Abstract
Background
Marked increases in fluconazole resistance in Candida tropicalis have been recently reported. In this study, the molecular mechanisms behind fluconazole resistance were investigated.
Methods
Twenty-two C. tropicalis clinical isolates, including 12 fluconazole-resistant isolates and 10 fluconazole-susceptible isolates, were collected from a tertiary care teaching hospital in Beijing between 2013 and 2017. Antifungal susceptibility testing, multilocus sequence typing, ERG11 amplification and sequencing, quantitative real-time reverse transcription-polymerase chain reaction (ERG11, UPC2, MDR1, and CDR1), and clinical data collection were performed for all C. tropicalis isolates.
Results
Multilocus sequence typing revealed that the 10 fluconazole-susceptible isolates and 12 fluconazole-resistant isolates were divided into nine and seven diploid sequence types, respectively. Of the 12 patients with fluconazole-resistant isolates, six had been previously exposed to azole and four had a fatal outcome. Y132F and S154F amino acid substitutions in Erg11p were found in all fluconazole-resistant isolates except one. MDR1 gene overexpression was identified in fluconazole-resistant isolates. In particular, seven high-level fluconazole resistant isolates (minimum inhibitory concentration ≥ 128 mg/L) and three pan-azole resistant isolates were identified. CDR1, ERG11, and UPC2 gene expression levels in fluconazole-resistant isolates were not significantly different from the control isolates (P = 0.262, P = 0.598, P = 0.114, respectively).
Conclusions
This study provides evidence that the combination of MDR1 gene overexpression and ERG11 missense mutations is responsible for high-level fluconazole resistance and pan-azole resistance in C. tropicalis clinical isolates. To the best of our knowledge, this is the first study investigating the relationship between MDR1 gene overexpression and increased fluconazole resistance.
Keywords: Candida tropicalis, Fluconazole resistance, MDR1
Background
In recent years, Candida species have emerged as important causes of nosocomial infections leading to high morbidity and mortality, particularly in immunocompromised patients [1–3]. In China, the prevalence of Candida tropicalis is similar to that of Candida parapsilosis, making it the second most common non-albicans Candida species [4].
Currently, azole antifungal agents are widely used in the battle against clinical Candida infections [5]. Due to the lower risk of side effects and low cost associated with fluconazole, it has become one of the most commonly used antifungals [6]. Until recently, C. tropicalis was susceptible to azole and other antifungal agents. However, azole resistance in C. tropicalis has been recently observed, possibly owing to the extensive use of antifungals in medical institutions [7]. Pan-azole-resistant C. tropicalis isolates, which pose serious clinical challenges, have also been found in multiple countries [8–10]. Indeed, the widespread use of azoles has been deemed to be an independent risk factor for fluconazole resistance in Candida species [11]. In addition, C. tropicalis appears to develop fluconazole resistance much more rapidly than other Candida species under in vitro selection [12].
Azole resistance in Candida species has been associated with mutations in or overexpression of the ERG11 gene or with upregulation of the CDR1 and MDR1 genes, the products of which are active transport pumps [13]. ERG11 gene mutations and overexpression have been demonstrated to be responsible for azole resistance in C. tropicalis clinical isolates in China [14]. However, there is still insufficient evidence of the role of the MDR1 and CDR1 genes in azole resistance of C. tropicalis clinical isolates. Therefore, in order to better understand the molecular mechanisms underlying azole resistance in C. tropicalis, we obtained a collection of fluconazole-resistant C. tropicalis isolates from a tertiary care teaching hospital and evaluated them for efflux pump gene expression level as well as ERG11 missense mutations. Our findings suggest that ERG11 missense mutations and increased MDR1 gene expression are highly correlated with fluconazole resistance in C. tropicalis clinical isolates.
Methods
Strains and clinical data collection
A total of 12 non-duplicated fluconazole-resistant (minimum inhibitory concentration [MIC], ≥8 mg/L) C. tropicalis isolates were retrospectively collected from hospitalized patients at Peking University People’s Hospital, a tertiary care teaching hospital with 1448 beds located in Beijing, China between 2013 and 2017. In the Peking University People’s Hospital, a high rate of resistance to fluconazole, with 20.30% resistant C. tropicalis isolates, was observed between 2013 and 2017. These isolates were recovered from various clinical specimens (excluding sputum samples) and presumptively identified by CHROMagar Candida (CHROMagar, Paris, France) and then confirmed by matrix-assisted laser desorption/ionization (MALDI-TOF) mass spectrometry (MS) (Bruker Daltonik GmbH, Bremen, Germany). Fluconazole-susceptible isolates (MIC, ≤2 mg/L) were randomly obtained from different wards between 2013 and 2017. All strains were stored at − 80 °C in 20% glycerol until use and maintained by biweekly passage on Sabouraud agar (Oxoid, Basingstoke, United Kingdom).
Clinical data and patient information were obtained retrospectively from medical records. Previous use of azole antifungal agents was defined as administration within 3 months prior to the isolation of C. tropicalis. Inpatient outcomes were assessed 30 days after C. tropicalis isolation. This study was approved by the Research Ethics Board at Peking University People’s Hospital.
Antifungal susceptibility testing
Antifungal susceptibility testing was performed using Sensititre YeastOne™ YO10 methodology (Thermo Scientific, Cleveland, OH, USA) in accordance with the manufacturer’s instructions. MICs were determined after 24 h of incubation at 35 °C and interpreted according to species-specific Clinical & Laboratory Standards (CLSI) breakpoints [15]. Candida krusei ATCC 6258 and C. parapsilosis ATCC 22019 were used as quality controls.
PCR amplification and sequencing of ERG11
Total genomic DNA was extracted from overnight cultures using the DNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s recommendations and used as the template for polymerase chain reaction (PCR) amplification of the ERG11 gene. The primers and conditions used for amplification and sequencing of ERG11 were described previously as shown in Table 1 [14]. Nucleotide sequences of the amplicon were aligned with the sequence of C. tropicalis MYA-3404 (GenBank accession number XM_002550136).
Table 1.
Primers used in this study
| Gene | DNA sequence (5′ to 3′) | Amplicon size (bp) | Reference |
|---|---|---|---|
| ERG11 for amplification | F:TGAAGAATATCCCACAGGCT | 1846 | This study |
| R:CTTAGCAAGAACTTCTAATGTT | |||
| ERG11 for RT-PCR | F:GAGATTTGATTGATTCCTTGTTGGT | 163 | This study |
| R:TGTGGTTGTTCAGCCAAATGC | |||
| CDR1 | F:CCAGAGGTTTGGATTCCGCT | 186 | This study |
| R:TGGCTTTGTCTGCTTTCCCA | |||
| MDR1 | F:GGGTGCATCATTCCAGCCTA | 189 | This study |
| R:GGGATGGCAATCATCACGAG | |||
| UPC2 | F:GAGTGGAACAACAACACAACAA | 208 | This study |
| R:TAAATCCCCTAAACCTGAAAGA | |||
| ACT1 | F:TTTACGCTGGTTTCTCCTTGCC | 322 | [14] |
| R:GCAGCTTCCAAACCTAAATCGG |
Multilocus sequence typing
Multilocus sequence typing (MLST) was performed as described on the PubMLST website (https://pubmlst.org/ctropicalis/). Each isolate was characterized as a diploid sequence type (DST) based on the sequence of six housekeeping genes [16]. Novel allelic profiles and DSTs were submitted to the C. tropicalis MLST database.
Relative quantification of gene expression
Total RNA extraction and real-time reverse transcription-PCR (RT-PCR) for the evaluation of expression levels of CDR1, MDR1, ERG11, and UPC2 were performed as descried previously [14, 17]. Total RNA was extracted from strains grown in yeast peptone dextrose media at mid exponential (log) phase using the RNeasy mini kit (Qiagen) according to the manufacturer’s instructions. Total RNA was quantified using a NanoDrop 2000C (Thermo Scientific) and reverse transcription was performed using PrimeScript™ RT Master Mix (Takara, Tokyo, Japan). Quantitative RT-PCR was performed in triplicate using an ABI 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA), SYBR® Premix Ex Taq™ II (Takara), and the primers listed in Table 1. The mRNA expression levels of target genes were normalized to the ACT1 gene using the relative standard curve method as described previously [14]. The transcript levels of fluconazole-resistant isolates were compared with the average expression level of the 10 fluconazole susceptible isolates.
Statistical analysis
All statistical analyses were performed using SPSS version 24.0 (IBM Corp., Armonk, NY, USA). Continuous variables were compared using the Mann-Whitney U test. P values < 0.05 were considered statistically significant.
Results
Antifungal susceptibility
Of the 22 C. tropicalis isolates, seven were determined to be high-level fluconazole-resistant isolates (MIC, ≥128 mg/L), five were low-level fluconazole-resistant isolates (MIC, 8−64 mg/L), and 10 were control isolates (MIC, ≤2 mg/L). After applying species-specific epidemiological cutoff values, three and twelve isolates were categorized as non-wild-type for itraconazole and posaconazole, respectively. Therefore, a total of three isolates (Ct07R, Ct09R, and Ct10R) had a non-wild-type phenotype or were resistant to the four azole antifungals tested. In addition, all isolates were sensitive to amphotericin B and 5-flucytosine. The results of antifungal susceptibility testing are shown in Table 2.
Table 2.
Amino acid substitutions, antifungal agent MICs, and patient’s characteristics for 22 Candida tropicalis clinical isolates
| Designation | Origin | Date of isolation | Age /Sex | Underlying disease | Ward | Substitution of amino acid residue (Erg11p) | DST | Antifungal agent MIC (mg/L) | Previous azole usea | Antifungal therapy | Thirty-day outcome | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FLU | VOR | ITR | POS | AMB | 5FC | |||||||||||
| Ct01R | Blood | 8/5/15 | 37/M | ALL | Hematology | Y132F, S154F | 225 | 128 | 4 | 0.25 | 0.25 | 0.5 | ≤0.06 | FLU, VOR | CAS, AMB | Survival |
| Ct02R | Pleural fluid | 10/4/14 | 65/M | Liver Transplantation | ICU | None | 639 | 8 | 0.5 | 0.25 | 0.25 | 0.5 | ≤0.06 | VOR | None | Death |
| Ct03R | Blood | 9/12/13 | 72/F | Chronic Renal Failure | Gastroenterological Surgery | Y132F, S154F | 639 | 256 | 4 | 0.5 | 0.5 | 0.5 | 0.25 | None | FLU, AMB | Death |
| Ct04R | Blood | 11/8/13 | 26/M | ALL | Hematology | Y132F, S154F | 329 | 64 | 2 | 0.25 | 0.25 | 0.5 | ≤0.06 | FLU | ITR | Survival |
| Ct05R | Blood | 2/22/15 | 40/M | CML | Hematology | Y132F, S154F | 615 | 128 | 4 | 0.25 | 0.5 | 0.5 | 0.12 | FLU, ITR | CAS | Survival |
| Ct06R | Blood | 4/6/17 | 38/M | AML | Hematology | Y132F, S154F | NT | 128 | 4 | 0.5 | 0.5 | 0.5 | 0.12 | FLU, VOR | CAS, AMB | Survival |
| Ct07R | Urine | 9/15/14 | 87/F | Cholangitis | ICU | Y132F, S154F | 225 | 256 | > 8 | 1 | 1 | 0.5 | ≤0.06 | None | None | Survival |
| Ct08R | Urine | 7/22/15 | 19/M | Nephrlithotomy | Urology | Y132F, S154F | 225 | 64 | 4 | 0.5 | 1 | 1 | ≤0.06 | None | VOR | Survival |
| Ct09R | Urine | 8/7/15 | 64/M | Pneumonia | Respiratory | Y132F, S154F | 225 | > 256 | 8 | 1 | 1 | 1 | ≤0.06 | None | None | Survival |
| Ct10R | Blood | 9/2/15 | 25/F | ALL | Hematology | Y132F, S154F | 506 | > 256 | 8 | 1 | 1 | 1 | ≤0.06 | ITR | CAS | Survival |
| Ct11R | Urine | 9/26/15 | 80/M | Pneumonia | ICU | Y132F, S154F | 508 | 64 | 2 | 0.25 | 0.25 | 0.5 | ≤0.06 | None | None | Death |
| Ct12R | Urine | 6/17/15 | 83/F | Adrenocortical Carcinoma | ICU | Y132F, S154F | 508 | 64 | 4 | 0.25 | 0.25 | 0.5 | ≤0.06 | None | None | Death |
| Ct01S | Cervix | 4/5/17 | 34/F | Premature Delivery | Obstetrics | None | NT | 2 | 0.12 | 0.12 | 0.12 | 0.25 | ≤0.06 | None | None | Survival |
| Ct02S | Sputum | 6/15/17 | 62/M | Respiratory Failure | Respiratory | None | NT | 1 | 0.12 | 0.12 | 0.12 | 0.25 | ≤0.06 | None | None | Survival |
| Ct03S | Urine | 6/23/17 | 16/F | Ewing’s Sarcoma | Musculoskeletal Tumor | None | NT | 1 | 0.06 | 0.12 | 0.12 | 0.25 | 0.12 | None | FLU | Survival |
| Ct04S | Catheter | 6/26/17 | 44/M | Hepatocellular Carcinoma | ICU | None | NT | 2 | 0.12 | 0.25 | 0.25 | 0.25 | ≤0.06 | None | None | Death |
| Ct05S | Blood | 11/27/14 | 50/M | Cirrhosis | Hepatology | None | 519 | 2 | 0.12 | 0.25 | 0.25 | 1 | ≤0.06 | None | CAS | Survival |
| Ct06S | Blood | 2/13/15 | 43/M | AML | Hematology | None | 169 | 1 | 0.12 | 0.25 | 0.5 | 0.5 | ≤0.06 | None | ITR | Death |
| Ct07S | Urine | 7/3/15 | 80/M | CHD | Cardiology | None | 343 | 1 | 0.12 | 0.25 | 0.25 | 0.5 | 0.25 | None | None | Survival |
| Ct08S | Blood | 9/25/15 | 42/M | AML | Hematology | None | 399 | 1 | 0.12 | 0.25 | 0.12 | 1 | 0.12 | None | VOR, AMB | Death |
| Ct09S | Blood | 10/31/14 | 51/F | AML | Hematology | None | 300 | 2 | 0.25 | 0.25 | 0.25 | 0.5 | 0.12 | None | VOR, AMB | Death |
| Ct10S | Urine | 7/26/17 | 23/M | HPS | ICU | None | 169 | 1 | 0.12 | 0.25 | 0.25 | 0.5 | ≤0.06 | None | None | Survival |
5FC 5-flucytosine, ALL acute lymphocytic leukemia, AMB amphotericin B, AML acute myeloid leukemia, CAS caspofungin, CHD coronary heart disease, CML chronic myeloid leukemia, FLU fluconazole, HPS hemophagocytic syndrome, ITR itraconazole, NT untyped, POS posaconazole, VOR voriconazole
awithin 3 months prior to the detection of C. tropicalis
ERG11 gene mutations
Sequence analysis of the ERG11 gene revealed no missense mutations in any control isolate. In contrast, two missense mutations, Y132F and S154F, were observed in all fluconazole-resistant isolates except isolate Ct02R (Table 2).
Multilocus sequence typing analysis
The 10 control isolates and 12 fluconazole-resistant isolates were grouped into nine and seven DSTs, respectively (Table 2). Fluconazole-susceptible isolates were more genetically diverse than fluconazole-resistant isolates. Four fluconazole-resistant isolates belonged to DST225. Two isolates belonged DST169, DST508, and DST639. In addition, the sequences of five isolates were not found in the C. tropicalis DST database and the allelic profiles and DSTs of these isolates were submitted to the C. tropicalis MLST database.
Expression levels of ERG11, UPC2, CDR1, and MDR1
Quantitative RT-PCR experiments revealed that expression levels of MDR1 were higher in the 12 fluconazole-resistant isolates than in the 10 control isolates. Moreover, seven high-level fluconazole resistant isolates had expression levels of MDR1 than five low-level resistant isolates. In addition, three pan-azole resistant isolates (Ct07R, Ct09R, and Ct10R) had higher expression levels of MDR1 than the other nine fluconazole-resistant isolates (Fig. 1a, b, c). In contrast, no significant difference was observed in the expression levels of ERG11, UPC2, or CDR1 between the 12 fluconazole-resistant isolates and the 10 control isolates (Fig. 1d).
Fig. 1.
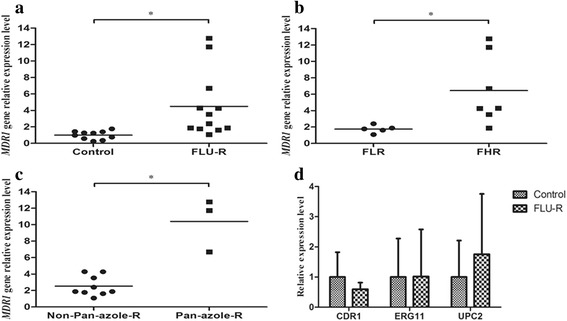
Expression levels of the MDR1, CDR1, ERG11, and UPC2gene in 22 C. tropicalis clinical isolates. a MDR1 gene expression levels were higher in fluconazole-resistant (FLU-R) isolates than in control isolates. b MDR1 gene expression levels were higher in high-level fluconazole-resistant (FHR) isolates than in low-level fluconazole-resistant (FLR) isolates. c MDR1 gene expression levels were higher in pan-azole-resistant (Pan-azole-R) isolates than in non-pan-azole-resistant isolates (Non-Pan-azole-R). d No significant difference was found in the expression levels of the CDR1, ERG11, or UPC2 genes between fluconazole-resistant (FLU-R) and control isolates. Error bars indicate standard deviations. *P < 0.05
Clinical information
Clinical information was gathered from chart reviews. Previous use of azole antifungals, history of antifungal therapy, and 30-day outcomes of all C. tropicalis infected patients are shown in Table 2. Of the 12 inpatients with fluconazole-resistant C. tropicalis, six had been exposed to azole within 3 months prior to the isolates being recovered. Four patients died from any cause in the 30 days after the detection of C. tropicalis isolates. In contrast, among the 10 patients from whom fluconazole-susceptible C. tropicalis isolates were recovered no patients had previously been exposed to azole and four patients died.
Discussion
C. tropicalis has received widespread attention in recent years due to an increase in fluconazole resistance and the high mortality rate associated with infections [7, 18]. In a global surveillance study conducted in 2013, 31.8% of non-susceptible C. tropicalis isolates worldwide were obtained from mainland China [19]. Moreover, the rate of fluconazole resistance of C. tropicalis increased from 11.2 to 42.7% between 2011 and 2015 in China [7]. However, understanding of the mechanisms underlying azole resistance in clinical C. tropicalis isolates is insufficient compared to that of other Candida species.
Studies have demonstrated that Erg11p amino acid substitutions in the 14α-sterol demethylase involved in the biosynthesis of sterol ergosterol results in decreased fluconazole susceptibility in C. tropicalis clinical isolates [9, 20, 21]. Jiang et al. demonstrated that two mutations, Y132F and S154F, are responsible for azole resistance by inserting them into wild-type isolate [14]. Similarly, the amino acid substitutions Y132F and S154F in Erg11p were found in all fluconazole-resistant isolates except one (Ct02R: MIC, 8 mg/L) in our study.
A previous study confirmed that the UPC2 gene, which encodes a global transcriptional regulator of the ERG11 gene, was up-regulated in azole-resistant C. tropicalis isolates [17]. Therefore, we examined UPC2 gene expression levels in 12 fluconazole-resistant isolates. However, we found that there was no difference in the expression of the UPC2 gene between fluconazole-resistant and control isolates. These results suggest that ERG11 overexpression, which results in up-regulation of target enzymes, is not involved in azole resistance in fluconazole-resistant isolates.
In addition to the targeted mutations and overexpression of the enzyme Erg11p, we found that overexpression of the efflux pumps Mdr1p and Cdr1p, encoded by the MDR1 and CDR1 genes, are involved in C. tropicalis azole resistance [22]. Consistent with this finding, pan-azole resistant C. tropicalis isolate, which overexpresses the MDR1 gene and contains the Y132F and S154F amino acid substitutions in Erg11p, has reportedly been isolated from a patient with acute lymphoblastic leukemia [8]. In the present study, we found that fluconazole-resistant isolates had significantly higher expression levels of the MDR1 gene than control isolates. Furthermore, high-level fluconazole-resistant isolates had higher levels of MDR1 expression than low-level fluconazole-resistant isolates. Most importantly, among the 12 fluconazole-resistant isolates, MDR1 expression levels were highest in the three pan-azole-resistant isolates. Hence, we conclude that overexpression of the MDR1 gene is associated with high-level fluconazole resistance in C. tropicalis clinical isolates. Additionally, these results indicate that the combination of MDR1 gene overexpression and the amino acid substitutions Y132F and S154F in Erg11p is an important mechanism of pan-azole resistance in C. tropicalis clinical isolates.
Of the 12 fluconazole resistant isolates obtained, four belonged to DST225, including two pan-azole resistant isolates (Ct07R and Ct09R). Interestingly, human and fruit samples have recently been found to contain DST225 C. tropicalis in Taiwan and these isolates have been shown to be less susceptible to azole drugs [23]. This suggests that C. tropicalis fluconazole resistance is related to specific genotypes, such as DST225.
Conclusion
In conclusion, specific mutations in the ERG11 gene and upregulation of drug efflux pumps are the most common mechanisms underlying azole resistance in C. tropicalis. This study finds that the MDR1 gene may have a role in determining a high-level of fluconazole resistance and pan-azole resistance in C. tropicalis clinical isolates. To the best of our knowledge, this is the first study investigating the relationship between MDR1 gene overexpression and high-level fluconazole resistance. However, further studies are needed to identify the mechanisms by which MDR1 is overexpressed. Homologs of gain-of-function mutations in the C. parapsilosis MRR1 gene, a regulator of the MDR1 gene [24], also need to be further examined in the future.
Acknowledgments
Funding
This work was supported by the National Natural Science Foundation of China (grant number 81625014).
Availability of data and materials
All data generated or analysed during this study are included in this published article. The nucleotide sequence data of ERG11 gene have been deposited in the GenBank database under accession numbers MH002313-MH002334.
Abbreviations
- DST
Diploid sequence type
- MALDI-TOF
Matrix-assisted laser desorption/ionization
- MIC
Minimum inhibitory concentration
- MLST
Multilocus sequence typing
- MS
Mass spectrometry
- PCR
Polymerase chain reaction
- RT-PCR
Real-time reverse transcription-PCR
Authors’ contributions
HW conceived and designed the study; LJ, ZC, QW and YW performed the experiments, analyzed the data and collected the clinical information; LJ and ZC drafted the manuscript; XW, HC and HW revised the manuscript critically for important intellectual content; All authors read and approved the final manuscript.
Ethics approval and consent to participate
In this study, patient sampling and data collection were approved by the research ethics board at Peking University People’s Hospital. Participants consent was not necessary as this study was retrospective and participants were anonymized.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Longyang Jin, Email: jly@bjmu.edu.cn.
Zhuorui Cao, Email: max_cao2016@163.com.
Qi Wang, Email: wangqi99887@sina.com.
Yichen Wang, Email: 1437809507@qq.com.
Xiaojuan Wang, Email: curliele@163.com.
Hongbin Chen, Email: swimmingfish01@163.com.
Hui Wang, Phone: +86-10-88326300, Email: whuibj@163.com, Email: wanghui@pkuph.edu.cn.
References
- 1.Perlin DS, Rautemaa-Richardson R, Alastruey-Izquierdo A. The global problem of antifungal resistance: prevalence, mechanisms, and management. Lancet Infect Dis. 2017;17(12):e383–e392. doi: 10.1016/S1473-3099(17)30316-X. [DOI] [PubMed] [Google Scholar]
- 2.Wu PF, Liu WL, Hsieh MH, et al. Epidemiology and antifungal susceptibility of candidemia isolates of non-albicans Candida species from cancer patients. Emerg Microbes Infect. 2017;6(10):e87. doi: 10.1038/emi.2017.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lortholary O, Renaudat C, Sitbon K, et al. The risk and clinical outcome of candidemia depending on underlying malignancy. Intensive Care Med. 2017;43(5):652–662. doi: 10.1007/s00134-017-4743-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu W, Tan J, Sun J, et al. Invasive candidiasis in intensive care units in China: in vitro antifungal susceptibility in the China-SCAN study. J Antimicrob Chemother. 2014;69(1):162–167. doi: 10.1093/jac/dkt330. [DOI] [PubMed] [Google Scholar]
- 5.Wiederhold NP. Antifungal resistance: current trends and future strategies to combat. Infect Drug Resist. 2017;10:249–259. doi: 10.2147/IDR.S124918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ou HT, Lee TY, Chen YC, et al. Pharmacoeconomic analysis of antifungal therapy for primary treatment of invasive candidiasis caused by Candida albicans and non-albicans Candida species. BMC Infect Dis. 2017;17:481. doi: 10.1186/s12879-017-2573-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan X, Xiao M, Liao K, et al. Notable increasing trend in azole non-susceptible Candida tropicalis causing invasive candidiasis in China (August 2009 to July 2014): molecular epidemiology and clinical azole consumption. Front Microbiol. 2017;8:464. doi: 10.3389/fmicb.2017.00464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.You L, Qian W, Yang Q, et al. ERG11 gene mutations and MDR1 upregulation confer pan-azole resistance in Candida tropicalis causing disseminated candidiasis in an acute lymphoblastic leukemia patient on posaconazole prophylaxis. Antimicrob Agents Chemother. 2017;61(7):e02496–e02416. doi: 10.1128/AAC.02496-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xisto MI, Caramalho RD, Rocha DA, et al. Pan-azole-resistant Candida tropicalis carrying homozygous erg11 mutations at position K143R: a new emerging superbug? J Antimicrob Chemother. 2017;72(4):988–992. doi: 10.1093/jac/dkw558. [DOI] [PubMed] [Google Scholar]
- 10.Álvarez-Pérez S, García ME, Cutuli MT, et al. Acquired multi-azole resistance in Candida tropicalis during persistent urinary tract infection in a dog. Med Mycol Case Rep. 2016;11:9–12. doi: 10.1016/j.mmcr.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Yang Q, Chen L, et al. Cross-resistance between voriconazole and fluconazole for non-albicans Candida infection: a case-case-control study. Eur J Clin Microbiol Infect Dis. 2017;36(11):2117–2126. doi: 10.1007/s10096-017-3034-4. [DOI] [PubMed] [Google Scholar]
- 12.Barchiesi F, Calabrese D, Sanglard D, et al. Experimental induction of fluconazole resistance in Candida tropicalis ATCC 750. Antimicrob Agents Chemother. 2000;44(6):1578–1584. doi: 10.1128/AAC.44.6.1578-1584.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi MJ, Won EJ, Shin JH, et al. Resistance mechanisms and clinical features of fluconazole-nonsusceptible Candida tropicalis isolates compared with fluconazole-less-susceptible isolates. Antimicrob Agents Chemother. 2016;60(6):3653–3661. doi: 10.1128/AAC.02652-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang C, Dong D, Yu B, et al. Mechanisms of azole resistance in 52 clinical isolates of Candida tropicalis in China. J Antimicrob Chemother. 2013;68(4):778–785. doi: 10.1093/jac/dks481. [DOI] [PubMed] [Google Scholar]
- 15.Pfaller MA, Diekema DJ. Progress in antifungal susceptibility testing of Candida spp. by use of clinical and laboratory standards institute broth microdilution methods, 2010 to 2012. J Clin Microbiol. 2012;50(9):2846–2856. doi: 10.1128/JCM.00937-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tavanti A, Davidson AD, Johnson EM, et al. Multilocus sequence typing for differentiation of strains of Candida tropicalis. J Clin Microbiol. 2005;43(11):5593–5560. doi: 10.1128/JCM.43.11.5593-5600.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang C, Ni Q, Dong D, et al. The role of UPC2 gene in azole-resistant Candida tropicalis. Mycopathologia. 2016;181(11–12):833–838. doi: 10.1007/s11046-016-0050-3. [DOI] [PubMed] [Google Scholar]
- 18.Chapman B, Slavin M, Marriott D, et al. Changing epidemiology of candidaemia in Australia. J Antimicrob Chemother. 2017;72(4):1103–1108. doi: 10.1093/jac/dkx047. [DOI] [PubMed] [Google Scholar]
- 19.Castanheira M, Messer SA, Rhomberg PR, et al. Antifungal susceptibility patterns of a global collection of fungal isolates: results of the SENTRY antifungal surveillance program (2013) Diagn Microbiol Infect Dis. 2016;85(2):200–204. doi: 10.1016/j.diagmicrobio.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 20.Forastiero A, Mesa-Arango AC, Alastruey-Izquiredo A, et al. Candida tropicalis antifungal cross-resistance is related to different azole target (Erg11p) modifications. Antimicrob Agents Chemother. 2013;57(10):4769–4781. doi: 10.1128/AAC.00477-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan J, Zhang J, Chen W, et al. The A395T mutation in ERG11 gene confers fluconazole resistance in Candida tropicalis causing candidemia. Mycopathologia. 2015;179(3–4):213–218. doi: 10.1007/s11046-014-9831-8. [DOI] [PubMed] [Google Scholar]
- 22.Whaley SG, Berkow EL, Rybak JM, et al. Azole antifungal resistance in Candida albicans and emerging non-albicans Candida species. Front Microbiol. 2017;7:2173. doi: 10.3389/fmicb.2016.02173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lo H-J, Tsai S-H, Chu W-L, et al. Fruits as the vehicle of drug resistant pathogenic yeasts. J Inf Secur. 2017;75(3):254–262. doi: 10.1016/j.jinf.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 24.Branco J, Silva AP, Silva RM, et al. Fluconazole and voriconazole resistance in Candida parapsilosis is conferred by gain-of-function mutations in MRR1 transcription factor gene. Antimicrob Agents Chemother. 2015;59(10):6629–6633. doi: 10.1128/AAC.00842-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this study are included in this published article. The nucleotide sequence data of ERG11 gene have been deposited in the GenBank database under accession numbers MH002313-MH002334.


