Ashker M, Melzer F, El-Diasty M, El-Beskawy M, Neubauer H. Use of serology and real time PCR to control an outbreak of bovine brucellosis at a dairy cattle farm in the Nile Delta region, Egypt. Ir. Vet. J. 2016;69:3. doi: 10.1186/s13620-016-0062-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Halling S.M, Tatum F.M, Bricker B.J. Sequence and characterization of an insertion sequence, IS711, fromBrucella ovis. Gene. 1993;133:123–127. doi: 10.1016/0378-1119(93)90236-v. [DOI] [PubMed] [Google Scholar]


